Abstract
Embryonic stem (ES) cells are pluripotent cells with the ability to self-renew indefinitely. These unique properties are controlled by genetic factors and chromatin structure. The exit from the self-renewing state is accompanied by changes in epigenetic chromatin modifications such as an induction in the silencing-associated histone H3 Lys 9 dimethylation and trimethylation (H3K9Me2/Me3) marks. Here, we show that the H3K9Me2 and H3K9Me3 demethylase genes, Jmjd1a and Jmjd2c, are positively regulated by the ES cell transcription factor Oct4. Interestingly, Jmjd1a or Jmjd2c depletion leads to ES cell differentiation, which is accompanied by a reduction in the expression of ES cell-specific genes and an induction of lineage marker genes. Jmjd1a demethylates H3K9Me2 at the promoter regions of Tcl1, Tcfcp2l1, and Zfp57 and positively regulates the expression of these pluripotency-associated genes. Jmjd2c acts as a positive regulator for Nanog, which encodes for a key transcription factor for self-renewal in ES cells. We further demonstrate that Jmjd2c is required to reverse the H3K9Me3 marks at the Nanog promoter region and consequently prevents transcriptional repressors HP1 and KAP1 from binding. Our results connect the ES cell transcription circuitry to chromatin modulation through H3K9 demethylation in pluripotent cells.
Keywords: Histone demethylase, embryonic stem cell, chromatin immunoprecipitation, self-renewal, pluripotency, Jumonji
Embryonic stem (ES) cells possess the remarkable properties of self-renewal and pluripotency. They may be cultured in vitro for an indefinite period of time while retaining the capacity to give rise to all cell types of the organism (Smith 2001; Loebel et al. 2003). For this reason, there has been much interest in ES cells as a source of differentiated cell types in cell replacement therapy (Donovan and Gearhart 2001). A key feature of the ES cell plasticity is the maintenance of the uncommitted “stemness” state, while being poised to enter lineage-specific differentiation programs.
ES cells exhibit unique chromatin features (Meshorer and Misteli 2006). In mouse ES cells, there are rapid exchanges of architectural chromatin proteins such as HP1 and histones H1, H2B, and H3, which might constitute a hyperdynamic and open chromatin environment (Meshorer et al. 2006). Upon differentiation, the binding dynamics of these factors are reduced, and they tend to be immobilized to the chromatin. In general, methylation of the Lys 4 residue of histone H3 (H3K4) correlates with active gene status, while methylation of histone H3K9 and K27 serve as repressive chromatin marks (Lachner and Jenuwein 2002; Turner 2002; Martin and Zhang 2005; Kouzarides 2007). Recent studies have reported the finding of “bivalent domains” in mouse ES cells (Azuara et al. 2006; Bernstein et al. 2006), where nucleosomes contain both the histone H3K4 trimethylation (H3K4Me3) mark as well as the histone H3K27 trimethylation (H3K27Me3) mark. The occurrence of these opposing chromatin marks is thought to keep lineage-specific genes repressed, yet keep them poised for activation upon differentiation. ES cell chromatin is also enriched in active marks (methylation of H3K4 and acetylation of H3 and H4) and deficient in silencing modifications (methylation of H3K9) (Lee et al. 2004; Meshorer and Misteli 2006). Differentiation of ES cells is accompanied by global changes in histone modifications and a transition to a transcriptionally less-permissive chromatin state characterized by a decrease in H3K4Me3 and an elevation of H3K9 methylation. These findings suggest that the dynamic repression of developmental pathways as well as the maintenance of transcriptional permissive chromatin with a low level of H3K9 methylation in ES cells by epigenetic processes is required for the maintenance of ES cells’ plasticity and pluripotency. However, the mechanisms and histone-modifying enzymes involved in maintaining this unique ES cell epigenetic state remain unclear. Hence, in addition to the identification of genetic factors that influence the decision between self-renewal and differentiation (Nichols et al. 1998; Avilion et al. 2003; Chambers et al. 2003; Mitsui et al. 2003; Elling et al. 2006; Ivanova et al. 2006; Loh et al. 2006; Matoba et al. 2006; Wang et al. 2006; Wu et al. 2006; Zhang et al. 2006; Galan-Caridad et al. 2007; Lim et al. 2007), the roles of epigenetic regulators are also of interest.
Jmjd1a, Jmjd2c, and several other JmjC domain-containing proteins have recently been shown to be histone demethylases (JHDMs) (Cloos et al. 2006; Fodor et al. 2006; Klose et al. 2006; Tsukada et al. 2006; Whetstine et al. 2006; Yamane et al. 2006). JHDMs catalyze oxidative demethylation reactions with iron and α-ketoglutarate as cofactors (Trojer and Reinberg 2006; Shi and Whetstine 2007). Jmjd1a can demethylate H3K9 mono- and dimethylation in vitro and functions as a coactivator for androgen receptor (AR) to demethylate chromatin of AR target genes (Yamane et al. 2006). Jmjd2c has a different specificity and is shown to convert H3K9 and H3K36 from trimethylation to dimethylation (Whetstine et al. 2006). Little is known about the role that these JHDMs play in modulating the chromatin structure of ES cells.
Oct4 is a POU domain-containing transcription factor encoded by Pou5f1. In the preimplantation embryo, Oct4 expression is restricted to the inner cell mass (ICM). Oct4 is also highly expressed in human and mouse ES cells (Palmieri et al. 1994). In the absence of Oct4, both in vivo (ICM) and in vitro (ES cells) pluripotent cells are induced to differentiate into the trophoblast lineage (Nichols et al. 1998). Thus Oct4 has an essential role in controlling cell fate decision and maintaining pluripotency in the early mammalian embryo and the ES cells. We previously mapped the Oct4-binding sites using chromatin immunoprecipitation (ChIP) coupled to a paired end ditag (PET) sequencing approach (Loh et al. 2006).
In this study, we showed that genes encoding for the histone demethylases Jmjd1a and Jmjd2c are bona fide targets of Oct4 in mouse ES cells. Using RNA interference (RNAi), we found that depletion of these JHDMs in ES cells resulted in cellular differentiation, providing evidence for their roles in the maintenance of self-renewal in ES cells. Furthermore, we have identified Tcl1 and Nanog to be downstream effectors of Jmjd1a and Jmjd2c, respectively. Our data support a model in which Oct4 up-regulates downstream histone demethylases, which, in turn, maintain permissive histone modifications with a low level of H3K9 methylation at the promoters of genes critical for the self-renewal of ES cells.
Results
Oct4 regulates the expression of histone modifiers Jmjd1a and Jmjd2c
We have previously mapped the in vivo binding sites of an ES cell transcription factor, Oct4 (Loh et al. 2006). We postulate that Oct4 controls the chromatin architecture of ES cells through downstream targets that encode for histone-modifying enzymes. Our previous ChIP-PET-binding site mapping study revealed Oct4-binding clusters within Jmjd1a and Jmjd2c genes (Supplementary Fig. 1A,B). We confirmed that Oct4 binds to Jmjd1a and Jmjd2c using the ChIP assay (Fig. 1A). Furthermore, depletion of Oct4 by RNAi led to decreased Jmjd1a and Jmjd2c expression (Fig. 1B). Depletion of two other transcription factors that have been implicated in ES cell self-renewal, Esrrb and Nanog, had no or little effect on Jmjd1a and Jmjd2c levels compared with Oct4 depletion (Fig. 1B; Supplementary Fig. 1C,D).
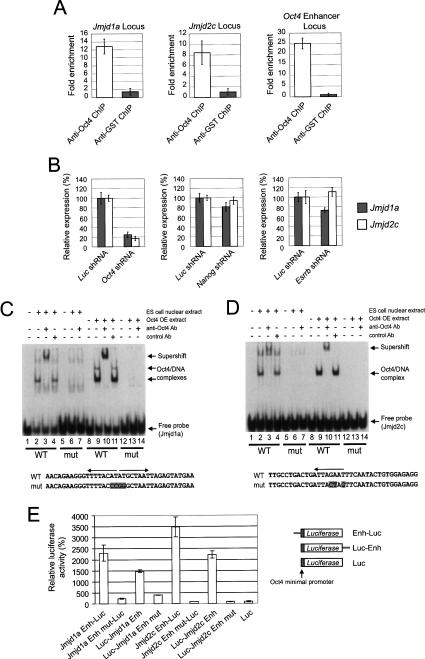
Figure 1.
Oct4 regulates the expression of Jmjd1a and Jmjd2c in pluripotent mouse ES cells. (A) Oct4 binds to the intronic regions of Jmjd1a and Jmjd2c. Real-time PCR detection of enriched fragments from ChIP assays in ES cells using Oct4 or control antibodies. Fold enrichment is the relative abundance of DNA fragments at the amplified region (see Supplementary Fig. 1) over a control amplified region. Validation of Oct4 ChIP was carried out using primers specific for known binding sites at the Oct4 enhancer locus. GST (glutathione S-transferase) antibody was used as a mock ChIP control. Data are presented as the mean ± SEM. (B) Oct4 knockdown down-regulated endogenous Jmjd1a and Jmjd2c mRNA levels. Oct4 suppression in ES cells by RNAi resulted in concomitant reductions in endogenous Jmjd1a and Jmjd2c. Nanog and Esrrb RNAi-transfected ES cells exhibited little or no reduction in Jmjd1a and Jmjd2c mRNA. cDNAs were prepared from the knockdown mouse ES cells and were analyzed using real-time PCR. The levels of the transcripts were normalized against control Luc (Luciferase) shRNA-transfected cells. After 24 h of transfection, the ES cells were selected for 3 d before harvest. Data are presented as the mean ± SEM. (C) Oct4 binds to the intronic sequences of Jmjd1a. EMSA was used to analyze the interactions between Oct4 and a 27-bp double-stranded DNA probe containing the oct elements (indicated by arrows). Both ES cell nuclear extracts and extracts from 293 cells overexpressing Oct4 (Oct4 OE extracts) were used for EMSAs. (Lanes 3,10) EMSA with the wild-type probe detected specific Oct4/DNA complexes as confirmed by supershift analysis. (Lanes 6,7,13,14) When mutant probe was used, no interaction was detected. The bottom panel shows the sequence of the oct elements and corresponding mutations (shaded) used in this study. (D) Oct4 binds to the intronic sequences of Jmjd2c. EMSA was used to analyze the interactions between Oct4 and a 27-bp double-stranded DNA probe containing the oct element (indicated by arrow). (Lanes 3,10) EMSA with the wild-type probe detected a specific Oct4/DNA complex as confirmed by supershift analysis. (Lanes 6,7,13,14) When mutant probe was used, no interaction was detected. The bottom panel shows the sequence of the oct element and corresponding mutations (shaded) used in this study. (E) The Oct4-bound intronic regions of Jmjd1a (a 655-bp fragment) or Jmjd2c (a 679-bp fragment) was inserted either upstream of (Enh-Luc) or downstream from (Luc-Enh) a luciferase gene driven by an Oct4 minimal promoter. Reporters were transiently transfected into ES cells for 3 d before measurement of luciferase activities. Enhancer constructs with mutated Oct4-binding sites (Jmjd1a/Jmjd2c Enh-Luc Mut or Jmjd1a/Jmjd2c Luc-Enh Mut) were also tested for enhancer activity in ES cells. For each transfection, we cotransfected a construct expressing Renilla luciferase driven by SV40 promoter to serve as an internal control. Data are presented as the mean ± SEM.
A probe was also designed based on the peak of the Oct4-binding profile at Jmjd1a intron (Supplementary Fig. 1A). This sequence contained two Oct4-binding sites and was used to test the interaction with Oct4 (Fig. 1C). Using the electrophoretic mobility shift assay (EMSA), we showed that Oct4 in ES cell nuclear extract bound to this probe (Fig. 1C, lanes 2–4) and that mutations introduced to the two Oct4-binding sites abolished the interaction (Fig. 1C, lanes 6,7). We were also able to demonstrate that ectopically expressed Oct4 in 293T cells bound to this probe through the Oct4-binding sites (Fig. 1C, lanes 9–11,13,14). A probe derived from the Jmjd2c intron contained an Oct4-binding site and was also bound by Oct4 (Fig. 1D).
Next, we cloned the Jmjd1a and Jmjd2c intronic DNA containing these Oct4-binding sites upstream of or downstream from a luciferase reporter to test for enhancer activity. We observed robust enhancer activity when these constructs were transfected into ES cells (Fig. 1E). Importantly, the same mutations that disrupted the in vitro Oct4/DNA interactions also abolished the enhancer activities (Fig. 1E). Taken together, these data show that Oct4 positively regulates the expression of these histone demethylase genes through the intronic Oct4 sites. Conversely, reprogramming of fibroblasts to induced pluripotent stem (iPS) cells is accompanied by increased expression of Jmjd1a and Jmjd2c (Supplementary Fig. 1E; Takahashi and Yamanaka 2006). Hence, the expression of Jmjd1a and Jmjd2c is positively correlated with the pluripotent state of ES and iPS cells.
Jmjd1a and Jmjd2c are critical regulators of ES cells
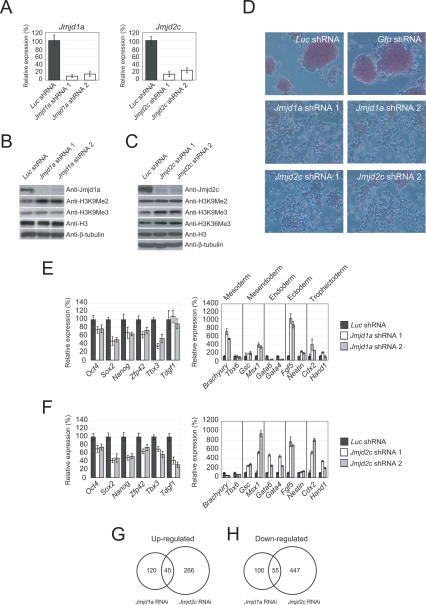
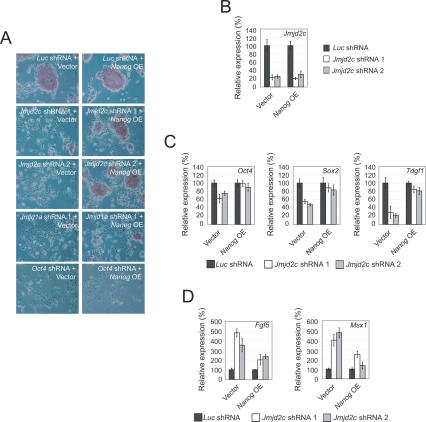
To confirm the activity of Jmjd1a and Jmjd2c in ES cells, we depleted their expression by RNAi. We used two short-hairpin RNA (shRNA) constructs targeting different regions of each transcript to ensure that the effects are specific. Both constructs were effective in reducing the RNA and protein levels (Fig. 2A–C). As expected, the level of H3K9Me2, but not H3K9Me3, of total cell histone H3, was increased upon Jmjd1a knockdown, indicating that Jmjd1a is involved in the reversal of H3K9Me2 of bulk chromatin in ES cells (Fig. 2B). Also as expected, Jmjd2c depletion increased H3K9Me3, but not H3K9Me2 (Fig. 2C). The level of H3K36Me3 was, however, not significantly affected by Jmjd2c depletion. The lack of change in H3K36 trimethylation could be due to redundancy of histone demethylases in ES cells or is reflective of the in vivo activity of Jmjd2c. These results indicate that Jmjd1a and Jmjd2c regulate the global levels of H3K9Me2 and H3K9Me3, respectively. Strikingly, the colony morphology of ES cells was changed dramatically, and the cells became flattened and fibroblast-like upon Jmjd1a or Jmjd2c knockdown (Fig. 2D). The alkaline phosphatase staining typical of undifferentiated ES cells was also reduced upon knockdown of both transcripts, indicative of differentiation (Fig. 2D). As targeting both transcripts by RNAi led to differentiation, we performed additional control experiments to establish the specificity of these shRNA constructs. Knockdown of Jmjd1a did not appreciably affect Jmjd2c and vice versa (Supplementary Fig. 2A,B). Therefore, each shRNA set was specific for targeting the intended transcript. To further substantiate the specificity of the Jmjd1a knockdown experiment, we mutated two nucleotide bases in our Jmjd1a shRNA constructs (Supplementary Fig. 3A). These mutations abolished their silencing effects, as the endogenous level of Jmjd1a was not affected (Supplementary Fig. 3B). Cells transfected with shRNA mutants also retained proper ES cell morphology and did not show appreciable changes in the ES cell markers (Supplementary Fig. 3C–E). As with Jmjd1a, introduction of two nucleotide mutations in the Jmjd2c shRNA constructs abolished their silencing effects (Supplementary Fig. 4A,B). The cells transfected with shRNA mutants also retained proper ES cell morphology, maintained the same levels of ES cell markers, and showed no induction of differentiation markers (Supplementary Fig. 4C–E). The differentiation phenotype is specific to Jmjd1a and Jmjd2c knockdown ES cells, as we did not observe morphology changes or any reduction in Oct4 expression when we depleted transcripts coding for other JmjC domain-containing proteins—Jarid2, Jarid1a, and Jhdm1 (Supplementary Fig. 5). To further characterize the Jmjd1a- and Jmjd2c-depleted ES cells, we analyzed their ability to form colonies in a replating assay. Transfected cells were dissociated with trypsin and replated to allow the ES cells to expand into colonies. Jmjd1a and Jmjd2c knockdown reduced the number of ES cell colony-forming units (CFUs) by fourfold to 19-fold compared with control knockdown (Supplementary Fig. 6). Taken together, our results indicate that Jmjd1a and Jmjd2c are critical for the maintenance of the self-renewal state of ES cells. Cellular differentiation induced by Jmjd1a or Jmjd2c knockdown was accompanied by a corresponding reduction in certain pluripotency markers and induction of genes associated with differentiation. Oct4, Sox2, and Nanog were down-regulated in response to the Jmjd1a or Jmjd2c depletion (Fig. 2E,F). It should be noted, however, that the reduction is modest compared with the knockdown of transcription factors such as Oct4. The expression of Tdgf1 was also not affected by Jmjd1a depletion. Time-course analyses indicated that Oct4, Sox2, and Nanog levels continued to decrease after an extended period of selection (Supplementary Fig. 7). We also examined the expression of lineage markers (Fig. 2E,F). Interestingly, Jmjd1a and Jmjd2c knockdown cells expressed markers of different lineages. For instance, Jmjd1a depletion induced the mesodermal marker Brachyury, while Jmjd2c depletion induced the endodermal markers Gata4 and Gata6. Knockdown of either transcript induced the expression of Msx1, Fgf5, and Cdx2 (Fig. 2E,F). Hence, the resulting cells were likely to be composed of multiple differentiated cell types. To understand the molecular basis of the JHDMs’ requirements in ES cell maintenance, we used Illumina gene expression microarrays to identify potential downstream target genes. One-hundred-sixty-five and 311 genes were up-regulated in Jmjd1a and Jmjd2c knockdown cells, respectively, while 45 of these genes were common between the two (Fig. 2G; Supplementary Table 1). On the other hand, 155 and 502 genes were down-regulated in Jmjd1a and Jmjd2c knockdown cells, respectively. Fifty-five of these genes were commonly down-regulated (Fig. 2H). Taken together, these data show that Jmjd1a and Jmjd2c may have overlapping but distinct roles in regulating gene expression in ES cells.
Figure 2.
Jmjd1a and Jmjd2c are required for the maintenance of self-renewal of ES cells. (A) Quantitative real-time PCR analysis of Jmjd1a and Jmjd2c expression after knockdown using two shRNA constructs targeting different regions of the respective transcripts. After 24 h of transfection, the ES cells were selected for 4 d before harvest. The levels of the transcripts were normalized against control Luc shRNA-transfected cells. Data are presented as the mean ± SEM. (B) Reduction of Jmjd1a after RNAi-mediated knockdown led to increased H3K9Me2. Western blot analyses of Jmjd1a knockdown and control ES cell lysates were carried out using anti-Jmjd1a, anti-H3K9Me2, or anti-H3K9Me3 antibodies. Anti-histone H3 and anti-β-tubulin antibodies were used as loading controls. (C) Reduction of Jmjd2c after RNAi-mediated knockdown led to increased H3K9Me3. Western blot analyses of Jmjd2c knockdown and control ES cell lysate were carried out using anti-Jmjd2c, anti-H3K9Me2, anti-H3K9Me3, or anti-H3K36Me3 antibodies. Anti-histone H3 and anti-β-tubulin antibodies were used as loading controls. (D) Jmjd1a and Jmjd2c knockdown led to differentiation. Flattened fibroblast-like cells were formed after Jmjd1a or Jmjd2c depletion. For control Luc or Gfp shRNA-transfected cells, distinct alkaline phosphatase-positive (red staining) ES cell colonies were maintained. The cells were stained after 4 d of puromycin selection. (E) Real-time PCR analysis of ES cell-associated gene expression (left panel) and lineage-specific marker gene expression (right panel) in Jmjd1a knockdown ES cells. The levels of the transcripts were normalized against control Luc shRNA-transfected cells. Data are presented as the mean ± SEM. (F) Real-time PCR analysis of ES cell-associated gene expression (left panel) and lineage-specific marker gene expression (right panel) in Jmjd2c knockdown ES cells. The levels of transcripts were normalized against control Luc shRNA-transfected cells. Data are presented as the mean ± SEM. (G) Venn diagram of overlapping and specifically up-regulated genes between Jmjd1a- and Jmjd2c-depleted ES cells. DNA microarrays were used to profile the gene expression of these cells. The levels of transcripts were compared with control Luc shRNA-transfected cells. The P-value for the overlap as computed using Monte Carlo simulation is <1e−08. (H) Venn diagram of overlapping and specifically down-regulated genes between Jmjd1a- and Jmjd2c-depleted ES cells. DNA microarrays were used to profile the gene expression of these cells. The levels of transcripts were compared with control Luc shRNA-transfected cells. The P-value for the overlap as computed using Monte Carlo simulation is <1e−08.
Jmjd1a regulates the expression of Tcl1 through demethylation of H3K9Me2
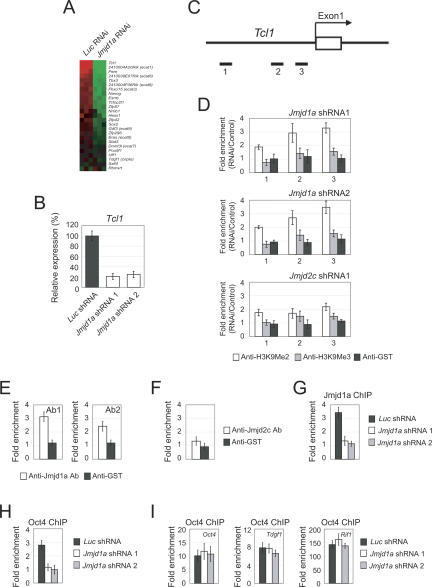
We reasoned that Jmjd1a and Jmjd2c could positively control the expression of candidate target genes through modulation of H3K9Me2 and H3K9Me3 levels. Because of the differentiation phenotype we observed upon Jmjd1a or Jmjd2c depletion, we also hypothesized that the genes controlled by Jmjd1a and Jmjd2c could be self-renewal regulators. Among the down-regulated genes identified by global gene expression profiling of Jmjd1a knockdown cells (Fig. 3A; Supplementary Table 1) are Tcl1, Tcfcp2l1, and Zfp57. These genes are preferentially expressed in pluripotent ES cells (Ivanova et al. 2002; Ramalho-Santos et al. 2002). We validated the down-regulation of these genes using real-time PCR (Fig. 3B; Supplementary Fig. 8). Tcl1, a gene encoding for a cofactor of the Akt1 kinase, is of particular interest as it has been shown to regulate self-renewal of ES cells (Ivanova et al. 2006; Matoba et al. 2006). We depleted Tcl1 using two different shRNA constructs (Supplementary Fig. 9). Consistent with a previous report, we found that Tcl1 is required to maintain the undifferentiated state of ES cells (Ivanova et al. 2006). Similar to Jmjd1a-depleted cells, differentiated markers (Fgf5, Mxs1, and Brachyury) were induced upon Tcl1 knockdown (Supplementary Fig. 9). To test if Jmjd1a regulates H3K9Me2 of chromatin associated with Tcl1, we used a ChIP assay to measure the level of H3K9Me2 at the promoter region. A series of primer pairs was used to interrogate this region (Fig. 3C). Interestingly, we detected an increase of H3K9Me2 at the Tcl1 promoter upon Jmjd1a depletion (Fig. 3D). Our two shRNA constructs showed the same effect and did not alter the level of H3K9Me3, indicating that this is a specific role of Jmjd1a (Fig. 3D). Knockdown of Jmjd2c did not alter the H3K9Me2 or H3K9Me3 level to the same extent as Jmjd1a depletion, further demonstrating the predominant modulation of H3K9Me2 at the Tcl1 promoter by Jmjd1a (Fig. 3D). As knockdown by RNAi will have indirect effects, we sought further evidence to confirm the action of Jmjd1a. To this end, we performed a ChIP assay using two independently generated anti-Jmjd1a antibodies. The result showed that Jmjd1a was bound to the Tcl1 promoter (Fig. 3E). In contrast, Jmjd2c showed only a very low or near background level of binding (Fig. 3F). Furthermore, the depletion of Jmjd1a using either shRNA construct abolished the Jmjd1a ChIP signal, indicating that the antibodies specifically recognized Jmjd1a (Fig. 3G). We and others have shown that the Tcl1 promoter is bound by Oct4 (Loh et al. 2006; Matoba et al. 2006). Next, we asked if the increase in H3K9Me2 affects Oct4 occupancy. The ChIP assay showed that the Oct4 binding at the Tcl1 promoter but not at the Oct4 enhancer, Tdgf1 and Rif1 loci, was abolished by Jmjd1a depletion (Fig. 3H,I). The loss of Oct4 binding is a likely cause for the down-regulation of Tcl1 upon Jmjd1a depletion.
Figure 3.
Jmjd1a regulates expression of Tcl1 through demethylation of H3K9Me2. (A) Microarray heat map depicting expression changes of selected ES cell-associated genes (Ivanova et al. 2002; Ramalho-Santos et al. 2002; Mitsui et al. 2003) after Jmjd1a knockdown. The gene expression levels were mean-centered to show their relative changes, and the genes were ordered according to their mean fold changes. (B) Jmjd1a positively regulates the expression of Tcl1. The expression of Tcl1 was analyzed after depletion of Jmjd1a using two shRNA constructs. After 24 h of transfection, the ES cells were selected with puromycin for 4 d before harvest. The levels of the transcripts were normalized against control Luc (Luciferase) shRNA-transfected cells. Data are presented as the mean ± SEM. (C) Schematic showing the locations of the amplicons (black bars labeled 1–3) used to detect ChIP-enriched fragments over the Tcl1 promoter. Amplicons are numbered in order relative to their sites along the gene. The open box represents an exon. (D) Analysis of H3K9Me2/Me3 modifications along the Tcl1 promoter by ChIP. ES cells were transfected with Luc (control) shRNA, Jmjd1a shRNA 1, Jmjd1a shRNA 2, or Jmjd2c shRNA 1. Fold enrichment is the relative abundance of DNA fragments detected by real-time PCR at the amplified region over a control amplified region and normalized with control Luc. GST antibody was used as a ChIP control. Data are presented as the mean ± SEM. (E) Jmjd1a interacts with the Tcl1 promoter region. ChIP assays were performed with two different anti-Jmjd1a antibodies. A primer pair targeting amplicon 3 was used. GST antibody was used as a ChIP control. Data are presented as the mean ± SEM. (F) Jmjd2c ChIP and real-time PCR showed no enrichment over the Tcl1 promoter region. Data are presented as the mean ± SEM. (G) Knockdown of Jmjd1a abolished the ChIP signal derived from anti-Jmjd1a antibody. ES cells were transfected with control Luc shRNA, Jmjd1a shRNA 1, or Jmjd1a shRNA 2. Data are presented as the mean ± SEM. (H) ChIP analysis showed that Oct4 binding to the Tcl1 promoter was diminished upon Jmjd1a depletion. ES cells were transfected with control Luc shRNA, Jmjd1a shRNA 1, or Jmjd1a shRNA 2. A primer pair targeting amplicon 3 was used. Data are presented as the mean ± SEM. (I) Oct4 binding at Oct4 enhancer and Tdgf1 and Rif1 loci was not affected in Jmjd1a depletion. Data are presented as the mean ± SEM.
Tcl1 is a downstream effector of Jmjd1a in regulating ES cells maintenance
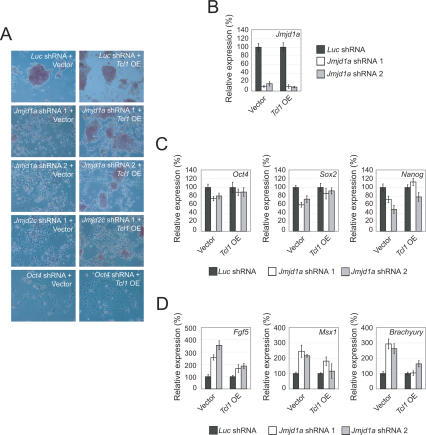
If Tcl1 is a key effector of Jmjd1a, one would predict that overexpression of Tcl1 will rescue the effects of Jmjd1a knockdown. To test this hypothesis, we cotransfected a Tcl1 expression plasmid with control, Jmjd1a, Jmjd2c, or Oct4 shRNA constructs. ES cells cotransfected with a vector control plasmid and Jmjd1a, Jmjd2c, or Oct4 shRNA constructs underwent differentiation based on the morphology changes, loss of ES cell colonies, and alkaline phosphatase staining (Fig. 4A). However, cells cotransfected with Tcl1 and Jmjd1a shRNA expression plasmids were found to retain ES cell morphology and alkaline phosphatase expression (Fig. 4A). The depletion of Jmjd1a was equally efficient in the control and Tcl1-overexpressing ES cells (Fig. 4B). This excludes the possibility that the phenotypic differences were due to insufficient depletion of Jmjd1a. In contrast, the degree of rescue by Tcl1 in Jmjd2c-depleted cells was less pronounced. Consistent with the morphology data, we observed a smaller reduction in ES cell markers and less induction of differentiation markers in Tcl1-rescued cells (Fig. 4C,D). Jmjd2c-depleted cells showed little restoration of Oct4, Sox2, or Nanog expression (Supplementary Fig. 10A). The expression of Msx1 but not Fgf5 was reduced, indicating incomplete rescue by Tcl1 (Supplementary Fig. 10B). Taken together, our Tcl1 rescue result indicates that Tcl1 can compensate for the loss of Jmjd1a. In addition, we screened the promoter regions of six other genes for Jmjd1a-dependent modulation of the H3K9Me2 level (Supplementary Table 2A) and detected Jmjd1a-dependent H3K9Me2 demethylation at Tcfcp2l1 and Zfp57. We also confirmed that Jmjd1a associates with these two promoters by ChIP assay (Supplementary Fig. 8). The data support a role of Jmjd1a in positively regulating these pluripotency-associated genes by demethylation of H3K9Me2 at their promoters. As we did not detect any changes in the H3K9Me2 level of Oct4 and Nanog promoter regions by ChIP, this indicates that not all differentially regulated genes are subject to Jmjd1a-mediated H3K9Me2 demethylation (Fig. 5; Supplementary Fig. 11). Using the two criteria of Jmjd1a occupancy and Jmjd1a-dependent change in the H3K9Me2 level, we conclude that Tcl1, Tcfcp2l1, and Zfp57 are direct targets of Jmjd1a.
Figure 4.
Tcl1 is the key downstream effector of Jmjd1a responsible for maintaining ES cells’ self-renewal. (A) Enforced Tcl1 overexpression (OE) could rescue the differentiation phenotype induced by Jmjd1a depletion. ES cells were transfected with a Tcl1-overexpressing vector and challenged with shRNA directing against various transcripts (Jmjd1a, Jmjd2c, or Oct4). The cells were stained for alkaline phosphatase activity, and the morphologies were examined by microscopy after 4 d of puromycin selection. Note the morphological rescue and the maintenance of alkaline phosphatase-positive colonies in Jmjd1a shRNA-treated cells. Little or no morphological rescue was observed when the cells were challenged with Jmjd2c or Oct4 shRNA, respectively. (B) Jmjd1a was similarly depleted both in Tcl1-overexpressing and control ES cells. Quantitative real-time PCR analysis of Jmjd1a expression after knockdown using two shRNA constructs cotransfected into ES cells with either control or Tcl1-overexpressing vector. The levels of the transcripts were normalized against control plasmid-transfected cells. Data are presented as the mean ± SEM. (C) Enforced Tcl1 overexpression reduced the down-regulation of Sox2 and Nanog upon Jmjd1a depletion. The levels of the transcripts were normalized against control plasmid transfected cells. Data are presented as the mean ± SEM. (D) Enforced Tcl1 overexpression compensated for the Jmjd1a loss of function by reducing the induction of differentiation markers Fgf5, Msx1, and Brachyury. Data are presented as the mean ± SEM.
Figure 5.
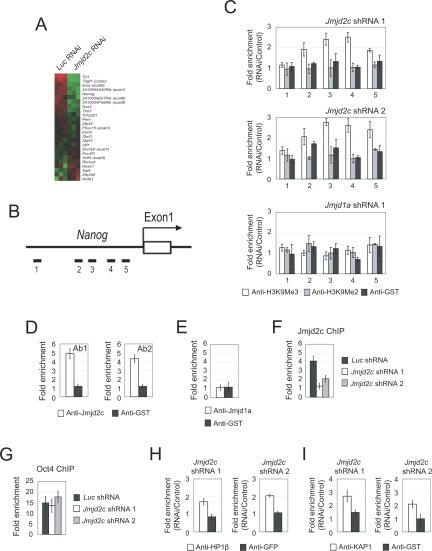
Jmjd2c regulates expression of Nanog through demethylation of H3K9Me3. (A) Microarray heat map plot depicting expression changes of selected ES cell-associated genes (Ivanova et al. 2002; Ramalho-Santos et al. 2002; Mitsui et al. 2003) after Jmjd2c knockdown. The gene expression levels were mean-centered to show their relative changes, and the genes were ordered according to their mean fold changes. (B) Schematic showing the location of the amplicons (black bars labeled 1–5) used to detect ChIP-enriched fragments over the Nanog promoter. Amplicons are numbered in order relative to their sites along the gene. The open box represents an exon. (C) Analysis of H3K9Me2/Me3 modifications along the Nanog promoter region by ChIP. ES cells were transfected with Luc (control) shRNA, Jmjd2c shRNA 1, Jmjd2c shRNA 2, or Jmjd1a shRNA 1. Fold enrichment is the relative abundance of DNA fragments detected by real-time PCR at the amplified region over a control amplified region and normalized with control Luc. GST antibody was used as a ChIP control. Data are presented as the mean ± SEM. (D) Jmjd2c associates with the Nanog promoter region. ChIP assays were performed with two different anti-Jmjd2c antibodies. A primer pair targeting amplicon 3 was used. GST antibody was used as a ChIP control. Data are presented as the mean ± SEM. (E) ChIP analysis showed no enrichment of Jmjd1a over the Nanog promoter region. A primer pair targeting amplicon 3 was used. Data are presented as the mean ± SEM. (F) Knockdown of Jmjd2c abolished the ChIP signal derived from anti-Jmjd2c antibody. ES cells were transfected with control Luc shRNA, Jmjd2c shRNA 1, or Jmjd2c shRNA 2. A primer pair targeting amplicon 3 was used. Data are presented as the mean ± SEM. (G) ChIP analysis showed Oct4 binding to the Nanog promoter remained unchanged upon Jmjd2c depletion. ES cells were transfected with control Luc shRNA, Jmjd2c shRNA 1, or Jmjd2c shRNA 2. A primer pair targeting amplicon 5 was used. Data are presented as the mean ± SEM. (H) ChIP analysis showed that HP1-β binding to the Nanog promoter was increased upon Jmjd2c depletion. ES cells were transfected with Luc shRNA (control), Jmjd2c shRNA 1, or Jmjd2c shRNA 2. A primer pair targeting amplicon 3 was used. Data are presented as the mean ± SEM. (I) ChIP analysis showed that KAP1 binding to the Nanog promoter was increased upon Jmjd2c depletion. ES cells were transfected with Luc shRNA (control), Jmjd2c shRNA 1, or Jmjd2c shRNA 2. A primer pair targeting amplicon 3 was used. Data are presented as the mean ± SEM.
Jmjd2c regulates the expression of Nanog through demethylation of H3K9Me3
To extend our study to Jmjd2c, we examined the H3K9Me3 status of Jmjd2c-regulated genes. Our marker gene analysis and microarray result showed that the expression of Nanog was reduced upon Jmjd2c depletion (Fig. 5A; Supplementary Table 1). Nanog is a key transcription factor important for the maintenance of pluripotency and self-renewal of ES cells (Chambers et al. 2003; Mitsui et al. 2003). Nanog expression was reduced to 50% upon Jmjd2c depletion (Fig. 2F), suggesting that there could be a Jmjd2c-dependent mechanism of regulating Nanog. Using a ChIP assay, we scanned the promoter region of Nanog (Fig. 5B). Upon knockdown using two shRNA constructs against Jmjd2c, we detected increased levels of H3K9Me3 to >2.5-fold at region 3 (Fig. 5C). This effect is specific to Jmjd2c, because knockdown of Jmjd1a did not result in an increase in H3K9Me2 or H3K9Me3 levels (Fig. 5C). The H3K9Me3 status at the promoter regions of nine other genes (Supplementary Table 2B) was, however, not affected by Jmjd2c depletion, indicating that Jmjd2c specifically regulates the chromatin associated with the Nanog promoter. Next, we asked if Jmjd2c is bound to the region that showed the greatest induction of H3K9Me3 upon Jmjd2c depletion. With two independently generated anti-Jmjd2c antibodies, we were able to detect Jmjd2c binding at the Nanog promoter (Fig. 5D). ChIP using an anti-Jmjd1a antibody showed no binding (Fig. 5E). The binding of Jmjd2c was abolished by either of our two Jmjd2c shRNA constructs, further showing that the ChIP signal is specific to Jmjd2c (Fig. 5F). Hence, the increase in H3K9Me3 level after Jmjd2c knockdown could be explained by the binding of Jmjd2c. As Nanog is a direct target gene for Oct4, we examined the Oct4 occupancy at the proximal promoter of Nanog. The ChIP assay showed that Oct4 binding at the Nanog promoter (Fig. 5G) was not affected by Jmjd2c depletion. H3K9Me3 can serve as a binding site for the transcriptional repressor protein HP1 in the silencing of gene expression (Bannister et al. 2001). We used a ChIP assay to test if the increase in H3K9Me3 levels over the Nanog promoter after Jmjd2c depletion leads to recruitment of the HP1-β/KAP1 corepressor complex (Ryan et al. 1999). Upon knockdown using two shRNA constructs against Jmjd2c, we detected an increase in HP1-β binding of up to twofold (Fig. 5H). Unlike Jmjd2c-depleted cells, we did not observe increased HP1-β after Jmjd1a knockdown in ES cells (data not shown). The ChIP assay also showed that the binding of corepressor KAP1 was induced after depletion of Jmjd2c (Fig. 5I). Taken together, these results suggest that Jmjd2c positively regulates Nanog by preventing H3K9 trimethylation and the binding of HP1/KAP1 complex on its promoter.
Nanog is a downstream effector of Jmjd2c in regulating ES cell maintenance
To determine if Nanog is a downstream effector of Jmjd2c in the maintenance of ES cells, we treated a Nanog-overexpressing ES cell line (Loh et al. 2006) with shRNAs against Jmjd2c. Interestingly, the two Jmjd2c shRNA constructs were unable to induce efficient differentiation as alkaline phosphatase-positive ES cell colonies were readily obtained (Fig. 6A). The depletion of Jmjd2c was equally efficient in the control and Nanog-overexpressing ES cells (Fig. 6B); this excludes the possibility that the phenotypic differences were due to insufficient depletion of Jmjd2c. Depletion of Jmjd1a in the Nanog-overexpressing ES cells showed more differentiation than Jmjd2c-depleted cells (Fig. 6A), suggesting that Nanog could not compensate for the loss of Jmjd1a. The rescued phenotype in Nanog-overexpressing cells was also supported by marker gene analyses. The levels of Oct4, Sox2, and Tdgf1 remained relatively unchanged (Fig. 6C), while the induction of differentiation markers such as Fgf5 and Msx1 was also reduced in Nanog-overexpressing ES cells as compared with control ES cells (Fig. 6D). Consistent with the morphology data, Jmjd1a knockdown cells showed no restoration of ES cell markers and little or no reduced expression of differentiation markers (Supplementary Figure 12). These results demonstrate that Nanog is able to rescue the knockdown effects of Jmjd2c depletion. Altogether, our finding provides mechanistic explanations for how Jmjd1a and Jmjd2c maintain the undifferentiated state of ES cells.
Figure 6.
Nanog is the key downstream effector of Jmjd2c responsible for maintaining ES cells’ self-renewal. (A) Overexpression of Nanog can rescue differentiation phenotype induced by Jmjd2c depletion. ES cells with constitutive Nanog overexpression (Loh et al. 2006) were challenged with shRNA directing against various transcripts (Jmjd2c, Jmjd1a, or Oct4). The cells were stained for alkaline phosphatase activity, and the morphologies were examined by microscopy. Note the morphological rescue and the maintenance of alkaline phosphatasepositive colonies in Jmjd2c shRNA-treated cells. Little or no morphological rescue was observed when the cells were challenged with Jmjd1a or Oct4 shRNA, respectively. (B) Jmjd2c was similarly depleted both in Nanog-overexpressing and control ES cells. Data are presented as the mean ± SEM. (C) Overexpression of Nanog reduced the down-regulation of Oct4, Sox2, and Tdgf1 upon Jmjd2c depletion. The levels of the transcripts were normalized against control plasmid-transfected cells. Data are presented as the mean ± SEM. (D) Enforced Nanog overexpression compensated for the Jmjd2c loss of function by reducing the induction of differentiation markers Fgf5 and Msx1. Data are presented as the mean ± SEM.
Discussion
Genetic network and epigenetic landscape in ES cells
During the process of ES cell division, the choice between self-renewal or differentiation is decided by the complex interplay between signaling pathways, transcription factor networks, and epigenetic processes. Recent studies have begun to define key players in the transcriptional factor networks of mouse ES cells (Nichols et al. 1998; Avilion et al. 2003; Chambers et al. 2003; Mitsui et al. 2003; Elling et al. 2006; Ivanova et al. 2006; Loh et al. 2006; Matoba et al. 2006; Wang et al. 2006; Wu et al. 2006; Zhang et al. 2006; Galan-Caridad et al. 2007; Lim et al. 2007). Alongside genetic factors, epigenetic mechanisms such as methylation of histones could also have important roles in maintaining self-renewal and pluripotency of ES cells.
In this study, we place two genes encoding JHDMs as downstream targets of Oct4, a critical regulator of pluripotency in ES cells. Oct4 binds to the regulatory regions of Jmjd1a and Jmjd2c as shown by the in vivo ChIP and in vitro EMSA assays. Significantly, both knockdown and reporter assays confirm that Oct4 is a positive regulator of the JHDMs. It is interesting to note that JMJD1A is also bound by OCT4 in human ES cells (Boyer et al. 2005); this could indicate evolutionary conservation of an important regulatory function. Apart from the JHDMs identified from the present study, Oct4 may also regulate genes encoding chromatin-modifying complexes in both human (e.g., SET, SMARCA, and MYST) (Boyer et al. 2005) and mouse ES cells (Ehmt1, Smarcad1, Myst2) (Loh et al. 2006). This suggests that Oct4 may govern the chromatin state of pluripotent ES cells by regulating the expression of genes directly involved in the epigenetic systems.
The repressive histone mark, H3K9 methylation, is maintained at a low level in ES cells. In contrast, differentiated cell types exhibit elevated levels of H3K9 methylation (Meshorer et al. 2006), which suggests a role for histone H3K9 demethylases in maintaining a transcriptionally permissive chromatin state. Removal of the JHDMs results in an elevation of global histone H3K9Me level, suggesting that the JHDMs play active roles in maintaining transcriptionally permissive chromatin in pluripotent ES cells. The up-regulation of these JHDMs in ES cells may explain the low level of repressive H3K9 methylation. However, it is not clear if the low global level of H3K9 methylation is required for the maintenance of the “stemness” state of ES cells.
Roles of Jmjd1a and Jmjd2c in the maintenance of the ES cell self-renewal
Previous studies have investigated the role of histone methylases in ES cells or early embryonic development. Setdb1 and Ezh2 knockout embryos show early embryonic lethality (embryonic day 3.5–4.5 [E3.5–E4.5]) and are defective in outgrowth of the ICM (O’Carroll et al. 2001; Dodge et al. 2004). It is not clear, however, if Setdb1 or Ezh2 is required to maintain the self-renewal of ES cells. G9a-null embryos survive until E9.5, but G9a knockout ES cells showed compromised differentiation processes (Tachibana et al. 2002). Similarly, while Suz12−/− ES cells (Pasini et al. 2007) can be established and expanded in tissue culture, the ability to give rise to proper differentiation is impaired.
In this study, we showed that ES cells depleted of Jmjd1a and Jmjd2c lost their distinctive colony morphologies and gave rise to fibroblast-like cells. Moreover, the knockdown cells were not able to efficiently form colonies in secondary replating assays. Differentiation induced by the depletion of these JHDMs led to the loss of pluripotency and self-renewal. Apart from these JHDMs, no other histone-modifying enzyme has been shown to be important in the maintenance of self-renewal of ES cells (Niwa 2007). Transcription regulators promote self-renewal through different mechanisms. These mechanisms may involve inhibition of differentiation or promoting proliferation (Niwa 2007). We provided evidence that Jmjd1a and Jmjd2c maintain the “stemness” state of ES cells through regulating downstream genes that encode for self-renewal regulators (Chambers et al. 2003; Mitsui et al. 2003; Ivanova et al. 2006; Matoba et al. 2006).
Previous work has shown that Jmjd1a demethylates H3K9Me2 of LamB1 and Stra6 in F9 embryonic carcinoma cells (Yamane et al. 2006). Knockdown of Jmjd1a in F9 cells also slightly reduces the expression of Oct4, Sox2, and Nanog (Yamane et al. 2006). We showed that Jmjd1a prevents the promoter regions of pluripotency-associated genes (Tcl1, Tcfcp2l1, and Zfp57) from H3K9 dimethylation (Fig. 7). An increase in H3K9 dimethylation is correlated with a reduction in the expression of the target genes, indicating that Jmjd1a positively regulates their expression. Oct4 recruitment to the Tcl1 promoter was reduced in the Jmjd1a knockdown ES cells. It is likely that the increased H3K9Me2 limited the access of Oct4 to the oct element of the Tcl1 promoter region. Thus we have identified a novel mechanism by which Jmjd1a maintains the pluripotent epigenetic state of a key regulator of ES cells. Tcl1, a gene encoding for a cofactor of the Akt1 kinase, is of interest as it has been shown to regulate self-renewal of ES cells (Ivanova et al. 2006; Matoba et al. 2006). Interestingly, forced expression of Tcl1 can rescue the differentiated phenotype brought about by the knockdown of Jmjd1a, but not Jmjd2c. Moreover, most of the pluripotent and differentiation markers’ levels were restored in the Tcl1 rescue experiments. This suggests that Tcl1 is the dominant target of Jmjd1a in regulating self-renewal. However, we observed that the Tcl1 rescue was only partial, as some differentiated cells were still observed. Thus it is possible that other Tcl1-independent mechanisms exist for Jmjd1a in regulating the self-renewal of ES cells. Further analysis using genome-wide ChIP will be important to define other regulatory targets of Jmjd1a that could be important in ES cell biology.
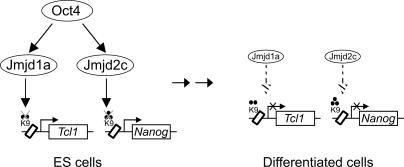
Figure 7.
Model for the maintenance of self-renewal of ES cells by Jmjd1a and Jmjd2c. Schematic showing the interplay of Oct4 with Jmjd1a and Jmjd2c in sustaining ES cells’ self-renewal. In ES cells, Oct4 up-regulates the levels of Jmjd1a and Jmjd2c. Jmjd1a and Jmjd2c maintain Tcl1 and Nanog by demethylation of the repressive H3K9Me2 and H3K9Me3 marks, respectively. Notably, Tcl1 and Nanog are both downstream targets of Oct4. With differentiation, the down-regulation of Jmjd1a and Jmjd2c (dashed arrows) results in an elevation of the repressive H3K9Me2/Me3 modifications and reduced expression of downstream genes.
Overexpression of the Jmjd2 family of JHDMs has been shown to demethylate H3K9Me3-enriched pericentric heterochromatin and cause delocalization of HP1-β (Cloos et al. 2006; Fodor et al. 2006; Klose et al. 2006). Here, we show that Jmjd2c is involved in regulating the euchromatin H3K9Me3 status of a key pluripotency gene, Nanog. Our result also suggests that the specific demethylation of H3K9Me3 by Jmjd2c at the Nanog promoter may prevent the binding of transcription corepressors such as HP1 and KAP1. Forced expression of Nanog can rescue the knockdown effects of Jmjd2c. However, as seen in the Tcl1 rescue, overexpressing Nanog does not completely rescue the Jmjd2c knockdown effect. This could indicate the presence of other Nanog-independent mechanisms downstream from Jmjd2c in the maintenance of self-renewal of ES cells.
A series of recent studies reported that ectopic expression of four transcription factors alone (Oct4, Sox2, c-Myc, and Klf4) is able to reprogram somatic cells to the pluripotent stem cell state (Takahashi and Yamanaka 2006; Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007). These remarkable studies demonstrate that the genetic program and epigenetic landscape of stem cells can be restored in differentiated cells. We postulate that some of these reprogramming factors may be able to up-regulate histone modifiers such as Jmjd1a and Jmjd2c to assist in the resetting of the epigenetic landscape of somatic cells. In the present study, we identified two novel nodes in the Oct4 transcription regulatory network, extending from Jmjd1a and Jmjd2c to Tcl1 and Nanog, respectively (Fig. 7). Hence, these state-specific demethylases appear to directly regulate a distinct set of genes. This is also the first example of an ES cell transcription factor regulating a novel pathway that specifies the epigenetic status of pluripotency-associated genes.
Materials and methods
Cell culture and transfection
Feeder-free E14 mouse ES cells were cultured at 37°C with 5% CO2. All cells were maintained on gelatin-coated dishes in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO), supplemented with 15% heat-inactivated fetal bovine serum (FBS; GIBCO), 0.055 mM β-mercaptoethanol (GIBCO), 2 mM L-glutamine, 0.1 mM MEM nonessential amino acid, 5000 U/mL penicillin/streptomycin, and 1000 U/mL LIF (Chemicon), as described previously (Chew et al. 2005). Transfection of shRNA and overexpression plasmids was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, 2 μg of plasmids were transfected into ES cells on 60-mm plates for RNA and protein extraction. For the ChIP assay, 18 μg of plasmids were transfected into ES cells on 150-mm plates. Puromycin (Sigma) selection was introduced 1 d after transfection at 1.0 μg/mL, and maintained for 4 d prior to harvesting. For the replating assay, after 3 d of puromycin selection, shRNA-transfected cells were trypsinized and resuspended in medium. Ten-thousand cells were plated onto newly gelatin-coated 60-mm plates to form secondary ES cell colonies. After 4 d, emerging colonies were stained for alkaline phosphatase activity. For all the data shown (unless indicated otherwise), the cells were harvested and analyzed after 4 d of puromycin selection. Detection of alkaline phosphatase, which is indicative of the undifferentiated state of ES cells, was carried out using a commercial ES Cell Characterization Kit from Chemicon (catalog no. SCR001).
RNAi assay
shRNA constructs were designed as described previously (Chew et al. 2005). Two shRNA constructs each for Jmjd1a and Jmjd2c were designed to target 19-base-pair (bp) gene-specific regions. The oligonucleotides used for Jmjd1a and Jmjd2c shRNA constructs are shown in Supplementary Figures 3A and 4A. These oligonucleotides were cloned into pSuperpuro (BglII and HindIII sites; Oligoengine). The pSuperpuro plasmid carries a puromycin gene driven by a PGK promoter. We used pSuperpuro constructs expressing shRNA against Luciferase (Firefly) or Green fluorescent protein (Gfp) as controls. These constructs were effective in knocking down coexpressed Luciferase or Gfp; therefore, they produced effective small interfering RNAs (siRNAs) in ES cells.
RNA isolation, reverse transcription, and real-time PCR analysis
Total RNA was extracted using Trizol (Invitrogen) and was purified with an RNeasy minikit (Qiagen). cDNA synthesis was performed with 1 μg of total RNA using the SuperScript II kit (Invitrogen) according to the manufacturer’s instructions. Endogenous mRNA levels were measured by real-time PCR analysis based on SYBR Green detection with the ABI Prism 7900HT machine (Applied Biosystems). Results were normalized with β-actin. The real-time PCR primers are available on request.
Protein extraction and Western blotting
Histones were extracted using the acid extraction method. Briefly, cells were scraped from culture dishes in chilled PBS, centrifuged, and washed once with ice-cold PBS. Cell pellets were then incubated in Triton extraction buffer (PBS, 0.5% Triton X-100, 2 mM PMSF) for 10 min on ice. Pellets were resuspended in 0.2 N HCl overnight for the extraction of histone. Total protein extracts were prepared by lysing cells in SDS loading buffer. Total protein (40 μg) or histone (5 μg) was separated by SDS-PAGE and transferred to PVDF membrane. The membrane was probed with either anti-H3K9Me2 (ab7312; Abcam), anti-H3K9Me3 (ab8898; Abcam), anti-Jmjd1a (amino acids 1–400 of mouse Jmjd1a raised in rabbit), or anti-Jmjd2c (amino acids 351–551 of mouse Jmjd2c raised in rabbit). Anti-H3 (ab1791; Abcam) or anti-β-tubulin was used as loading control.
Microarray
mRNAs derived from Jmjd1a shRNA 1-, Jmjd2c shRNA 1-, and Luc shRNA-treated ES cells were reverse-transcribed, labeled, and analyzed using the Illumina microarray platform (Sentrix Mouse-6 Expression BeadChip version 1.0). Arrays were processed as per the manufacturer’s instructions. Three biological repeats of the profiles (each for control and knockdown of the two genes) were used to generate statistically significant gene lists. Rank Invariant normalization was used to normalize the microarrays. Significance analysis of microarrays (SAM) was used to select differentially expressed genes. The differentially expressed genes were selected based on the following three criteria: fold change (FC) > 1.5 for up-regulated, FC < 0.6 for down-regulated; q value < 2%; and detection probability >0.99 in at least all three samples of any one group (control or treatment). Microarray data will be uploaded to a public microarray resource site. To compute the nominal P-value for the overlapping gene lists, we performed Monte Carlo simulation as described previously (Loh et al. 2006)
ChIP assay
ChIP assay was carried out as described previously (Loh et al. 2006). Briefly, cells were cross-linked with 1% (w/v) formaldehyde for 10 min at room temperature, and formaldehyde was then inactivated by the addition of 125 mM glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-Oct4 (sc-8628; Santa Cruz Biotechnology), anti-Sox2 (sc-17320; Santa Cruz Biotechnology), anti-H3K9Me2 (ab7312; Abcam), anti-H3K9Me3 (ab8898; Abcam), anti-Jmjd1a Ab1 (amino acids 1–300 of mouse Jmjd1a raised in rabbit), anti-Jmjd1a Ab2 (amino acids 1–400 of mouse Jmjd1a raised in rabbit), anti-Jmjd2c Ab1 (amino acids 523–702 of mouse Jmjd2c raised in rabbit), anti-Jmjd2c Ab2 (amino acids 351–551 of mouse Jmjd2c raised in rabbit), anti-HP1-β (MAB3448; Chemicon), or anti-KAP1 (ab22553; Abcam) antibodies. Anti-GST (sc-459; Santa Cruz Biotechnology) or anti-GFP (sc-9996; Santa Cruz Biotechnology) antibodies were used as mock ChIP controls. Quantitative PCR analyses were performed in real time using the ABI PRISM 7900 sequence detection system and SYBR green master mix. Threshold cycles (Ct) were determined for both immunoprecipitated DNA and known amount of DNA from input sample for different primer pairs. Relative occupancy values (also known as fold enrichments) were calculated by determining the immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and were normalized to the level observed at a control region, which was defined as 1.0. The coordinates for the control region, which is downstream from the Nanog gene, is chr6:123352993–123353158 (mm5 genome build). For all the primers used, each gave a single product of the right size, as confirmed by agarose gel electrophoresis and dissociation curve analysis. The real-time PCR primers are available on request.
Acknowledgments
We acknowledge Ching-Aeng Lim and Katty Kuay for technical help. We thank Andrew Hutchins, Linda Lim, Ching-Aeng Lim, Neil Clarke, Keh-Chuang Chin, Bing Lim, Larry Stanton, Paul Robson, and Edwin Chueng for critical comments on the manuscript. We are grateful to the Biomedical Research Council of Agency of Science, Technology, and Research (A*STAR) for funding. Y.H.L. is supported by the A*STAR graduate scholarship. W.Z. is supported by the National University of Singapore graduate scholarship. This work is supported in part by the Singapore Stem Cell Consortium.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1588207
References
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Perez L., Vivian N., Lovell-Badge R., Vivian N., Lovell-Badge R., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Casanova M., Warnes G., Merkenschlager M., Warnes G., Merkenschlager M., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Zegerman P., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Miska E.A., Allshire R.C., Kouzarides T., Allshire R.C., Kouzarides T., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Fry B., Meissner A., Wernig M., Plath K., Meissner A., Wernig M., Plath K., Wernig M., Plath K., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A., Robertson M., Nichols J., Lee S., Tweedie S., Smith A., Nichols J., Lee S., Tweedie S., Smith A., Lee S., Tweedie S., Smith A., Tweedie S., Smith A., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chew J.L., Loh Y.H., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Loh Y.H., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Li P., Ang Y.S., Lim B., Robson P., Ang Y.S., Lim B., Robson P., Lim B., Robson P., Robson P., et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Rappsilber J., Antal T., Hansen K.H., Helin K., Antal T., Hansen K.H., Helin K., Hansen K.H., Helin K., Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Dodge J.E., Kang Y.K., Beppu H., Lei H., Li E., Kang Y.K., Beppu H., Lei H., Li E., Beppu H., Lei H., Li E., Lei H., Li E., Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan P.J., Gearhart J., Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414:92–97. doi: 10.1038/35102154. [DOI] [PubMed] [Google Scholar]
- Elling U., Klasen C., Eisenberger T., Anlag K., Treier M., Klasen C., Eisenberger T., Anlag K., Treier M., Eisenberger T., Anlag K., Treier M., Anlag K., Treier M., Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc. Natl. Acad. Sci. 2006;103:16319–16324. doi: 10.1073/pnas.0607884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Opravil S., Mechtler K., Schotta G., Jenuwein T., Mechtler K., Schotta G., Jenuwein T., Schotta G., Jenuwein T., Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Caridad J.M., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B., Doetsch F.K., Mirny L.A., Reizis B., Mirny L.A., Reizis B., Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., Lemischka I.R., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., Lemischka I.R., Schaniel C., Hackney J.A., Moore K.A., Lemischka I.R., Hackney J.A., Moore K.A., Lemischka I.R., Moore K.A., Lemischka I.R., Lemischka I.R. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R., DeCoste C., Schafer X., Lun Y., Lemischka I.R., Schafer X., Lun Y., Lemischka I.R., Lun Y., Lemischka I.R., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tempst P., Wong J., Zhang Y., Wong J., Zhang Y., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M., Jenuwein T., Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Hart S.R., Skalnik D.G., Hart S.R., Skalnik D.G., Skalnik D.G. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lim L.S., Loh Y.H., Zhang W., Li Y., Chen X., Wang Y., Bakre M., Ng H.H., Stanton L.W., Loh Y.H., Zhang W., Li Y., Chen X., Wang Y., Bakre M., Ng H.H., Stanton L.W., Zhang W., Li Y., Chen X., Wang Y., Bakre M., Ng H.H., Stanton L.W., Li Y., Chen X., Wang Y., Bakre M., Ng H.H., Stanton L.W., Chen X., Wang Y., Bakre M., Ng H.H., Stanton L.W., Wang Y., Bakre M., Ng H.H., Stanton L.W., Bakre M., Ng H.H., Stanton L.W., Ng H.H., Stanton L.W., Stanton L.W. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell. 2007;18:1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel D.A., Watson C.M., De Young R.A., Tam P.P., Watson C.M., De Young R.A., Tam P.P., De Young R.A., Tam P.P., Tam P.P. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev. Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Chen X., Bourque G., George J., Leong B., Liu J., Bourque G., George J., Leong B., Liu J., George J., Leong B., Liu J., Leong B., Liu J., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Yachechko R., Tchieu J., Jaenisch R., Tchieu J., Jaenisch R., Jaenisch R., et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Matoba R., Niwa H., Masui S., Ohtsuka S., Carter M.G., Sharov A.A., Minoru K., Niwa H., Masui S., Ohtsuka S., Carter M.G., Sharov A.A., Minoru K., Masui S., Ohtsuka S., Carter M.G., Sharov A.A., Minoru K., Ohtsuka S., Carter M.G., Sharov A.A., Minoru K., Carter M.G., Sharov A.A., Minoru K., Sharov A.A., Minoru K., Minoru K. Dissecting oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Misteli T., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T., George E., Scambler P.J., Brown D.T., Misteli T., Scambler P.J., Brown D.T., Misteli T., Brown D.T., Misteli T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Takahashi K., Maruyama M., Maeda M., Yamanaka S., Maruyama M., Maeda M., Yamanaka S., Maeda M., Yamanaka S., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Klewe-Nebenius D., Chambers I., Schöler H., Smith A., Chambers I., Schöler H., Smith A., Schöler H., Smith A., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T., Pagani M., Barton S.C., Surani M.A., Jenuwein T., Barton S.C., Surani M.A., Jenuwein T., Surani M.A., Jenuwein T., Jenuwein T. The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S., Ichisaka T., Yamanaka S., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Palmieri S.L., Peter W., Hess H., Schöler H.R., Peter W., Hess H., Schöler H.R., Hess H., Schöler H.R., Schöler H.R. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K., Bracken A.P., Hansen J.B., Capillo M., Helin K., Hansen J.B., Capillo M., Helin K., Capillo M., Helin K., Helin K. The Polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A., Matsuzaki Y., Mulligan R.C., Melton D.A., Mulligan R.C., Melton D.A., Melton D.A. ‘Stemness’: Transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Ryan R.F., Schultz D.C., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J., Schultz D.C., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J., Friedman J.R., Fredericks W.J., Rauscher F.J., Fredericks W.J., Rauscher F.J., Rauscher F.J. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: A potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Whetstine J.R., Whetstine J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Smith A.G. Embryo-derived stem cells: Of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Fukuda M., Takeda N., Niida H., Kato H., Takeda N., Niida H., Kato H., Niida H., Kato H., Kato H., et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trojer P., Reinberg D., Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Borchers C.H., Tempst P., Zhang Y., Tempst P., Zhang Y., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Turner B.M. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H., Levasseur D.N., Theunissen T.W., Orkin S.H., Theunissen T.W., Orkin S.H., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R., Hochedlinger K., Bernstein B.E., Jaenisch R., Bernstein B.E., Jaenisch R., Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Spooner E., Li E., Zhang G., Colaiacovo M., Li E., Zhang G., Colaiacovo M., Zhang G., Colaiacovo M., Colaiacovo M., et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wu Q., Chen X., Zhang J., Loh Y.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Chen X., Zhang J., Loh Y.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Zhang J., Loh Y.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Loh Y.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H., Zhang W., Sze S.K., Lim B., Ng H.H., Sze S.K., Lim B., Ng H.H., Lim B., Ng H.H., Ng H.H. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tempst P., Wong J., Zhang Y., Wong J., Zhang Y., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tam W.L., Tong G.Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Tam W.L., Tong G.Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Tong G.Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Lou Y., Yang J., Ma Y., Chai L., Yang J., Ma Y., Chai L., Ma Y., Chai L., Chai L., et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]