Abstract
Two forms of RNA Polymerase IV (PolIVa/PolIVb) have been implicated in RNA-directed DNA methylation (RdDM) in Arabidopsis. Prevailing models imply a distinct function for PolIVb by association of Argonaute4 (AGO4) with the C-terminal domain (CTD) of its largest subunit NRPD1b. Here we show that the extended CTD of NRPD1b-type proteins exhibits conserved Argonaute-binding capacity through a WG/GW-rich region that functionally distinguishes Pol IVb from Pol IVa, and that is essential for RdDM. Site-specific mutagenesis and domain-swapping experiments between AtNRPD1b and the human protein GW182 demonstrated that reiterated WG/GW motifs form evolutionarily and functionally conserved Argonaute-binding platforms in RNA interference (RNAi)-related components.
Keywords: RNAPIV; CTD; WG/GW motifs, Argonaute; RNA silencing
In eukaryotes, small RNAs (sRNAs) of ∼20–30 nucleotides (nt) are essential effectors of RNA silencing, a conserved pathway that leads to suppression of gene expression and plays important roles in many biological processes (Baulcombe 2004). sRNAs act as guides to direct mRNA degradation, translational repression, heterochromatin formation, and DNA elimination. In most RNA interference (RNAi) pathways, production of sRNAs is initiated by Dicer-like ribonucleases acting on double-stranded RNAs (dsRNAs) of various origins. miRNAs are generated from imperfect dsRNA regions originating from hairpin-shaped precursors (Baulcombe 2004), whereas small interfering RNAs (siRNAs) are produced from long dsRNAs derived from either convergent transcription or the action of an RNA-dependent RNA polymerase (RdRP) on endogenous single-stranded templates (Brodersen and Voinnet 2006). To accomplish their regulatory functions, sRNAs are incorporated into effector complexes by association with one of the Argonaute (AGO) family members. MicroRNA/siRNAs associate with AGOs to form RNA-induced silencing complexes (RISCs), which elicit decay or translational repression of complementary mRNA targets, a process known as post-transcriptional gene silencing (PTGS) (Peters and Meister 2007). Heterochromatic siRNAs interact with AGOs to form RNA-induced initiation of transcriptional silencing complexes (RITS), which direct heterochromatin formation on DNA targets, a process known as transcriptional gene silencing (TGS) (Zaratiegui et al. 2007). Both RISC- and RITS-associated AGO components have slicing activity that initiates the silencing process by cleaving mRNA/nascent transcript targets at sRNA-complementary sites (Peters and Meister 2007; Zaratiegui et al. 2007). However, slicing is not a prerequisite for silencing, as animal miRNAs predominantly repress gene expression at the translational level or affect mRNA stability by promoting mRNA deadenylation and decapping in cellular processing bodies (Eulalio et al. 2007).
Plants have evolved a sophisticated network of RNAi-related pathways producing distinct types of sRNAs that function in both PTGS and TGS (Brodersen and Voinnet 2006). RNA-directed DNA methylation (RdDM) is the prominent nuclear silencing pathway in plants, characterized by the biogenesis of a specific class of 24-nt sRNAs presumed to guide formation of transcriptionally silent heterochromatin at repeated DNA loci (Brodersen and Voinnet 2006). Forward and reverse genetic approaches have implicated conserved RNAi proteins, such as DICER-like 3 (DCL3), RNA-dependent RNA polymerase 2 (RDR2) and Argonaute4 (AGO4), in generating and utilizing sRNAs in the RdDM pathway (Zilberman et al. 2003; Xie et al. 2004). More recently, a plant-specific eukaryotic-type RNA polymerase, RNA Polymerase IV (Pol IV), has been implicated in biosynthesis of the 24-nt sRNA class in Arabidopsis and shown to be essential for RdDM (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005; Zhang et al. 2007). Unlike Pol I, Pol II, and Pol III, Pol IV exists in two forms, Pol IVa and Pol IVb, which share the same second largest subunit, NRPD2a, but differ in their largest subunit, named NRPD1a and NRPD1b, respectively (Kanno et al. 2005; Pontier et al. 2005). Analysis of nrpd mutants suggested that Pol IVa and Pol IVb play complementary roles in RdDM, robust silencing at repeated DNA loci requiring sequential engagement of both forms (Kanno et al. 2005; Pontier et al. 2005). PolIVa/RDR2/DCL3 were proposed to play an upstream role in RdDM by producing the 24-nt sRNAs with Pol IVb/AGO4/ sRNAs acting downstream in targeting DNA methylation (Chan et al. 2006; Li et al. 2006; Pontes et al. 2006).
Although the exact compositions of Pol IVa and Pol IVb remain to be determined, the functional distinction between the two forms probably depends on differences in their specific largest subunit. Indeed, one striking difference between NRPD1a and NRPD1b is the presence of an extended, repeat-containing C-terminal domain (CTD) in NRPD1b (Pontier et al. 2005) that has been shown to interact with AGO4 (Li et al. 2006). This partnership could endow NRPD1b/PolIVb with an enhanced capacity to couple transcription to post-transcriptional silencing events. To determine to what extent the CTD of Pol IVb is essential for RdDM, and the nature of the molecular determinants controlling the binding of AGO4 to this region, we have undertaken a biochemical and functional analysis of this domain. Here we show that AGO4 indeed binds the reiterated specific domain of the NRPD1b CTD, and that this region functionally distinguishes Pol IVb from Pol IVa, conferring an essential role for the former in RdDM. Sequence analysis of the AGO4-binding region shows the presence of WG/GW repeat signatures that are also found in otherwise divergent proteins from a wide variety of organisms. Mutagenesis and domain-swapping experiments between the WG/GW-rich regions of AtNRPD1b and human RNAi-related protein GW182 confirm that these domains form evolutionarily and functionally conserved AGO-binding platforms. Taken together, our results indicate that the WG/GW are motifs of ancient origin that have coevolved with AGO proteins throughout the eukaryotic lineage to fulfill the role of AGO-binding determinants that somehow confer additional/novel functions to specific proteins.
Results and Discussion
The reiterated region of the Pol IVb CTD binds AGO4 and is required for RdDM
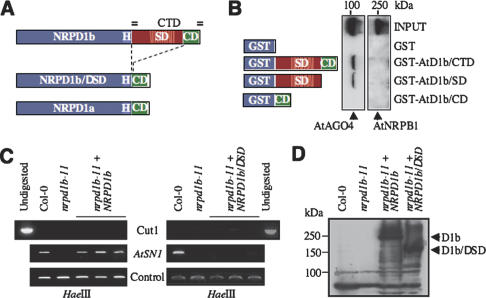
The Arabidopsis genome encodes two forms of the largest subunit of Pol IV (NRPD1a and NRPD1b), both containing a C-terminal DCL-like domain (common domain, CD), while NRPD1b is characterized by an additional, reiterated hydrophilic domain (specific domain, SD) (Fig. 1A; Kanno et al. 2005; Pontier et al. 2005). Two truncated fusion proteins (GST-AtD1b/SD and GST-AtD1b/CD) (Fig. 1B), in which one or other of these subdomains was removed, were used to define precisely the region of the NRPD1b CTD necessary for interaction with AGO4, using GST and GST-AtD1b/CTD constructs as controls. Figure 1B shows that GST-AtD1b/SD, but not GST-AtD1b/CD, was able to specifically interact with AGO4, indicating that it recognizes the SD. This interaction is conserved as AGO4 also interacts with the SD of spinach (Spinacia oleracea, So) NRPD1b (Fig. 2C, lane GST-SoD1b/SD). This result is fully consistent with our previous data indicating that AGO4 associates specifically with NRPD1b/Pol IVb in vivo (Li et al. 2006).
Figure 1.
The SD of the NRPD1b CTD binds AGO4 and is required for RdDM in Arabidopsis. (A) AtNRPD1a/b and NRPD1b/DSD proteins are shown schematically. The SD and CD are shown in red and green, respectively. The conserved box H and the CTD repeats are indicated. (B) Myc-AGO4 extract was applied onto equimolar amounts of GST and GST-based fusion protein beads, and the bound proteins were detected by immunoblotting with anti-Myc and anti-RPB1 antibodies. (C) AtSN1 DNA methylation in nrpd1b-11 is rescued by transgenes expressing NRPD1b but not NRPD1b/DSD (three T1 transgenic lines for each construct are shown). AtSN1 retroelement and controls (Cut 1 and control lanes) were amplified from HaeIII-digested genomic DNA as described (Pontier et al. 2005). (D) Proteins were extracted from pooled samples of the T1 transgenic lines shown in C, quantified, and subjected to immunoblot analysis using the anti-Flag M2 monoclonal antibodies (Sigma-Aldrich).
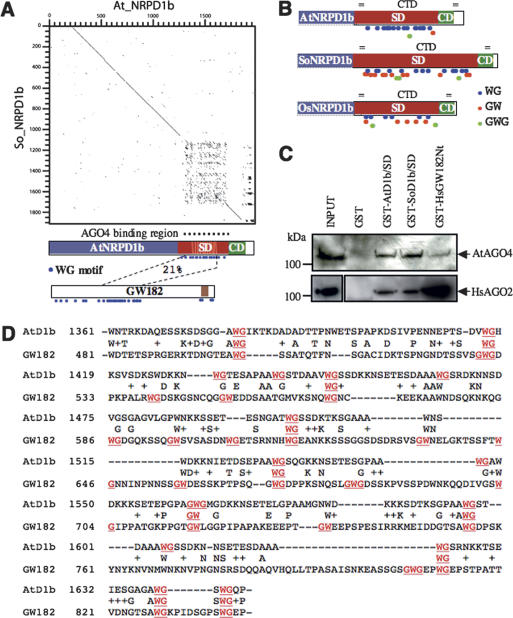
Figure 2.
The SD of the NRPD1b CTD has a WG/GW repeat signature sequence. (A) Dot plot analysis of AtNRPD1b versus SoNRPD1b highlighting the divergent region of the CTD. Positions of the divergent and AGO4-binding regions are indicated in relation to a schematic representation of AtNRPD1b. The SD shows sequence identity to the N-terminal region of HsGW182. The WG motifs are indicated with blue dots. The RNA recognition motif (RRM) domain of HsGW182 is indicated in brown. (B) Relative distribution of the WG/GW/GWG motifs within the SDs of plant NRPD1b proteins. (C) Myc-AGO4 or HA-AGO2 extracts were applied onto equimolar amounts of GST and GST-based fusion protein beads, and bound proteins were detected by immunoblotting with anti-Myc and anti-HA antibodies. (D) Alignment of AtNRPD1b and HsGW182. The WG/GW repeats are highlighted in red.
To investigate the importance of the AGO4-binding region for Pol IVb function in vivo, we tested the ability of a truncated NRPD1b protein lacking SD (NRPD1b/DSD) (Fig. 1A) to restore de novo DNA methylation at AtSN1 and 5S rDNA loci in the nrpd1b-11 mutant (Pontier et al. 2005). To minimize detrimental side effects of deletion of the SD on the general architecture of NRPD1b, the length of the linker separating the CD from the evolutionary conserved box H was kept similar in NRPD1b/DSD compared with NRPD1a (Supplementary Fig. S1A). Analysis of the genomic construct transgenes expressing epitope-tagged NRPD1b and NRPD1b/DSD proteins indicated that, in contrast to the clear capacity of NRPD1b to rescue its corresponding mutation and restore DNA methylation, NRPD1b/DSD had no effect on the methylation status of the nrpd1b-11 mutant (Fig. 1C; Supplementary Fig. S1B) despite confirmation of its expression level by Western blotting (Fig. 1D). Collectively, our results indicate that the SD of the NRPD1b CTD binds AGO4 and confers an essential role for NRPD1b/Pol IVb in RdDM.
The SD of the NRPD1b CTD has a WG/GW repeat signature sequence
To learn more about molecular determinants in the SD of AtNRPD1b that are involved in AGO4 binding, we compared the AtNRPD1b sequence with proteins from other plants. A dot plot of AtNRPD1b against the partial sequence of SoNRPD1b surprisingly shows that the SD regions are highly divergent, in contrast to considerable sequence conservation in the rest of the proteins (Fig. 2A; Supplementary Fig. S2). BLAST alignment indicates that the only conserved sequences between the two proteins are neighboring tryptophan–glycine/glycine–tryptophan residues (WG, GW, or GWG, henceforth referred to as WG/GW) present throughout the SDs (Supplementary Fig. S2B). Interestingly, nine out of 17 WG present in the SD of AtNRPD1b are located in a highly conserved region of the imperfect 16-amino-acid repeats identified previously in the extended CTD (Supplementary Fig. S3; Pontier et al. 2005). Alignment of the Arabidopsis sequence with rice (Oryza sativa, Os) NRPD1b shows similar organization (Supplementary Fig. S4), the Arabidopsis SD having only 23% amino acid identity with both spinach and rice sequences, while the rest of the proteins are ∼60% identical in both cases. In summary, our sequence analysis indicates that WG/GW motifs are evolutionarily conserved and widely distributed within the SDs of three Pol IV NRPD1b subunits (Fig. 2B).
These observations prompted us to look for proteins in other organisms having similar WG/GW-rich domains. Given the biased amino acid composition of this region, we used the PSI-BLAST program (Altschul et al. 1997) to compare the repeat domain with sequences in the nonredundant protein database. This analysis identified a human protein, HsGW182 (also known as HsTNRC6A), that contains an ∼350-amino-acid domain having a similar WG/GW-rich composition (Fig. 2A,D). Furthermore, an analysis of the amino acid content of the aligned regions from the Arabidopsis and human proteins showed a highly similar bias in composition (Supplementary Fig. S5), the two domains being rich in glycine, serine, and tryptophan, and to a lesser extent, glutamic acid, aspartic acid, and asparagine. On the other hand, certain residues are present at very low levels, notably cysteine, phenylalanine, histidine, methionine, and tyrosine. We were particularly intrigued by this sequence similarity as HsGW182 was first described as a component of cytoplasmic bodies (Eystathioy et al. 2002) that have been linked to RNAi (Jakymiw et al. 2005; Liu et al. 2005), and has been shown to interact with HsAGO1 and HsAGO2 (Jakymiw et al. 2005; Liu et al. 2005). Taken together, these data point to a possible functional convergence as AGO-binding domains of both WG/GW-rich regions of the silencing-related proteins AtNRPD1b and HsGW182 and suggest these regions could be interchangeable in terms of AGO-binding capacity. To explore this possibility further, we generated a fusion protein containing the WG/GW-rich region of HsGW182 (GST-HsGW182Nt) and monitored the ability of this construct to interact with AtAGO4 or HsAGO2 proteins compared with the plant NRPD1b-derived constructs, GST-AtD1b/SD and GST-SoD1b/SD. Both AtAGO4 and HsAGO2 were able to interact with the three WG/GW-rich constructs regardless of their origins with, however, a slight preference for plant over human WG/GW-rich sequences for AtAGO4 and, conversely, a stronger preference for human over plant WG/GW-rich sequences for HsAGO2 (Fig. 2C).
Reiterated WG/GW motifs form evolutionarily conserved AGO-binding platforms
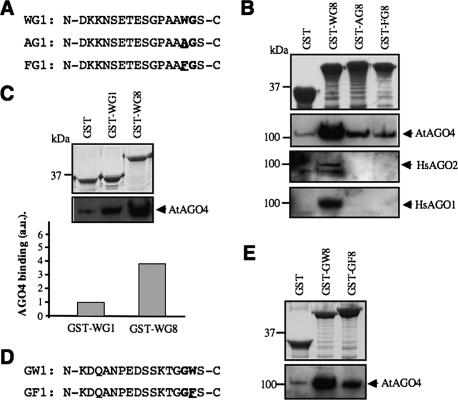
Having established that WG/GW-rich regions have conserved AGO-binding capacity, we asked whether these motifs are directly involved in this binding. In both NRPD1b- and GW182-type proteins, most tryptophan residues are in degenerate repeats (Eystathioy et al. 2002; Pontier et al. 2005), comprised primarily of apparently randomly distributed WGs. To assess the role of these motifs in binding AGOs, we generated several fusion constructs in which wild-type or mutant versions of the AtNRPD1b consensus monomer (Fig. 3A), referred to hereafter as WG1/AG1/FG1, were multimerized to eight copies at the C-terminal end of the GST moiety, generating GST-WG8/AG8/FG8 constructs (Fig. 3B). Consensus repeat mutants contained tryptophan-to-alanine (WG1 → AG1) and tryptophan-to-phenylalanine (WG1 → FG1) substitutions (Fig. 3A), two mutations allowing direct assessment of the contribution of the tryptophan side chain to the affinity of AGOs for the GST fusion proteins. Pull-down assays confirmed that the GST-WG8 interacts with AtAGO4 with high affinity, whereas both GST-AG8/FG8 exhibit strongly reduced affinity (Fig. 3B), indicating that WG motifs are critical determinants in the binding of AtAGO4 to the SD of AtNRPD1b. Consistent with a conserved AGO-binding capacity, both HsAGO1 and HsAGO2, two phylogenetically distant AGOs, also show tryptophan-dependent binding to GST-WG8 (Fig. 3B). AtAGO4 also binds specifically to GST-WG1, indicating that a single motif is sufficient to direct binding (Fig. 3C). Quantitative analysis of AtAGO4 binding to GST-WG1 compared with GST-WG8 also suggested that multiple AGOs can bind the latter protein (Fig. 3C). Likewise, pull-down assays indicated that a GST-GW8 fusion protein (derived from a SoNRPD1b loose consensus monomer) interacts with AtAGO4, whereas GST-GF8 mutant protein has strongly reduced affinity (Fig. 3D,E). Collectively, our results indicate that reiterated WG/GW motifs are critical determinants in the binding of AtAGO4 to the SD and form evolutionarily conserved AGO-binding platforms.
Figure 3.
Reiterated WG/GW motifs form conserved AGO-binding platform. (A) Sequences of wild-type and mutant AtNRPD1b consensus monomers. (B) Myc-AGO4 and HA-AGO1/2 extracts were applied onto equimolar amounts of GST and GST-based fusion protein beads, and bound proteins were detected by immunoblotting with anti-Myc and anti-HA antibodies. (C) Myc-AGO4 extract was applied onto equimolar amounts of GST, GST-WG1/WG8 beads, and bound proteins were detected by immunoblotting and quantified using ImageJ software. (D) Sequences of wild-type and mutant SoNRPD1b consensus monomers. (E) Myc-AGO4 extract was applied onto equimolar amounts of GST, GST-GW8/GF8 beads, and bound proteins were detected by immunoblotting with anti-Myc antibodies.
Conserved AGO-binding platforms are functional in vivo
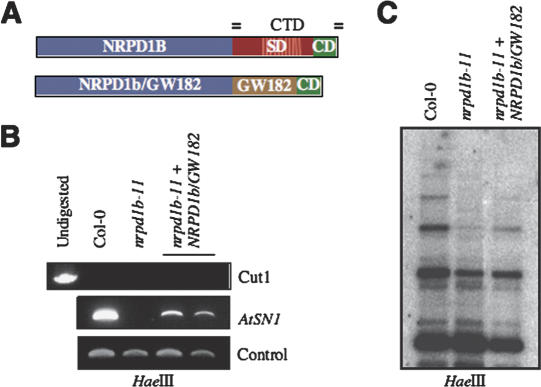
To further assess whether the presence of reiterated WG/GW motifs is sufficient for SD function in AtNRPD1b, we constructed a mutant, NRPD1b/GW182 (Fig. 4A), in which the SD of AtNRPD1b was substituted by its homologous region in HsGW182, two protein domains whose primary sequence similarity is limited to similar WG/GW-rich composition (Fig. 2D). The ability of NRPD1b/GW182 to restore de novo DNA methylation in our in vivo complementation system was tested on several T1 transgenes expressing the domain-swapping mutant in nrpd1b-11. As shown for two representative T1 transgenic lines, rescue experiments indicate that NRPD1b/GW182 partially restored de novo DNA methylation at both AtSN1 and 5S rDNA loci (Fig. 4B,C) as compared with the wild-type NRPD1b protein, suggesting that the SD of NRPD1b and the N-terminal region of HsGW182 form functionally equivalent domains. This finding highlights the importance of the SD of the NRPD1b CTD in RdDM, and supports a model in which AGO binding to WG/GW-rich protein platforms is evolutionarily and functionally conserved in RNAi-related proteins.
Figure 4.
Reiterated WG/GW motifs form functionally conserved AGO-binding platforms. (A) AtNRPD1b and NRPD1b/GW182 mutant are shown schematically. The N-terminal WG/GW-rich region of HsGW182 is shown in brown. (B) AtSN1 DNA methylation in nrpd1b-11 mutant is partially restored by transgenes expressing NRPD1b/GW182 (two T1 transgenic lines are shown). (C) 5S DNA methylation in nrpd1b-11 mutant is partially restored by transgenes expressing NRPD1b/GW182 (DNA was extracted from pooled samples of four T1 transgenic lines).
Widespread distribution of WG platforms
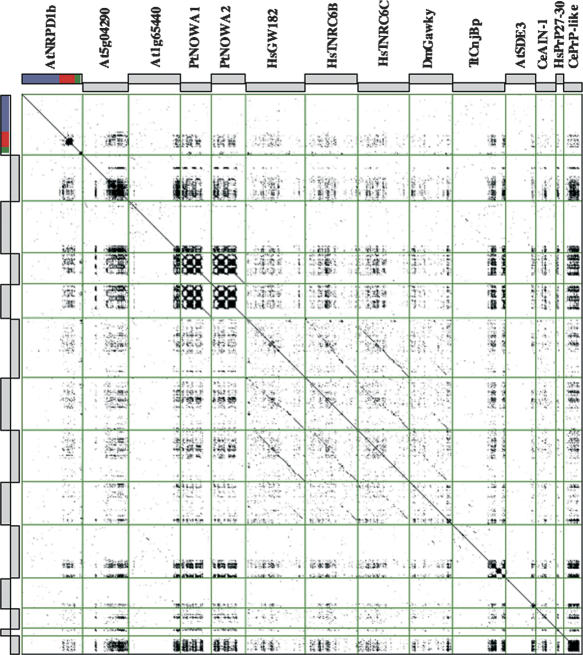
Despite their presence in a number of proteins from widely divergent organisms, WG/GW domains, first described in HsGW182 (Eystathioy et al. 2002), often warranted fleeting mention at best. The low sequence similarity between repeats from even closely related species means that they are not easily detected by sequence comparison or motif search algorithms. Targeted genus and species-specific PSI-BLAST alignment identified proteins harboring at least five contiguous WG/GW repeats in organisms from humans to Tetrahymena thermophila (Fig. 5). The complete sequences were compared by an all-against-all dot plot that clearly shows that, for the majority of these proteins, sequence similarity is only observed in the WG/GW domains (Fig. 5).
Figure 5.
Exhaustive search for conserved WG/GW platforms. Proteins containing conserved WG/GW motifs from different organisms were identified by PSI-BLAST alignment of the SD from AtNRPD1b against protein databases. An all-against-all alignment of the identified proteins was carried out using the Dotter program. The figure presents a twofold mirror image of the comparison of each sequence against all of the others, the diagonal representing self-versus-self alignments highlighting intramolecular repetitions. Sequences used were Arabidopsis thaliana NRPD1b; conceptual translations of Arabidopsis genes At5g04290 and At1g65440; P. tetraurelia; NOWA1/2; Homo sapiens proteins GW182, TNRC6B, and TNRC6C; Drosophila melanogaster protein GAWKY; T. thermophila cnjB; A. thaliana SDE3; C. elegans AIN-1; H. sapiens PrP27-30; and C. elegans PrP-like.
Interestingly, almost all the proteins identified by our analysis have proposed functions that are related to targeted nucleic acid modification, with several of them being implicated in RNAi-related pathways. HsGW182, which is discussed above, and HsTNRC6B have been shown to interact with AGOs, while the similar human protein, TNRC6C, has also been localized in cytoplasmic GW-bodies (Schneider et al. 2006). The Caenorhabditis elegans developmental timing regulator, AIN-1, has been proposed to target the AGO protein ALG-1 to GW-bodies (Ding et al. 2005). The Nowa proteins from Paramecium tetraurelia, identified as containing short repeats similar to those of the PrP27 prion, were shown to be essential components of a trans-nuclear cross-talk mechanism that is responsible for epigenetic programming of genome rearrangements (Nowacki et al. 2005). No clear role has yet been described for the T. thermophila conjugation-specific cnjB protein (Taylor and Martindale 1993), although RNAi has since been implicated in conjugation in this organism (Yao et al. 2003). Interestingly, the C-terminal repeat region of TtcnjB shows an amino acid compositional bias similar to that of NRPD1b, being rich in tryptophan, glycine, serine, and asparagine residues. A puzzling WG/GW platform candidat inferred from sequence similarity is the human major prion protein precursor PrP27-30 (accession no. P04156) that has an N-terminal region containing several GWG octapeptide repeats (Prusiner 1998). Although a role in nucleic acid metabolism has been suggested for this protein (Gabus et al. 2001), no direct link has yet been described between prions and RNA silencing. Similarly, a C. elegans putative protein referred to as PrP-like Q/N-rich protein 29 (pqn-29, accession no. NP_497948) was identified as containing a large central WG/GW platform, but more studies will be necessary in order to reach any definitive conclusion concerning the role of this putative protein. In Arabidopsis, proteins encoded by At5g04290 and At1g65440 are members of the SPT5 and SPT6 families of transcription elongation factors but differ from known SPTs by the presence of large C-terminal extension with WG repeats (Ponting 2002), while SDE3, carrying a GW C-terminal extension, is an RNA helicase required for PTGS (Dalmay et al. 2001). Pairwise alignments confirm that only the repeated WG/GW motifs show similarity between these divergent proteins (results not shown). Overall, our observations strongly suggest the presence of compositionally biased WG/GW platforms in many proteins from widely divergent species.
We show here in the extended CTD of the plant NRPD1b proteins the presence of an evolutionarily conserved WG/GW-rich AGO4-binding platform that is essential for RdDM and for the specific function of PolIVb compared with PolIVa. An AGO-associated role for WG/GW-rich proteins was first suggested for human GW182, and also subsequently has been shown for the paralogous and homologous sequences TNRC6B, TNRC6C, GAWKY, and AIN-1 (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005; Behm-Ansmant et al. 2006; Schneider et al. 2006). All these regions are characterized by low sequence similarity and the presence of conserved GW/WG motifs. Our analysis shows that WG/GW-rich region are interchangeable in terms of AGO-binding capacity, and suggests that they are capable of interaction with multiple AGOs in a tryptophan-dependent manner. The importance of tryptophan residues was recently suggested by Partridge et al. (2007), who showed that mutation of a WG motif in the central region of the Schizosaccharomyces pombe TAS3 protein abolishes TAS3–AGO1 binding, which is essential for the establishment of centromeric heterochromatin. Although our results do not allow a precise definition of the region of Argonaute involved in interaction with the WG platform, it is interesting to note that the WG/GW-rich subregion of Drosophila GAWKY shown by Behm-Ansmant et al. (2006) to interact with the PIWI domain of Drosophila AGO1 is the most similar to both the Arabidopsis NRPD1 SD and the human GW182 fragment used here. As a whole, our observations suggest the existence of an ancestral AGO-binding platform whose association with other functional motifs in proteins from widely divergent species confers different specificities in AGO-related functions. The conclusion that the platforms confer additional functions to specific proteins is reinforced by the presence in Arabidopsis (and other plants) of proteins homologous with NRPD1b, At5g04290, and At1g65440 but lacking WG/GW motifs (Pontier et al. 2005; data not shown). The presence of potential platforms in proteins for which no AGO-related function has yet been shown raises the intriguing possibility of a role for such platforms in a number of other cellular functions.
Materials and methods
Methods in sequence analysis
Sequence alignments were carried out using either the BLASTN or PSI-BLAST programs (Altschul et al. 1997). The Dotter program (Sonnhammer and Durbin 1995) was used for dot plots.
Generation of the transgenic plants
A 1-kb NRPD1b promoter region was amplified with Phusion polymerase (New England Biolabs) using primers 354/466 and cloned into a 2xFlag-containing pCambia 1300 vector. NRPD1b genomic region was PCR-amplified using primers 456/457 and inserted into this vector. NRPD1b/DSD was obtained by cloning first the CD genomic region (amino acids 1708–1874, primers 581/582) and then the N-terminal RPB1-like region (amino acids 1–1410, primers 456/658) into the 2xFlag vector. Fusion PCR was performed to join the HsGW182 homologuous fragment (amino acids 455–762) with the CD domain using primers 582, 621, 622, and 623. The fusion was inserted into the pCambia1300 derivative to obtain NRPD1b/GW182. nrpd1b-11 and nrpd1a-4 plants were transformed by the floral dipping method using Agrobacterium tumefaciens GV3121 transformed strains.
Transient transfection of human AGO constructs in 293 cells
Cells were plated (80% confluence) in T150 tissue culture dishes for 1 d. Transient transfection assays were performed by the calcium phosphate coprecipitation method (Chen and Okayama 1987) using 15 μg of DNA per plate. After 48 h of incubation, cells were harvested, extracted in 1 mL of lysis buffer (20 mM Tris-HCl at pH 7.9, 0.25 M KCl, 10% glycerol, 1 mM EDTA, 5 mM DTT, 0.5% NP40), and used in GST pull-down assays.
Multimer construction
In order to allow a cloning scheme that doubles the number of repeats with each cloning cycle, a BglII site was introduced between SacI and BamHI in the pBluescript SK vector. The primers corresponding to each monomer (545/546 for WG1, 547/548 for AG1, 579/580 for FG1, 661/662 for GW1, and 663/664 for GF1) were annealed and subcloned into the BglII–BamHI-digested vector. Then, using the BglII, BamHI, and HindIII enzymes and the strategy described by Robinett et al. (1996), we doubled the number of repeats lying between BglII and BamHI sites at each cloning cycle. After three rounds of cloning, the multimers were introduced into a BamHI–HindIII-digested pET41a(+) vector (Novagen).
Plant protein extraction and immunoblot
Total plant protein extracts were obtained following the method described in Pontier et al. (2005). Protein extracts from the transgenic lines expressing epitope-Flag NRPD1b-based proteins were pooled, quantified, and subjected to immunoblot analysis using a dilution of 1/1000 of anti-Flag M2 Monoclonal antibody (Sigma-Aldrich).
GST-CTD binding experiments
The AtD1b/CTD (amino acids 1410–1874, primers 204/205), AtD1b/SD (amino acds 1410–1728, primers 204/458), AtD1b/CD (amino acids 1708–1874, primers 518/205), SoD1b/SD (amino acids 1112–1363 of partial SoNRPD1b, primers 557/558), and the HsGW182Nt (amino acids 455–762, primers 543/544) cDNA fragments were RT–PCR-amplified, cloned into pET41a(+), and expressed in Escherichia coli BL21. The GST fusion proteins were purified on glutathione Sepharose 4B beads (Amersham Biosciences) and used in pull-down assays as described (Li et al. 2006). AGO4 binding was quantified using ImageJ software.
Methylation detection assays were performed as described in the Supplemental Material.
Acknowledgments
We thank J.-R. Pagés for his help with artwork and O. Voinnet for fruitful discussion. Lagrange laboratory research was supported by CNRS and the ANR grant NT05-3_45717, Cooke laboratory work was supported by the CNRS, and Hakimi laboratory work was supported by the INSERM and the CNRS (ATIP). S.E.J. is a Howard Hughes Medical Institute Investigator. M.E.S. was supported by an ANR fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.451207
References
- Altschul S.F., Thomas L.D., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Thomas L.D., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipmann D.J., Zhang J., Zhang Z., Miller W., Lipmann D.J., Zhang Z., Miller W., Lipmann D.J., Miller W., Lipmann D.J., Lipmann D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Doerks T., Stark A., Bork P., Izaurralde E., Stark A., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O., Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Zhang X., Bernatavichute Y.V., Jacobsen S.E., Zhang X., Bernatavichute Y.V., Jacobsen S.E., Bernatavichute Y.V., Jacobsen S.E., Jacobsen S.E. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:1923–1933. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Horsefield R., Braunstein T.H., Baulcombe D.C., Horsefield R., Braunstein T.H., Baulcombe D.C., Braunstein T.H., Baulcombe D.C., Baulcombe D.C. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;17:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., Spencer A., Morita K., Han M., Morita K., Han M., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the AGO protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E., Behm-Ansmant I., Izaurralde E., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J., Keene J.D., Griffith K., Fritzler M.J., Griffith K., Fritzler M.J., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C., Derrington E., Leblanc P., Chnaidermann J., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Derrington E., Leblanc P., Chnaidermann J., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Leblanc P., Chnaidermann J., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Chnaidermann J., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P., Morillas M., Surewicz W.K., Marc D., Nandi P., Surewicz W.K., Marc D., Nandi P., Marc D., Nandi P., Nandi P., et al. The prion protein has RNA binding and chaperoning characteristic of nucleocapsid protein NCP7 of HIV-1. J. Biol. Chem. 2001;276:19301–19309. doi: 10.1074/jbc.M009754200. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C., Jensen M.B., Dalmay T., Baulcombe D.C., Dalmay T., Baulcombe D.C., Baulcombe D.C. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:115–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Hamel J.C., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Daxinger L., Kreil D.P., Matzke M., Matzke A.J., Kreil D.P., Matzke M., Matzke A.J., Matzke M., Matzke A.J., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E., Lagrange T., Pikaard C.S., Jacobsen S.E., Pikaard C.S., Jacobsen S.E., Jacobsen S.E. An Argonaute4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Yates J.R., Parker R., Hannon G.J., Parker R., Hannon G.J., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M., Zagorski-Ostoja W., Meyer E., Zagorski-Ostoja W., Meyer E., Meyer E. Nowa1p and Nowa2p: Novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S., Ream T., Nunes P.C., Pontes O., Pikaard C.S., Nunes P.C., Pontes O., Pikaard C.S., Pontes O., Pikaard C.S., Pikaard C.S. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Partridge J.F., DeBeauchamp J.L., Kosinski A.M., Ulrich D.L., Hadler M.J., Noffsinger V.J., DeBeauchamp J.L., Kosinski A.M., Ulrich D.L., Hadler M.J., Noffsinger V.J., Kosinski A.M., Ulrich D.L., Hadler M.J., Noffsinger V.J., Ulrich D.L., Hadler M.J., Noffsinger V.J., Hadler M.J., Noffsinger V.J., Noffsinger V.J. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol. Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Peters L., Meister G., Meister G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S., Vitins A., Jacobsen S.E., Pikaard C.S., Jacobsen S.E., Pikaard C.S., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–82. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Bulski A., Saez-Vasquez J., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Saez-Vasquez J., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Hakimi M.-A., Lerbs-Mache S., Colot V., Lagrange T., Lerbs-Mache S., Colot V., Lagrange T., Colot V., Lagrange T., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. Prions. Proc. Natl. Acad. Sci. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S., Willhelm C., Sudlow G., Murray A., Belmont A.S., Sudlow G., Murray A., Belmont A.S., Murray A., Belmont A.S., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using Lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.D., Najand N., Chaker S., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Najand N., Chaker S., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Chaker S., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J., Hobman T.C., Locke J., Simmonds A.J., Locke J., Simmonds A.J., Simmonds A.J. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J. Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., Durbin R., Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein analysis. Gene. 1995;167:1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Taylor F.M., Martindale D.W., Martindale D.W. Retroviral-type zinc fingers and glycine-rich repeats in a protein encoded by cnjB, a Tetrahymena gene active during meiosis. Nucleic Acids Res. 1993;25:4610–4614. doi: 10.1093/nar/21.19.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Zilberman D., Jacobsen S.E., Carrington J.C., Jacobsen S.E., Carrington J.C., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–652. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.C., Fuller P., Xi X., Fuller P., Xi X., Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A., Irvine D.V., Martienssen R.A., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E., Henderson I.R., Lu C., Green P.J., Jacobsen S.E., Lu C., Green P.J., Jacobsen S.E., Green P.J., Jacobsen S.E., Jacobsen S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E., Cao X., Jacobsen S.E., Jacobsen S.E. Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]