Abstract
A significant proportion of colorectal cancer (CRC) develops through the serrated neoplasia pathway. Such tumours show a distinctive molecular phenotype of somatic BRAF mutations and widespread concordant methylation events in CpG islands (CIMP). These features are also observed in the polyps developing in individuals with hyperplastic polyposis syndrome (HPS). In HPS, multiple serrated polyps develop in the colorectum, and approximately 50% of cases present with at least one CRC. Observations of rare affected sibships including identical twins, suggest a recessive or co‐dominant mode of inheritance. In addition, up to 50% of individuals with HPS report a family history of CRC. At a population level, recent work has demonstrated that patients with serrated pathway cancers characterised by BRAF mutation are four times more likely to have a family history of CRC than patients with common CRC. These findings suggest an increased genetic predisposition for serrated pathway CRC in the wider population. We propose that HPS results from the inheritance of two putative co‐dominant alleles in approximately 1 in 2000 individuals. Therefore carriers of one co‐dominant allele may number up to 1 in 25, and it is likely that these carriers are at increased risk of CRC, accounting for, at least in part, the burden of serrated pathway CRC in the population. This proposition may have important implications for screening and prevention of CRC in individuals who have an increased risk for development of serrated pathway cancers, namely those with multiple, proximal, large or advanced serrated polyps, and their relatives.
During the past decade a quiet revolution has been taking place in the way we view the development of colorectal cancer. One of the most important seeds for this change has been the recognition of a serrated polyp pathway‐type colorectal tumorigenesis associated with somatic BRAF mutation and widespread gene promoter hypermethylation as an alternative to the adenoma‐carcinoma sequence.1 In this paper, we review the evidence that the development of advanced serrated polyps is a genetic trait with implications for CRC screening and prevention in the wider population.2
Heterogeneous nature of colorectal cancer
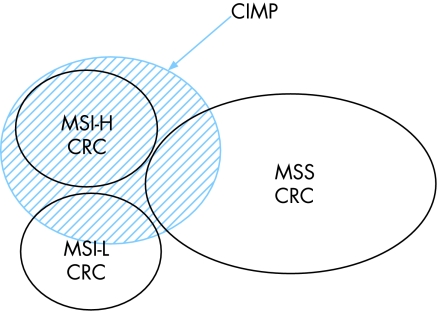
Like many epithelial malignancies, colorectal cancer (CRC) is a heterogeneous disease with respect to tumour phenotype, risk factors, genetic predisposition, response to treatment and outcome and can be classified into groups with clinical relevance using molecular pathology features.3,4 Identifying genetic and environmental risk factors for subsets within this disorder is likely to improve our understanding of aetiology, and thereby contribute to CRC prevention by targeting of screening and other preventative measures to those most at risk. Hypermethylation of CpG islands in gene promoters is a well‐accepted mechanism for expression silencing of tumour suppressor genes in a wide variety of human cancers. However, a distinct phenomenon associated with widespread and concordant CpG island methylation events was first described in 1999 in gastrointestinal tumours.5 CRCs with this CpG island methylator phenotype (CIMP CRC) demonstrate particular mutation profiles, proximal location in the colon, and increased mucinous and poorly‐differentiated histology.6 Previous observations support the notion that CIMP tumours in the colon represent a distinct sub‐type of CRC.7,8,9 For example, using multiple CpG island markers, 295 CRC and cluster analysis, Weisenberger and colleagues were able to convincingly show that CIMP CRCs possess a discrete molecular trait which includes consistent and quantitatively‐based hypermethylation of a subset of 5 CpG island markers from an original panel of 195. Subsequent validation was achieved using this novel 5‐marker panel9 in an independent series of CRC. In keeping with several previous reports, somatic BRAF mutation and CIMP were strongly associated with each other (odds ratio for association = 203), and with cancers situated in the proximal colon.9,10,11 CIMP CRCs included almost all sporadic cases with microsatellite instability (MSI) and, importantly, a further proportion of CRCs that were microsatellite stable (fig1).
Figure 1 The Distribution of CIMP and MSI in Sporadic Colorectal Cancer. Diagram shows the distribution of MSI and CIMP in sporadic CRC. CIMP accounts for all MSI‐H sporadic CRC, but only a proportion of MSI‐L and MSS subtypes.
The epidemiology of CIMP cancers
Studies investigating the epidemiology of CIMP CRCs have also provided evidence that they represent a separate entity with their own genetic and environmental risk factors.7,8,9 An association with proximal location, female sex and advanced age was observed in a case‐series study of 396 CRCs.12 The association with female gender however, was only evident when microsatellite unstable tumours were included. The epidemiology of CIMP CRC was subsequently investigated in a large population‐based panel of over 800 cases from North America.8 In this study, CIMP was unequivocally demonstrated within the population,7 occurring in 30% of CRCs, and, consistent with previous studies, showing a tight association with somatic BRAF mutation (odds ratio for association = 39). In a further analysis of this cohort, there was a tendency for CIMP microsatellite stable tumours to have a positive family history of colorectal cancer, though this was not statistically significant. Interestingly, when 911 cases from the same population cohort were analysed for somatic BRAF mutation, family history of colon cancer was significantly associated with BRAF mutation positive microsatellite stable cancers (OR 4.2; 95% CI 1.65–10.84), suggesting a genetic predisposition to develop CRC with BRAF mutation,13 ie, to develop CRC with the molecular genetic hallmarks of the serrated neoplasia pathway.
Studies on the effect of diet, particularly folate intake, on the propensity to develop CRC with CIMP and BRAF mutation have produced no consistent findings.14 However, smoking has been recently demonstrated to have an effect on the development of CIMP CRC. Samowitz and colleagues analysed the large North American population described above for a previously reported finding of an association between MSI and smoking.15 In this more recent study, smoking was significantly associated with CIMP and with BRAF mutation irrespective of MSI status.16 Individuals who smoked >20 cigarettes per day had a 2‐fold increased risk of CIMP CRC but no increased risk of CIMP negative CRC. Parallel findings were returned when tumours were analysed according to BRAF mutation status.
Serrated pathway precursor lesions
An effective approach to CRC screening and prevention is the removal of benign precursor lesions (polyps) which have a high malignant potential. The two most common epithelial polyp types in the colorectum are adenomas and serrated (hyperplastic or metaplastic) polyps. For decades following the description of the adenoma‐carcinoma developmental progression, it was believed that almost all CRCs evolved from advanced adenomatous polyps. In recent years, however, convincing evidence has emerged that a significant proportion of CRC develops within a small subset of serrated polyps, notably examples that are large, proximally located and demonstrating atypical architecture. In particular, cancers with CIMP and BRAF mutation arise in a sub‐type of serrated polyps called sessile serrated adenomas (SSA).17 The incidence of SSAs amongst lesions removed from patients undergoing colonoscopy ranges from 2–9%.18,19 Further, Spring and colleagues19 found that the presence of at least one SSA was associated with increased polyp burden consistent with an underlying predisposition. In this study, an association between family history of CRC and the presence of advanced serrated polyps was also observed, though fell short of statistical significance. In accordance with their status as a precursor lesion, SSAs demonstrate a high level of BRAF mutation,10 CIMP and a proximal predilection.19 In a study of sporadic serrated polyps, O'Brien and colleagues also demonstrated CIMP was more prevalent in larger and more proximally located lesions. Interestingly, Lazarus et al found that advanced serrated lesions recurred at twice the rate of adenomas20 after resection.
Hyperplastic polyposis syndrome and families with serrated neoplasia
Serrated polyps are a common finding in an ageing population, and the overwhelming majority are diminutive and innocuous lesions occurring in the distal colon.21 However, serrated polyps are also seen in a condition called hyperplastic polyposis syndrome (HPS). HPS has been phenotypically defined by Burt and Jass as demonstrating (1) at least five histologically diagnosed hyperplastic polyps proximal to the sigmoid colon, two of which are greater than 10 mm in diameter, or (2) any number of hyperplastic polyps occurring proximal to the sigmoid colon in an individual who has a first‐degree relative with hyperplastic polyposis, or (3) more than 30 hyperplastic polyps of any size but distributed throughout the colon.22 The condition represents a human model for the serrated pathway of CRC development analogous to familial adenomatous polyposis as a model for the sporadic adenoma‐carcinoma sequence. It was in a patient with HPS and six synchronous CRC, that the serrated pathway was first recognised at a molecular level.23 Most cases of HPS are identified in the sixth or seventh decade, though several case reports, as well as the experience of the authors, indicate that the disorder may be evident much earlier.24,25,26,27 The serrated pathway results in cancers that are characterised by somatic BRAF mutation and CIMP, features that in turn demonstrate a high rate of concordance within individual lesions in those with HPS.28,29 Further, in a large multi‐ethnic patient cohort attending a gastroenterology service in New Zealand, all cases of HPS originated from the European component, which comprised only 50% of patients (Parry et al, unpublished observations).
Though HPS may present without synchronous cancer, it is estimated that in over half of all reported cases, the findings will include at least one CRC.30,31,32 However, several series of individuals with HPS have been reported where no CRC has developed.33,34 Because HPS is relatively rare, it is difficult to assess the actual age‐related incidence of CRC in this condition due to possible ascertainment bias in the reporting of the most phenotypically interesting cases. Therefore the disparity in cancer incidence between series and case reports is likely to continue. In reports of series where CRC was present, it appeared that the risk of a synchronous CRC was higher in those with atypical or large serrated polyps, and with adenomatous or dysplastic changes.31,35
An explanation for the variability in development of CRC in HPS may lie in genetic heterogeneity. The issue of heterogeneity in HPS was first raised with the publication of six cases by Torlakovic and Snover27 in 1996. In their report, they described the polyps as more like serrated adenomas than those seen in classical cases of HPS, and suggested that this sub‐type should be noted given the possible malignant potential of the lesions. Such lesions are now referred to as SSAs, can also occur in apparently sporadic settings, and are thought to be the major precursor lesions underlying the serrated pathway.36,37 Further reports have described two sub‐types of HPS, one with multiple but not necessarily large hyperplastic polyps, and another with multiple lesions which include a diversity of polyp sub‐types including hyperplastic polyps, SSAs, traditional adenomas, and polyps with mixed elements.38,39 The latter sub‐type is more likely to have polyps >1 cm, dysplastic changes, to involve the proximal colon and to be associated with the presence of CRC.31 Such patients are more likely to demonstrate somatic BRAF mutation,28 as opposed to the KRAS mutations seen in small typical hyperplastic polyps, particularly in the distal colon.38,40 Whether the two sub‐types differ at the fundamental level of a germline mutation, or whether they are the result of a common mutation interacting with different genetic backgrounds or environmental modifiers remains to be elucidated.
Though it was initially considered to be a condition that was not associated with familial risk, HPS in a family setting has now been reported on several occasions. Descriptions of a familial syndrome with origins in the serrated pathway were initially reported from New Zealand in 1996, and in 1997.41,42 Further families with HPS were subsequently described by others.30,38,43 In addition, familial cancer syndromes associated with BRAF mutation‐bearing tumours have been described from Australia (where 2 of 11 CRC families included cases of HPS),44 and Sweden.13 As well as the presence of HPS in multiple family members,30,38,41,42,43 CRC occurs in the relatives of up to 50% of HPS cases.30,35 Further, Azimuddin and colleagues described 16 cases of large atypical hyperplastic polyps from a series of colonoscopies. All but one lesion occurred in the proximal colon, and 9 of 16 cases had a family history of CRC. Interestingly, a family history of CRC was more likely to occur where the polyps demonstrated dysplastic changes.45
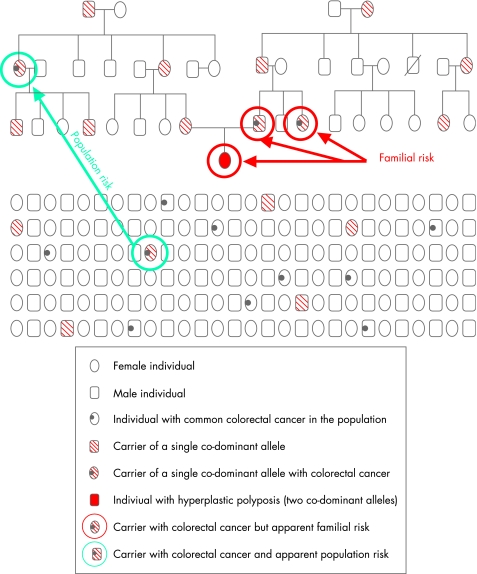
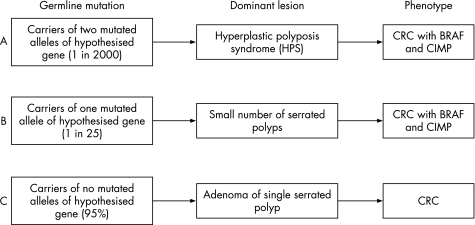
A phenotype of multiple serrated polyps, and occasional affected sibships including consanguineous kindreds30 and identical twins, suggest an autosomal recessive or co‐dominant mechanism as the most likely mode of inheritance. In a co‐dominant mode of inheritance, an intermediate phenotype may/may not be evident when the co‐dominant allele is paired with a normal allele. When paired with a recessive allele, however, the phenotype produced will differ significantly from the intermediate phenotype. It has been estimated that HPS (individuals with two putative co‐dominant alleles) occurs in the UK population at a rate of 1 in 2000.46 Given that the carriers of one co‐dominant allele may therefore number up to 1 in 25, and that up to 50% of HPS individuals report a family history of CRC,30,31 it is likely that the carriers of one allele may account, at least in part, for the burden of serrated pathway CRC in the population (fig 2). The presence of such cancers in the population therefore could be explained by the presence of common less penetrant co‐dominant alleles.47 Such individuals may develop a small number of serrated polyps and a subset of these may go on to develop a cancer bearing a BRAF mutation (fig 3).
Figure 2 Schematic of Serrated Pathway CRC in Families and in the Population. A hypothetical but typical family is depicted in the upper panel of the diagram. Co‐dominantly inherited HPS (individual with solid red symbol) assumes that both parents carry a single co‐dominant allele, and that one each of their parents in turn also carries a single allele co‐dominant allele. A subset of the carriers in a family may develop CRC, as has been reported previously.30,35,43 A simulated segment of the population is depicted in the lower panel. Given the frequency of HPS in the United Kingdom is 1 in 2000,46 carriers of one co‐dominant allele, most of whom are likely to be asymptomatic, would be distributed throughout the population at rates approaching 1 in 25. A subset of the population will develop CRC, and less than one in 10 of these cases will be non‐MSI‐H serrated pathway CRC. The individuals circled in red are likely to be identified as a family at risk due to the number of cases with colorectal neoplasms. However, the carrier in the family with CRC (circled in green) would appear as an isolated case of serrated pathway CRC in the population indistinguishable from the population‐based CRC which is characterised by BRAF mutation and microsatellite stability (also circled in green).
Figure 3 Model for the Hypothesis of the Relationship between HPS and Serrated Pathway Cancer in the Population. Diagram demonstrates the phenotypic consequences of A. Two putative co‐dominant HPS alleles; B. One putative co‐dominant HPS allele; C. Non‐carrier. Note: In B and C most will not express the phenotype.
Though MSI‐H BRAF mutation‐bearing CRC may be present in HPS,23 and in CRC families with serrated neoplasia,44 CRC in HPS are more likely to be non‐MSI‐H.30 Importantly, in the North American population, it was the non‐MSI‐H BRAF mutation‐bearing CRC which showed the strongest association with a family history of CRC.48 The association of female gender with CIMP cancers is largely confined to the MSI‐H subset,12 and individuals of advanced age. However, there is no female predominance in HPS, suggesting that associations between female gender and CIMP may be due to genetic and environmental factors outside what is proposed here.
Family cancer considerations
A major risk factor for the development of CRC is a family history of the disease, with on average 15% of all CRC cases having an affected first‐degree relative.49,50,51 Risk of CRC in relatives is also strongly related to the age of onset in the proband.49 Individuals with an affected first‐degree relative have an increased risk for developing CRC from 2–4 fold.52,53 The proportion of CRC which involves a genetic susceptibility has been estimated at about 30%, and in a significant proportion of these families, the number and distribution of affected individuals suggests that a single dominantly inherited mutation is implicated.51 The magnitude of residual familial risk is currently unknown, however it has been proposed that at least 30% of excess risk in relatives is not explained by known syndromes,49 and may involve a spectrum of modes of inheritance including autosomal dominant and co‐dominant or recessive types.
It is important to understand the genetic basis for inherited susceptibility to CRC for design of both gene discovery and family screening programs.49 Several studies have attempted to define the inheritance of the residual familial CRC risk. Aaltonen and colleagues have shown that a model in which the familial clustering of CRC was attributed to chance was dismissed with a probability of less than 0.001.49 Further, they suggested that polygenic inheritance is the most likely model to explain the excess CRC risk in the remaining families as this model performed marginally better in their study after excluding known loci. Lindor and colleagues described a cohort of non‐Lynch Syndrome familial CRC with an autosomal dominant mode of inheritance which has been confirmed by subsequent studies,54,55,56 whilst in a population‐based dataset, Hemminki and colleagues proposed a recessive mode of inheritance of proximal cancers.57 A further study by Jenkins et al supported a recessive mode of inheritance for residual genetic risk.58 In summary, there is increasing support in the literature for common lower penetrance cancer susceptibility alleles present at increased frequency in CRC cases with strong family histories compared to consecutive case series. In such families it may be difficult to determine whether the mode of inheritance is incomplete dominance or co‐dominance, and though a co‐dominant mode of inheritance is proposed in this review, the role of genetic background in incomplete dominance cannot be excluded as a possibility. In addition, confounding by somatic mutation in the population may affect the validity of a co‐dominant model.
Parallels with MUTYH‐associated polyposis
A precedent for an alternative mode of inheritance in families with a genetic susceptibility to CRC development occurs in MUTYH polyposis. In 2002, several groups discovered that biallelic mutation of a base excision repair gene MUTYH (a mutation in each of the two MUTYH alleles), predisposed to multiple adenomatous polyps, and increased the risk of CRC, in an autosomal recessively‐inherited manner.59,60 However, subsequent studies have thrown into doubt a completely recessive mode of inheritance for MUTYH mutations in the causation of CRC. Jenkins and colleagues,61 using a family‐based design have shown a higher prevalence of CRC among monoallelic carriers (those with mutation in only one MUTYH allele) compared with non‐carriers. These findings are consistent with those of other groups,62,63,64,65,66 suggesting an additional dominant effect in the inheritance of MUTYH mutation. This premise has been further supported by reports of increased LOH at the MUTYH locus in tumours where a single germline mutation is present.62,64 In examining the possibility of an additional dominant effect in MUTYH inheritance, a co‐dominant model (homozygote risk > heterozygote risk > wild‐type risk) gave a significantly better fit than did a purely recessive model. The analysis of Jenkins showed that biallelic mutation carriers were 52 times more likely to develop CRC, and monoallelic carriers are three times more likely to develop CRC, than non‐carriers. Family‐based studies are more efficient for the study of rare variants than case‐control designs,61 due to the density of potential gene carriers, and the opportunity to observe vertical transmission. Peterlongo reported three biallelic MUTYH mutation carriers in CRC families with pedigree structures suggesting dominant inheritance,67,68 whilst Croitoru et al found biallelic and monoallelic carriers were more likely to have first and second degree relatives with CRC.62 These findings are consistent with those of other groups who have used a case‐control design to compare frequency of mutations in cases to that of non‐cases. Most recently, Tenesa and colleagues69 showed in both their own data and in a meta‐analysis an additional dominant effect increasing risk 1.4 fold in the inheritance of an MUTYH mutation. Although an additional dominant effect for heterozygous mutation carriers has been found in multiple studies,61,62,63,64,66,68,69,70,71 several reports have suggested that these findings are of borderline significance, and that the question of risk in monoallelic carriers remains unanswered.67,72,73
Clinical implications of serrated neoplasia in the population
Colorectal cancer is a cause of significant cancer‐associated mortality and morbidity in Western populations. The implications of a genetic predisposition to serrated neoplasia are significant.2 Unlike the greater portion of colorectal cancers which evolve from an adenomatous polyp, cancers with CIMP and BRAF mutation arise in SSAs.17 These lesions are common in individuals with HPS, and therefore it is possible that SSAs, a lesion with a high frequency of BRAF mutation,10 and a proximal predilection,19 may be more common in European populations. It has been reported that malignant transformation in the serrated pathway may be unusually rapid in some clinical settings. Hyman and colleagues report that three cases of HPS developed CRC despite 2‐yearly colonoscopy.43 Azimuddin and colleagues presented evidence that 3‐yearly colonoscopy was inadequate for some families with atypical serrated polyps.45 Further, larger studies such as that carried out by Lazarus and colleagues suggested that serrated neoplasms are more likely to account for the occurrence of interval cancers.20 Interval cancers have been found to be three times more likely to occur in the proximal colon,74 and almost four times more likely to have high‐level microsatellite instability.75 However, there is currently no large body of data which confirms the rapid evolution to cancer of advanced serrated polyps. It is also not known for certain whether interval cancers could be explained by a flat or sessile lesion which may have been missed at colonoscopy, and if so, whether chromo‐endoscopy would address this question. Until these issues are resolved, recommendations for frequency and modality of CRC screening in those most at risk for the development of serrated neoplasia, particularly individuals with HPS and their families, remain undefined.76
It is worth speculating on the clinical implications should the hypothesised genetic causes of HPS be identified. For example if we assume: (i) a homozygous mutation in a single gene is a cause for HPS; (ii) between 1 in 1,000 and 1 in 4,000 of the population are homozygous carriers (based on a single published estimate of HPS frequency of 1 in 2000;46 and (iii) heterozygote carriers have a 1.5‐fold lifetime risk of colorectal cancer (∼10% cumulative risk); then this mutation will cause between 1 in 160 and 1 in 400 of the population to develop CRC in their lifetime because of being a mutation carrier, which is about 8%–15% of all CRC. Interestingly, somatic BRAF mutation is observed in 9% of CRC at the population level.8 The identification of the genetic variant associated with HPS will be a necessary first step in the examination of this hypothesis. Sequence variants in MYH and EPHB2 have been reported in rare HPS cases though these did not account for the majority of cases seen in the respective studies.30,77
Summary and conclusion
The proposition that a subset of serrated polyps may give rise to CRC is approaching worldwide acceptance. In Western populations, a significant proportion of CRC has its origins in serrated precursor lesions. However, it is also likely that the development of advanced serrated polyps has its basis in a relatively common genetic predisposition. Description of families and individuals with multiple serrated polyps not withstanding, the increased prevalence of HPS in Europeans, and the significantly increased risk of family history in population‐based cases of BRAF mutation‐bearing CRC both support the existence of a genetic predisposition to develop advanced serrated lesions. Cases with HPS may represent the most clinically apparent manifestation of a widespread predisposition in the population.
Acknowledgments and compliances
The authors thank Professor Bryan Parry, Professor Finlay Macrae, Mr Daniel Buchanan and Mr Michael Walsh for helpful discussions in the preparation of this manuscript.
JY is supported by Cancer Council Queensland, the National Health and Medical Research Council of Australia and National Cancer Institute Grant UO1‐ U‐01‐74778
Abbreviations
CRC - Colorectal Cancer
CIMP - CpG Island Methylator Phenotype
HPS - Hyperplastic Polyposis Syndrome
MSI - Microsatellite Instability
SSA - Sessile Serrated Adenoma
Footnotes
Competing interests: None.
References
- 1.Jass J R, Whitehall V L, Young J.et al Emerging concepts in colorectal neoplasia. Gastroenterology 2002123862–876. [DOI] [PubMed] [Google Scholar]
- 2.Young J, Jass J R. The case for a genetic predisposition to serrated neoplasia in the colorectum: hypothesis and review of the literature. Cancer Epidemiol Biomarkers Prev 2006151778–1784. [DOI] [PubMed] [Google Scholar]
- 3.Jass J R. Towards a molecular classification of colorectal cancer. Int J Colorectal Dis 199914194–200. [DOI] [PubMed] [Google Scholar]
- 4.Jass J R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 200750113–130. [DOI] [PubMed] [Google Scholar]
- 5.Toyota M, Ahuja N, Ohe‐Toyota M.et al CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999968681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rijnsoever M, Grieu F, Elsaleh H.et al Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut 200251797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issa J P, Shen L, Toyota M. CIMP, at last. Gastroenterology 20051291121–1124. [DOI] [PubMed] [Google Scholar]
- 8.Samowitz W S, Albertsen H, Herrick J.et al Evaluation of a large, population‐based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005129837–845. [DOI] [PubMed] [Google Scholar]
- 9.Weisenberger D J, Siegmund K D, Campan M.et al CpG island methylator phenotype underlies sporadic MSI and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 200638787–793. [DOI] [PubMed] [Google Scholar]
- 10.Kambara T, Simms L A, Whitehall V L.et al BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004531137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Rijnsoever M, Elsaleh H, Joseph D.et al CpG island methylator phenotype is an independent predictor of survival benefit from 5‐fluorouracil in stage III colorectal cancer. Clin Cancer Res 200392898–2903. [PubMed] [Google Scholar]
- 12.Hawkins N, Norrie M, Cheong K.et al CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 20021221376–1387. [DOI] [PubMed] [Google Scholar]
- 13.Vandrovcova J, Lagerstedt‐Robinsson K, Pahlman L.et al Somatic BRAF‐V600E Mutations in Familial Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 2006152270–2273. [DOI] [PubMed] [Google Scholar]
- 14.Slattery M L, Curtin K, Sweeney C.et al Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer 2007120656–663. [DOI] [PubMed] [Google Scholar]
- 15.Slattery M L, Samowtiz W, Ma K.et al CYP1A1, cigarette smoking, and colon and rectal cancer. Am J Epidemiol 2004160842–852. [DOI] [PubMed] [Google Scholar]
- 16.Samowitz W S, Albertsen H, Sweeney C.et al Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst 2006981731–1738. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein N S. Clinical significance of (sessile) serrated adenomas: Another piece of the puzzle. Am J Clin Pathol 2005123329–330. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi T, Sugihara K, Jass J R. Demographic and pathological characteristics of serrated polyps of colorectum. Histopathology 20054732–40. [DOI] [PubMed] [Google Scholar]
- 19.Spring K J, Zhao Z Z, Karamatic R.et al High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology 20061311400–1407. [DOI] [PubMed] [Google Scholar]
- 20.Lazarus R, Junttila O E, Karttunen T J.et al The risk of metachronous neoplasia in patients with serrated adenoma. Am J Clin Pathol 2005123349–359. [DOI] [PubMed] [Google Scholar]
- 21.Lin O S, Schembre D B, McCormick S E.et al Risk of proximal colorectal neoplasia among asymptomatic patients with distal hyperplastic polyps. Am J Med 20051181113–1119. [DOI] [PubMed] [Google Scholar]
- 22.Burt R, Jass J R. Hyperplastic polyposis. In: Hamilton SR, Aaltonen LA, eds. Pathology and Genetics of Tumours of the Digestive System. Lyon: IARC Press 2000135–136.
- 23.Jass J R, Iino H, Ruszkiewicz A.et al Neoplastic progression occurs through mutator pathways in hyperplastic polyposis of the colorectum. Gut 20004743–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengoechea O, Martinez‐Penuela J M, Larrinaga B.et al Hyperplastic polyposis of the colorectum and adenocarcinoma in a 24‐year‐old man. Am J Surg Pathol 198711323–327. [DOI] [PubMed] [Google Scholar]
- 25.Cohen S M, Brown L, Janower M L.et al Multiple metaplastic (hyperplastic) polyposis of the colon. Gastrointest Radiol 19816333–335. [DOI] [PubMed] [Google Scholar]
- 26.Keljo D J, Weinberg A G, Winick N.et al Rectal cancer in an 11‐year‐old girl with hyperplastic polyposis. J Pediatr Gastroenterol Nutr 199928327–332. [DOI] [PubMed] [Google Scholar]
- 27.Torlakovic E, Snover D C. Serrated adenomatous polyposis in humans. Gastroenterology 1996110748–755. [DOI] [PubMed] [Google Scholar]
- 28.Beach R, Chan A O, Wu T T.et al BRAF mutations in aberrant crypt foci and hyperplastic polyposis. Am J Pathol 20051661069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan A O, Issa J P, Morris J S.et al Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 2002160529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow E, Lipton L, Lynch E.et al Hyperplastic polyposis syndrome: phenotypic presentations and the role of MBD4 and MYH. Gastroenterology 200613130–39. [DOI] [PubMed] [Google Scholar]
- 31.Leggett B A, Devereaux B, Biden K.et al Hyperplastic polyposis: association with colorectal cancer. Am J Surg Pathol 200125177–184. [DOI] [PubMed] [Google Scholar]
- 32.Rubio C A, Stemme S, Jaramillo E.et al Hyperplastic polyposis coli syndrome and colorectal carcinoma. Endoscopy 200638266–270. [DOI] [PubMed] [Google Scholar]
- 33.Ferrandez A, Samowitz W, DiSario J A.et al Phenotypic characteristics and risk of cancer development in hyperplastic polyposis: case series and literature review. Am J Gastroenterol 2004992012–2018. [DOI] [PubMed] [Google Scholar]
- 34.Williams G T, Arthur J F, Bussey H J.et al Metaplastic polyps and polyposis of the colorectum. Histopathology 19804155–170. [DOI] [PubMed] [Google Scholar]
- 35.Lage P, Cravo M, Sousa R.et al Management of Portuguese patients with hyperplastic polyposis and screening of at‐risk first‐degree relatives: a contribution for future guidelines based on a clinical study. Am J Gastroenterol 2004991779–1784. [DOI] [PubMed] [Google Scholar]
- 36.Torlakovic E, Skovlund E, Snover D C.et al Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol 20032765–81. [DOI] [PubMed] [Google Scholar]
- 37.Torlakovic E, Snover D C. Sessile serrated adenoma: a brief history and current status. Crit Rev Oncog 20061227–39. [DOI] [PubMed] [Google Scholar]
- 38.Rashid A, Houlihan P S, Booker S.et al Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology 2000119323–332. [DOI] [PubMed] [Google Scholar]
- 39.Renaut A J, Douglas P R, Newstead G L. Hyperplastic polyposis of the colon and rectum. Colorectal Dis 20024213–215. [DOI] [PubMed] [Google Scholar]
- 40.O'Brien M J, Yang S, Clebanoff J L.et al Hyperplastic (serrated) polyps of the colorectum: relationship of CpG island methylator phenotype and K‐ras mutation to location and histologic subtype. Am J Surg Pathol 200428423–434. [DOI] [PubMed] [Google Scholar]
- 41.Jass J R, Cottier D S, Pokos V.et al Mixed epithelial polyps in association with hereditary non‐polyposis colorectal cancer providing an alternative pathway of cancer histogenesis. Pathology 19972928–33. [DOI] [PubMed] [Google Scholar]
- 42.Jeevaratnam P, Cottier D S, Browett P J.et al Familial giant hyperplastic polyposis predisposing to colorectal cancer: a new hereditary bowel cancer syndrome. J Pathol 199617920–25. [DOI] [PubMed] [Google Scholar]
- 43.Hyman N H, Anderson P, Blasyk H. Hyperplastic polyposis and the risk of colorectal cancer. Dis Colon Rectum 2004472101–2104. [DOI] [PubMed] [Google Scholar]
- 44.Young J, Barker M A, Simms L A.et al Evidence for BRAF mutation and variable levels of microsatellite instability in a syndrome of familial colorectal cancer. Clin Gastroenterol Hepatol 20053254–263. [DOI] [PubMed] [Google Scholar]
- 45.Azimuddin K, Stasik J J, Khubchandani I T.et al Hyperplastic polyps: “more than meets the eye”? Report of 16 cases. Dis Colon Rectum 2000431309–1313. [DOI] [PubMed] [Google Scholar]
- 46.Lockett M J, Atkin W S. Hyperplastic polyposis: prevalence and cancer risk. Gut 200148A4 [Google Scholar]
- 47.Kampman E. A First‐Degree Relative with Colorectal Cancer: What Are We Missing? Cancer Epidemiol Biomarkers Prev 2007161–3. [DOI] [PubMed] [Google Scholar]
- 48.Samowitz W S, Sweeney C, Herrick J.et al Poor survival associated with the BRAF V600E mutation in microsatellite‐stable colon cancers. Cancer Res 2005656063–6069. [DOI] [PubMed] [Google Scholar]
- 49.Aaltonen L, Johns L, Jarvinen H.et al Explaining the Familial Colorectal Cancer Risk Associated with Mismatch Repair (MMR)‐Deficient and MMR‐Stable Tumors. Clin Cancer Res 200713356–361. [DOI] [PubMed] [Google Scholar]
- 50.Carstensen B, Soll‐Johanning H, Villadsen E.et al Familial aggregation of colorectal cancer in the general population. Int J Cancer 199668428–435. [DOI] [PubMed] [Google Scholar]
- 51.Houlston R S, Collins A, Slack J.et al Dominant genes for colorectal cancer are not rare. Ann Hum Genet. 1992;56(Pt 2)99–103. [DOI] [PubMed]
- 52.Johns L E, Houlston R S. A systematic review and meta‐analysis of familial colorectal cancer risk. Am J Gastroenterol 2001962992–3003. [DOI] [PubMed] [Google Scholar]
- 53.Johns L E, Kee F, Collins B J.et al Colorectal cancer mortality in first‐degree relatives of early‐onset colorectal cancer cases. Dis Colon Rectum 200245681–686. [DOI] [PubMed] [Google Scholar]
- 54.Lindor N M, Rabe K, Petersen G M.et al Lower cancer incidence in Amsterdam‐I criteria families without mismatch repair deficiency: familial colorectal cancer type X. Jama 20052931979–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llor X, Pons E, Xicola R M.et al Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res 2005117304–7310. [DOI] [PubMed] [Google Scholar]
- 56.Mueller‐Koch Y, Vogelsang H, Kopp R.et al Hereditary non‐polyposis colorectal cancer: clinical and molecular evidence for a new entity of hereditary colorectal cancer. Gut 2005541733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemminki K, Chen B. Familial risks for colorectal cancer show evidence on recessive inheritance. Int J Cancer 2005115835–838. [DOI] [PubMed] [Google Scholar]
- 58.Jenkins M A, Baglietto L, Dite G S.et al After hMSH2 and hMLH1‐‐what next? Analysis of three‐generational, population‐based, early‐onset colorectal cancer families. Int J Cancer 2002102166–171. [DOI] [PubMed] [Google Scholar]
- 59.Al‐Tassan N, Chmiel N H, Maynard J.et al Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat Genet 200230227–232. [DOI] [PubMed] [Google Scholar]
- 60.Sieber O M, Lipton L, Crabtree M.et al Multiple colorectal adenomas, classic adenomatous polyposis, and germ‐line mutations in MYH. N Engl J Med 2003348791–799. [DOI] [PubMed] [Google Scholar]
- 61.Jenkins M A, Croitoru M E, Monga N.et al Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population‐based case‐family study. Cancer Epidemiol Biomarkers Prev 200615312–314. [DOI] [PubMed] [Google Scholar]
- 62.Croitoru M E, Cleary S P, Di Nicola N.et al Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst 2004961631–1634. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Baudhuin L M, Boardman L A.et al MYH mutations in patients with attenuated and classic polyposis and with young‐onset colorectal cancer without polyps. Gastroenterology 20041279–16. [DOI] [PubMed] [Google Scholar]
- 64.Kambara T, Whitehall V L, Spring K J.et al Role of inherited defects of MYH in the development of sporadic colorectal cancer. Genes Chromosomes Cancer 2004401–9. [DOI] [PubMed] [Google Scholar]
- 65.Fleischmann C, Peto J, Cheadle J.et al Comprehensive analysis of the contribution of germline MYH variation to early‐onset colorectal cancer. Int J Cancer 2004109554–558. [DOI] [PubMed] [Google Scholar]
- 66.Enholm S, Hienonen T, Suomalainen A.et al Proportion and phenotype of MYH‐associated colorectal neoplasia in a population‐based series of Finnish colorectal cancer patients. Am J Pathol 2003163827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterlongo P, Mitra N, Chuai S.et al Colorectal cancer risk in individuals with biallelic or monoallelic mutations of MYH. Int J Cancer 2005114505–507. [DOI] [PubMed] [Google Scholar]
- 68.Peterlongo P, Mitra N, Sanchez de Abajo A.et al Increased frequency of disease‐causing MYH mutations in colon cancer families. Carcinogenesis 2006272243–2249. [DOI] [PubMed] [Google Scholar]
- 69.Tenesa A, Farrington S M, Dunlop M G. Re: Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst. 2005;97:320–1; author reply 321–2. [DOI] [PubMed]
- 70.Croitoru M E, Cleary S P, Berk T.et al Germline MYH mutations in a clinic‐based series of Canadian multiple colorectal adenoma patients. J Surg Oncol 2007 [DOI] [PubMed]
- 71.Farrington S M, Tenesa A, Barnetson R.et al Germline susceptibility to colorectal cancer due to base‐excision repair gene defects. Am J Hum Genet 200577112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balaguer F, Castellvi‐Bel S, Castells A.et al Identification of MYH mutation carriers in colorectal cancer: a multicenter, case‐control, population‐based study. Clin Gastroenterol Hepatol 20075379–387. [DOI] [PubMed] [Google Scholar]
- 73.Webb E L, Rudd M F, Houlston R S. Colorectal cancer risk in monoallelic carriers of MYH variants. Am J Hum Genet. 2006;79:768–71; author reply 771–2. [DOI] [PMC free article] [PubMed]
- 74.Farrar W D, Sawhney M S, Nelson D B.et al Colorectal cancers found after a complete colonoscopy. Clin Gastroenterol Hepatol 200641259–1264. [DOI] [PubMed] [Google Scholar]
- 75.Sawhney M S, Farrar W D, Gudiseva S.et al Microsatellite instability in interval colon cancers. Gastroenterology 20061311700–1705. [DOI] [PubMed] [Google Scholar]
- 76.Winawer S J, Zauber A G, Fletcher R H.et al Guidelines for colonoscopy surveillance after polypectomy: a consensus update by the US Multi‐Society Task Force on Colorectal Cancer and the American Cancer Society. Gastroenterology 20061301872–1885. [DOI] [PubMed] [Google Scholar]
- 77.Kokko A, Laiho P, Lehtonen R.et al EPHB2 germline variants in patients with colorectal cancer or hyperplastic polyposis. BMC Cancer 20066145. [DOI] [PMC free article] [PubMed] [Google Scholar]