Abstract
Background and aims
SPARC (secreted protein acidic, rich in cysteine) is a matricellular protein that has been found to be activated in a number of human cancers. More recently, it has been shown to be upregulated in human gastric and colorectal cancer. We therefore wished to address the functional importance of SPARC upregulation to intestinal tumorigenesis in vivo.
Methods
SPARC upregulation was determined in intestinal adenomas of tumour‐prone ApcMin/+ mice at both the RNA and the protein level. To determine the functional importance of SPARC for intestinal tumorigenesis we then intercrossed Sparc knockout mice with ApcMin/+ mice (n = 20). Intestinal enterocyte migration was examined using bromodeoxyuridine labelling studies.
Results
Levels of murine Sparc and several related proteins were upregulated in adenomas arising in ApcMin/+ mice. A deficiency of Sparc strongly suppressed adenoma formation in ApcMin/+ mice (p⩾0.0001). Importantly, a deficiency of Sparc also accelerated enterocyte migration (p = 0.01), as perturbed slow epithelial migration may underpin adenoma formation in the intestine.
Conclusions
These data implicate Sparc in both cell migration and tumour formation, and identify Sparc as a potential therapeutic target for colorectal cancer.
SPARC is the prototype of a matricellular protein family, which includes Hevin/Sc1, Spock1, Spock2 and Spock3 (also known as Testicans 1–3), Smoc1 and Smoc2. Matricellular proteins are those proteins that associate with the extracellular matrix (ECM) but do not have a structural function, rather being thought to modulate cell–ECM interactions. SPARC has been shown to be expressed in many late stage cancers,1 and has been associated with numerous functions including proliferation, survival, adhesion, migration and invasion.1,2 More recently, SPARC upregulation has been associated with gastric, oesophageal and colorectal cancer.3,4,5 In both gastric and oesophageal cancer this upregulation has now been observed at early stages of tumorigenesis, and in gastric cancer high SPARC levels have been shown to correlate with poor prognosis.4 These data indicate that SPARC may play an important role not only for tumour invasion in colorectal cancer, but also for tumour initiation and tumour growth. Other members of the SPARC family have also been implicated in the suppression or promotion of tumorigenesis. Downregulation (or loss of the chromosomal region containing Hevin) has thus been noted in many types of cancer;1 and raised expression of Smoc1 has been found in grade II–IV astrocytomas.6 Testican2 has also been implicated in the invasive behaviour of astrocytic tumours,7 possibly by blocking the Testican1/3‐mediated inactivation of membrane type matrix metalloproteinases. Furthermore, Testican1 has been found to be downregulated in the embryonal‐rhabdomyosarcoma cell line RD, suggesting that the loss of its expression may be related to the progression of malignancy.8 We have therefore investigated the expression of Sparc family members in adenomatous and normal epithelium from a well‐known model of spontaneous intestinal tumorigenesis, the ApcMin/+ mouse. Following confirmation of Sparc upregulation in adenomas, we then directly addressed the role Sparc plays in adenoma formation by crossing the SparcTm1Cam null allele9 (subsequently referred to as Sparc−) into the ApcMin/+ background and studying enterocyte cell migration and subsequent adenoma development.
Methods
Assay for intestinal tumorigenesis
SparcTm1Cam mice segregating equally for C57Bl6 and Ola129 genomes were mated to ApcMin/+ mice maintained on an inbred C57Bl/6J background. Progeny from this cross were then interbred to generate cohorts of ApcMin/+Sparc+/+ and ApcMin/+Sparc−/− mice. These cohorts were therefore segregating for C57/Bl6 (75%) and 129/Ola (25%) genomes. All mice were genotyped for the Mom‐1 allele via polymerase chain reaction (PCR) analysis.10 Mice were culled at 230 days and the intestinal tumour burden was determined by removing the entire intestine and mounting en face. Preparations were fixed in methacarn (4:2:1 methanol, chloroform, and glacial acetic acid), and lesion number and size were scored macroscopically. For migration analyses, mice were injected with 0.25 ml bromodeoxyuridine (BrdU; Amershaml/GE Healthcare UK Ltd, Little Chalfont, Buckinghamshire, UK) and were then killed at the appropriate timepoint. Intestines were flushed with water and fixed overnight in methacarn before rolling the gut for sectioning. All fixed samples were embedded in paraffin and sectioned to 5–6 μm on poly‐l‐lysine slides for BrdU immunohistochemical analysis (1:50 serotec).
Immunohistochemical analysis for SPARC
Sparc immunohistochemical analysis (IHC) was performed using anti‐Sparc antibody (Abcam #ab19528) at a concentration of one part in 1000 after antigen retrieval in citrate buffer in a pressure cooker. This antibody only recognises SPARC and does not crossreact with any other family members. IHC was performed on formalin‐fixed tumours and normal tissue from ApcMin/+Sparc+/+ and ApcMin/+Sparc−/− mice. For tumour microarray analysis, the MaxArray human colon carcinoma tissue microarray (Zymed, Invitrogen, Paisley, UK) was used. Each slide contains 60 tissue cores from different colon carcinomas. They consist of 49 adenocarcinomas, seven mucinous carcinomas, three signet‐ring adenocarcinomas and one adenosquamous carcinoma. The slides were stained as described above.
RNA extraction and reverse transcription
Colon and tumour samples were dissected from the intestines of ApcMin/+Sparc+/+ and ApcMin/+Sparc−/− mice, placed in RNAlater (Ambion/Applied Biosystems, Warrington, UK) and stored at −20°C. Samples were subsequently placed in TRIzol (Invitrogen, Paisley, UK) and homogenized. RNA was extracted according to the manufacturer's protocol. RNA samples were quantitated using both formaldehyde gel electrophoresis and spectrophotometry (Camspec, Cambridge, UK). An aliquot of 100 μg of each RNA was further purified using RNeasy columns (Qiagen, Crawley, UK), then quantitated again as previously described; 15 μg RNA was DNase treated (DNAfree; Ambion), according to the manufacturer's protocol; 5 μg of each RNA was reverse transcribed using the SuperscriptII First Strand Synthesis System for reverse transcriptase (RT)–PCR (Invitrogen), according to the manufacturer's instructions. A “no RT” control corresponding to each sample was also produced, these were treated in exactly the same way as the RT samples except that no SuperscriptII enzyme was added. After RT, samples were diluted from a 20 μl final volume to 50 μl.
Semi‐quantitative PCR
Low cycle PCR of the mouse Gapdh and β‐actin genes were used to standardize the concentration of RNA samples. Experimental samples were then tested at varying cycles until band intensity differences were noted or PCR products were no longer visible. The primers used are listed below in table 1. PCR band intensities were estimated via image analysis (Scion Image; Scion Corp., Frederick, Maryland, USA). Sparc−/−ApcMin+/− mice all showed an absence of Sparc when Sparc RT–PCR were carried out. These results corresponded in all cases with results obtained from genotyping (data not shown).
Table 1 Primer sequences.
| Primer | Sequence |
|---|---|
| GapdhF | ACCACAGTCCATGCCATCAC |
| GapdhR | TCCACCACCCTGTTGCTGTA |
| β‐ActinF | CGTGGGCCGCCCTAGGCACCA |
| β‐ActinR | TTGGCCTTAGGGTTCAGGGGG |
| SparcF | GCTGTGTTGGAAACGGAGTTG |
| SparcR | CTTGCCATGTGGGTTCTGACT |
| Sc1F | GGATGAGGAGGATGGAGATGG |
| Sc1R | TGGGTTTGCTTGCTGCTTTC |
| TesticanF | ACGGTTGTGTGGGAGAAAGG |
| TesticanR | GCTGGGAACCATGCTGTAGG |
| Testican2F | GGTTCCAGCTCCTTCGTGAG |
| Testican2R | AGACGCGGCCATCTTTGTAG |
| Testican3F | TGGAACAAATTCCGAGACGAAG |
| Testican3R | GCAGGTAGATGACGGGAGACC |
| Smoc1F | AGAGTGACGCCAGAGCCAAG |
| Smoc1R | TCGCTGCTGTTGCTATCCAG |
| Smoc2F | CCTTCCTGTCCCGATGTGAG |
| Smoc2R | TGATGGGTCTTCCATTTGGTG |
Results and discussion
Sparc and a number of related proteins are upregulated in adenomas from the ApcMin/+ mouse
Our microarray analysis using the Affymetrix MOE430_2 chip comparing adenomas from the ApcMin/+ mouse to normal epithelium showed a twofold upregulation of SPARC in adenomas. Similarly, a recent array paper has found SPARC to be upregulated in adenomas from the ApcMin/+ mouse.12 To confirm this, we used semi‐quantititative RT–PCR for SPARC in matched RNA from adenomatous and normal epithelium harvested from eight different animals at 230 days (supplementary fig 1; supplementary fig 1 can be viewed on the Gut website at http://gut.bmj.com/supplemental). Quantitative image analysis, in comparison with β‐actin and Gapdh levels followed by statistical testing showed an increase in SPARC RNA levels in the adenomas (average fold change 3.05, t‐test, p = 0.0155). This is consistent with previous studies in humans showing elevated levels of SPARC in colorectal and gastric carcinomas.4,5
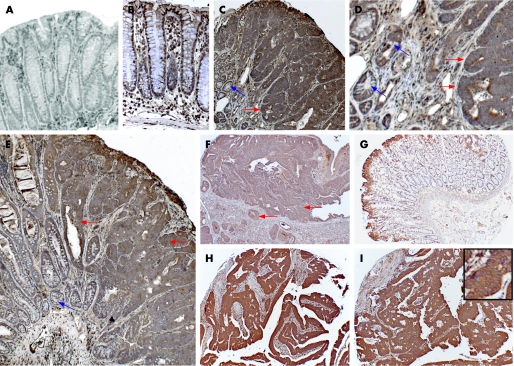
Figure 1 Upregulation of Sparc during colorectal tumorigenesis. (A–B) Sparc immunohistochemical analysis (IHC) on colon from ApcMin/+Sparc−/− (A) and ApcMin/+Sparc+/+ (B) mice, note lack of epithelial staining in Sparc−/− null colon. (C–E) Sparc IHC on adenomas arising in Apc ApcMin/+Sparc+/+ mice. Red arrows denote a higher expression of SPARC in adenomas than surrounding normal colonic crypts (blue arrows).This is highlighted in panel D, which is a close‐up of the edge of a tumour shown in C. (F) Sparc is upregulated in kidney tumours formed after Apc loss in renal epithelium.11 (G) Sparc IHC of normal human colonic epithelium. (H–I) Upregulation of Sparc in human colorectal adenocarcinoma. Inset in I shows SPARC localised to cell membranes.
The microarray analysis also showed results close to significance for a number of other Sparc family members. We therefore tested all of these (Hevin/Sc1, Testican1–3, Smoc1 and 2) via semi‐quantitative RT–PCR (supplementary fig 1). Hevin/Sc‐1 was downregulated in adenomas compared with normal epithelium (average fold change, 0.45, t‐test, p = 0.0004), whereas Smoc1 (average fold change 6.95, t‐test, p = 0.0359) and Testican2 (average fold change 47.83, t‐test, p = 0.0494) were upregulated. Testican 1 (t‐test, p = 0.6715), Testican 3 (t‐test, p = 0.6614), and Smoc 2 (t‐test, p = 0.1435) were not differentially regulated in adenomas compared with normal epithelium.
Taken together, our data and that of others1,6,7,8,12 point to a significant deregulation for at least four members of the Sparc family within adenomas; Sparc itself, Hevin, Smoc1 and Testican2. We found no evidence, however, that Hevin, Smoc1 or Testican2 compensate for the absence of Sparc in ApcMin/+Sparc−/− mice, as the relative deregulation of these genes was comparable in adenomas arising in either the Sparc+/+ or Sparc−/− backgrounds.
To confirm that this upregulation of SPARC messenger RNA translated into increased levels of SPARC protein, we next performed IHC for SPARC protein in normal and adenomatous epithelium from the ApcMin/+ mouse. Consistent with the microarray and semi‐quantitative RT–PCR analysis, we observed the upregulation of SPARC protein in all the adenomas we examined from the ApcMin/+ mouse (n = 20) (fig 1A–E). We also found SPARC upregulated in renal lesions and carcinoma caused by the loss of Apc in murine renal epithelium (fig 1F), implying that Sparc upregulation may occur in all Apc‐deficient tumours. To confirm previous reports that SPARC is still upregulated at later stages of tumorigenesis,3,4,5 we stained a human tumour microarray that contained 49 colorectal adenocarcinomas. All adenocarcinomas had appreciably higher levels of SPARC than control colonic epithelium (fig 1G–I). Taken together, these data suggest that SPARC is upregulated at earlier stages of colorectal cancer than previously thought, and may therefore play a role in these earlier stages.
Sparc deficiency suppresses intestinal tumorigenesis in the ApcMin/+ mouse
One of the difficulties with expression data is interpreting the in‐vivo significance of the changes. For example, the upregulation of a given gene may reflect altered cellular composition rather than altered transcriptional status. To address this directly, we wished to investigate the in‐vivo consequences of Sparc deficiency upon adenoma formation using mice deficient for Sparc. Two lines of Sparc null mice have been generated independently, with broadly similar phenotypes.9,13 Mice lacking Sparc are viable and fertile and live a normal lifespan. They develop subcortical posterior cataract at varying ages, depending on the strain background. Further phenotypic abnormalities include loss of bone density, skin laxity, increased adiposity, and kinked tails.1Sparc null mice do not show an increased incidence of spontaneous tumours, and indeed show mixed responses to implanted tumours. The transplantation of Lewis lung carcinoma and pancreatic cancer cells into Sparc null mice thus resulted in increased tumour growth, possibly as a result of a more permissive ECM environment for tumour progression.14,15 Conversely, the injection of tumour material from a mammary carcinoma model into Sparc null animals resulted in reduced tumour growth.16 In order to assess the effect of Sparc deficiency upon intestinal adenoma formation, we crossed the SparcTm1Cam null allele onto the ApcMin/+ background.
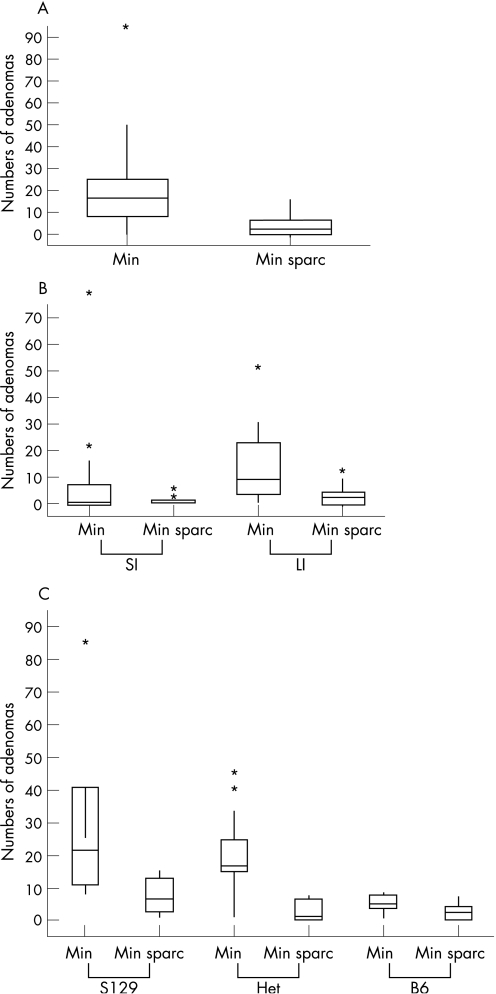
Mice were intercrossed and aged for 230 days and then adenoma number and distribution was scored (fig 2A). ApcMin/+Sparc+/+ had a median of 16 (n = 31) intestinal adenomas, whereas ApcMin/+ Sparc−/− had a median of just three (n = 21, p>0.0001 Mann–Whitney). Recent experiments have shown that heterozygosity for Bub1R accelerates colonic tumorigenesis in the ApcMin/+ mouse while suppressing small intestinal tumorigenesis.17 Therefore, we next analyzed tumour distribution to examine if suppression occurred equally between the large and small intestine. Figure 2B shows that SPARC deficiency significantly reduced adenoma formation in both the small and large intestine (small intestine p = 0.0023, Large intestine p = 0.0001, Mann–Whitney).
Figure 2 Sparc deficiency suppresses intestinal tumorigenesis at 230 days. (A) Boxplots show numbers of adenomas per mouse at 230 days. The horizontal boxed line represents the median. ApcMin/+Sparc−/− mice (n = 21) have significantly fewer tumours at 230 days than ApcMin/+Sparc+/+ mice (n = 31). (B) Sparc deficiency suppresses intestinal tumorigenesis in both the large and small intestine. Boxplots show the numbers of adenomas per mouse at 230 days. (C) Sparc deficiency suppresses intestinal tumorigenesis independently of the Mom‐1 locus. Boxplots show the numbers of adenomas per mouse at 230 days. On all three Mom‐1 backgrounds ApcMin/+Sparc−/− mice have significantly fewer tumours at 230 days than ApcMin/+Sparc+/+ mice. Box plots express the first (Q1) and third (Q3) quartiles within a given dataset by the upper and lower horizontal lines in a rectangular box, inside which is a horizontal line showing the median. The whiskers extend upwards and downwards to the highest or lowest observation within the upper (Q3+1.5 × interquartile range) and lower (Q1−1.5 × interquartile range) limits. Values outside the upper and lower limits are “outliers” and are shown by individual symbols.
This study was carried out using a second generation backcross to the C57Bl/6J background, and mice were segregating for the S129 and C57/BL6J alleles at the Mom‐1 locus. Given the significant impact of the Mom‐1 locus upon Apc‐mediated intestinal tumorigenesis,18 we therefore examined whether Sparc deficiency had suppressed tumorigenesis on all three possible Mom‐1 backgrounds. Consistent with previous studies,18 we show that mice that have the Mom‐1C57Bl6J locus developed fewer tumours than mice with the Mom‐1S129 locus (ApcMin/+Sparc+/+Mom‐1S129/S129 median 22 adenomas (n = 6), ApcMin/+ Sparc+/+Mom‐1BL6J/BL6J median five adenomas (n = 6); p = 0.003 Mann–Whitney). Irrespective of this, figure 2C shows that Sparc deficiency significantly reduced the number of adenomas at 230 days independently of Mom‐1 status (ApcMin/+Sparc+/+Mom‐1S129/S129 median 22 adenomas (n = 6) versus ApcMin/+Sparc−/−Mom‐1S129/S129 median 6.5 adenomas (n = 6); p = 0.018 Mann–Whitney: ApcMin/+Sparc+/+Mom‐1S129/BL6J median 17 adenomas (n = 19) versus ApcMin/+Sparc−/−Mom‐1S129/BL6J median one adenoma (n = 7); p = 0.0006 Mann–Whitney: ApcMin/+Sparc+/+Mom‐1BL6J/BL6J median five adenomas (n = 6) vs ApcMin/+Sparc−/−Mom‐1BL6J/BL6J median two adenomas (n = 8), p = 0.03 Mann–Whitney).
Sparc deficiency made no difference to adenoma size (small intestine adenomas; ApcMin/+Sparc+/+ median 9 mm2 (n = 64), ApcMin/+Sparc−/− median 7.5 mm2 (n = 10) p = 0.98, Mann–Whitney; large intestine adenomas; ApcMin/+Sparc+/+ median 9.0 mm2 (n = 305), ApcMin/+Sparc−/− median 12.0 mm2 (n = 65) p = 0.08, Mann–Whitney). This observation implies either that Sparc deficiency modifies adenoma initiation and is not directly relevant to adenoma growth; or that those adenomas that do arise in the absence of Sparc grow as quickly as their wild‐type counterparts after compensatory deregulation of another protein. No gross difference was seen in tumour progression or invasion, with no evidence of either adenocarcinoma or metastasis in either ApcMin/+Sparc+/+ or ApcMin/+Sparc−/− mice at 230 days.
ApcMin/+ mice are also predisposed to small pancreatic lesions, which progress to pancreatic adenocarcinoma in the additional absence of p53.19 Given that Sparc deficiency has previously been implicated in pancreatic tumorigenesis,15 we also screened for adenocarcinoma development in all cohorts. No pancreatic adenocarcinomas were observed in any mice at 230 days, a timepoint at which ApcMin/+p53−/− mice show tumorigenesis. Sparc deficiency therefore does not grossly alter the predisposition to pancreatic adenocarcinoma in this model.
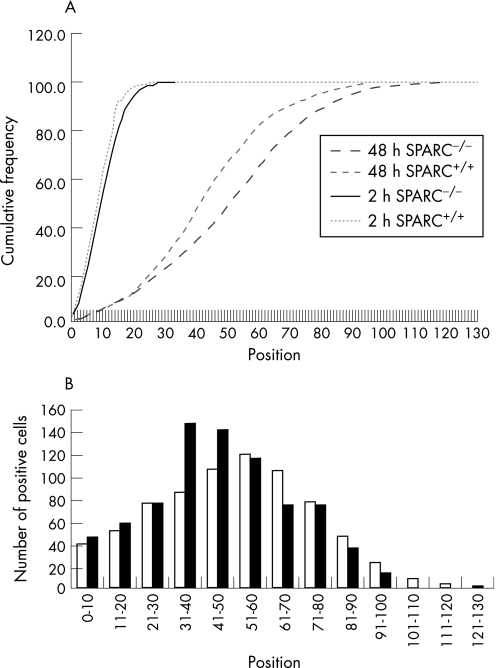
We therefore showed that Sparc deficiency protects against intestinal adenoma formation in the mouse. The mechanism underlying this protection may relate to its recognised role in mediating cell adhesion,20 which may impact upon rates of cell migration along the crypt–villus axis. Several chemopreventive agents have established roles in increasing rates of migration, which is presumed to lead to the deletion of deleterious cells, thereby suppressing adenoma formation.21,22,23 We have therefore assessed migration rates by scoring the position of BrdU‐positive cells in mice 2 and 48 hours after exposure to BrdU. Sparc deficiency did not influence either the position or number of BrdU‐positive cells 2 hours after exposure (Kolomorov–Smirnov, p = 0.3) (fig 3). At 48 hours, however, Sparc‐deficient enterocytes had moved to a higher position on the crypt villus axis (p = 0.01). This increase mimics the increase reported for aspirin, sulindac and curcumin, and therefore provides a mechanistic basis for the adenoma suppression we observed.21,22,23 This is also consistent with recent studies showing that Sparc (and Sparc thrombospondin‐2 double mutants) showed increased wound healing as a consequence of increased dermal fibroblast migration.24
Figure 3 Sparc deficiency accelerates enterocyte movement along the crypt villus axis. (A) Cumulative frequency plot showing the distribution of bromodeoxyuridine (BrdU)‐labelled cells along the length of the crypt–villus axis 2 and 48 hours after exposure to BrdU. At 2 hours there is no difference in distribution between Sparc−/−ApcMin/+ and Sparc+/+ApcMin/+ mice (Kolmogorov–Smirnov, p = 0.3). At 48 hours, enterocytes in the Sparc−/−ApcMin/+ mice have moved significantly further along the axis, indicated by a shift in the graph to the right (Kolmogorov–Smirnov, p = 0.01). (B) The distribution at 48 hours; open bars, Sparc−/−ApcMin/+, closed bars, Sparc+/+ApcMin/+. For all data, a minimum of three mice were used of each genotype.
Taken together our data highlight a novel role for Sparc, and possibly other Sparc family members, in adenoma formation. We have shown that Sparc is upregulated at the early stages of intestinal tumorigenesis and is required for efficient tumorigenesis. Given that Sparc deficiency does not lead to embryonic lethality, these results raise the possibility that Sparc may represent a good target for therapeutic intervention in colorectal cancer.
Supplementary figure 1 can be view on the Gut website at http://gut.bmj.com/supplemental.
Copyright © 2007 BMJ Publishing Group & British Society of Gastroenterology
Abbreviations
APC - Adenomatous polyposis coli
BrdU - bromodeoxyuridine
ECM - extracellular matrix
IHC - immunohistochemical analysis
PCR - polymerase chain reaction
SPARC - secreted protein acidic, rich in cysteine
Footnotes
Grants: This work was funded by Cancer Research UK.
Conflict of interest: None declared.
Supplementary figure 1 can be view on the Gut website at http://gut.bmj.com/supplemental.
References
- 1.Framson P E, Sage E H. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem 200492679–690. [DOI] [PubMed] [Google Scholar]
- 2.Schiemann B J, Neil J R, Schiemann W P. SPARC inhibits epithelial cell proliferation in part through stimulation of the transforming growth factor‐beta‐signalling system. Mol Biol Cell 2003143977–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brabender J, Lord R V, Metzger R.et al Differential SPARC mRNA expression in Barrett's oesophagus. Br J Cancer 2003891508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C S, Lin K H, Chen S L.et al Overexpression of SPARC gene in human gastric carcinoma and its clinic‐pathologic significance. Br J Cancer 2004911924–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takemasa I, Higuchi H, Yamamoto H.et al Construction of preferential cDNA microarray specialized for human colorectal carcinoma: molecular sketch of colorectal cancer. Biochem Biophys Res Commun 20012851244–1249. [DOI] [PubMed] [Google Scholar]
- 6.Boon K, Edwards J B, Eberhart C G.et al Identification of astrocytoma associated genes including cell surface markers. BMC Cancer 2004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakada M, Miyamori H, Yamashita J.et al Testican 2 abrogates inhibition of membrane‐type matrix metalloproteinases by other testican family proteins. Cancer Res 2003633364–3369. [PubMed] [Google Scholar]
- 8.Genini M, Schwalbe P, Scholl F A.et al Isolation of genes differentially expressed in human primary myoblasts and embryonal rhabdomyosarcoma. Int J Cancer 199666571–577. [DOI] [PubMed] [Google Scholar]
- 9.Gilmour D T, Lyon G J, Carlton M B.et al Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40 develop normally but show severe age‐onset cataract formation and disruption of the lens. EMBO J 1998171860–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reed K R, Sansom O J, Hayes A J.et al PPARdelta status and Apc‐mediated tumourigenesis in the mouse intestine. Oncogene 200423(55)8992–8996. [DOI] [PubMed] [Google Scholar]
- 11.Sansom O J, Griffiths D F, Reed K R.et al Apc deficiency predisposes to renal carcinoma in the mouse. Oncogene 2005248205–8210. [DOI] [PubMed] [Google Scholar]
- 12.Martinez C, Bhattacharya S, Freeman T.et al Expression profiling of murine intestinal adenomas reveals early deregulation of multiple matrix metalloproteinase (Mmp) genes. J Pathol 2005206100–110. [DOI] [PubMed] [Google Scholar]
- 13.Norose K, Clark J I, Syed N A.et al SPARC deficiency leads to early‐onset cataractogenesis. Invest Ophthalmol Vis Sci 1998392674–2680. [PubMed] [Google Scholar]
- 14.Brekken R A, Puolakkainen P, Graves D C.et al Enhanced growth of tumors in SPARC null mice is associated with changes in the 17 ECM. J Clin Invest 2003111487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puolakkainen P A, Brekken R A, Muneer S.et al Enhanced growth of pancreatic tumors in SPARC‐null mice is associated with decreased deposition of extracellular matrix and reduced tumor cell apoptosis. Mol Cancer Res 20042215–224. [PubMed] [Google Scholar]
- 16.Sangaletti S, Stoppacciaro A, Guiducci C.et al Leukocyte, rather than tumor‐produced SPARC, determines stroma and collagen type IV deposition in mammary carcinoma. J Exp Med 20031981475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao C V, Yang Y M, Swamy M V.et al Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A 20051024365–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormier R T, Hong K H, Halberg R B.et al Secretory phospholipase Pla2g2a confers resistance to intestinal tumorigenesis. Nat Genet 19971788–91. [DOI] [PubMed] [Google Scholar]
- 19.Clarke A R, Cummings M C, Harrison D J. Interaction between murine germline mutations in p53 and APC predisposes to pancreatic neoplasia but not to increased intestinal malignancy. Oncogene 1995111913–1920. [PubMed] [Google Scholar]
- 20.Murphy‐Ullrich J E. The de‐adhesive activity of matricellular proteins: is intermediate cell adhesion an adaptive state? J Clin Invest 2001107785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmoud N N, Dannenberg A J, Mestre J.et al Aspirin prevents tumors in a murine model of familial adenomatous polyposis. Surgery 1998124225–231. [PubMed] [Google Scholar]
- 22.Mahmoud N N, Bilinski R T, Churchill M R.et al Genotype–phenotype correlation in murine Apc mutation: differences in enterocyte migration and response to sulindac. Cancer Res 199959353–359. [PubMed] [Google Scholar]
- 23.Fenton J I, Wolff M S, Orth M W.et al Membrane‐type matrix metalloproteinases mediate curcumin‐induced cell migration in non‐tumorigenic colon epithelial cells differing in Apc genotype. Carcinogenesis 200261065–1070. [DOI] [PubMed] [Google Scholar]
- 24.Puolakkainen P A, Bradshaw A D, Brekken R A.et al SPARC‐thrombospondin‐2‐double‐null mice exhibit enhanced cutaneous wound healing and increased fibrovascular invasion of subcutaneous polyvinyl alcohol sponges. J Histochem Cytochem 200553571–581. [DOI] [PubMed] [Google Scholar]