Abstract
microRNAs (miRNAs) silence gene expression by suppressing protein production and/or by promoting mRNA decay. To elucidate how silencing is accomplished, we screened an RNA interference library for suppressors of miRNA-mediated regulation in Drosophila melanogaster cells. In addition to proteins known to be required for miRNA biogenesis and function (i.e., Drosha, Pasha, Dicer-1, AGO1, and GW182), the screen identified the decapping activator Ge-1 as being required for silencing by miRNAs. Depleting Ge-1 alone and/or in combination with other decapping activators (e.g., DCP1, EDC3, HPat, or Me31B) suppresses silencing of several miRNA targets, indicating that miRNAs elicit mRNA decapping. A comparison of gene expression profiles in cells depleted of AGO1 or of individual decapping activators shows that ∼15% of AGO1-targets are also regulated by Ge-1, DCP1, and HPat, whereas 5% are dependent on EDC3 and LSm1–7. These percentages are underestimated because decapping activators are partially redundant. Furthermore, in the absence of active translation, some miRNA targets are stabilized, whereas others continue to be degraded in a miRNA-dependent manner. These findings suggest that miRNAs mediate post-transcriptional gene silencing by more than one mechanism.
Keywords: Argonaute, decapping activators, decapping, miRNAs, mRNA decay, P-bodies, varicose
Post-transcriptional gene regulation plays a central role in biological processes as diverse as development, differentiation, stress response, and growth control. In particular, regulations of mRNA half-lives and of translation are key mechanisms by which the expression of many genes can be rapidly changed in response to extracellular signals (for review, see Wilusz and Wilusz 2004). Post-transcriptional regulation is often mediated by specific RNA-binding proteins that recognize control elements in the mRNA 3′ untranslated regions (UTRs) and promote (or antagonize) mRNA degradation or translational repression. mRNA decay or translational repression can also be triggered by short interfering RNAs (siRNAs) or microRNAs (miRNAs), two classes of noncoding RNAs that associate with proteins of the Argonaute family, to regulate the expression of fully or partially complementary target mRNAs (for review, see Bartel 2004).
Degradation of bulk mRNA in eukaryotes is normally initiated by the gradual shortening of the poly(A) tail (deadenylation) (for review, see Parker and Song 2004). In Saccharomyces cerevisiae and Drosophila melanogaster cells, the major deadenylase activity is associated with the CAF1:CCR4:NOT complex (Parker and Song 2004). Deadenylated mRNA can then be exonucleolytically digested from the 3′ end by the exosome and cofactors (Parker and Song 2004). Alternatively, decay proceeds by the removal of the cap structure by the decapping enzyme DCP2 and subsequent 5′-to-3′ exonucleolytic degradation by XRN1 (Parker and Song 2004).
In S. cerevisiae, several proteins stimulate decapping by DCP2 including DCP1, EDC3 (enhancer of decapping 3), the LSm1–7 complex, Pat1, and the RNA helicase Dhh1. These proteins are generically termed decapping activators, although they may activate decapping by different mechanisms (Parker and Song 2004). In human cells, DCP2 is part of a multimeric protein complex that includes DCP1, EDC3, the RNA helicase RCK/p54 (the human ortholog of Dhh1), and Ge-1 (also known as human enhancer of decapping large subunit, Hedls), a protein that has no orthologs in S. cerevisiae (Fenger-Grøn et al. 2005; Yu et al. 2005).
Recent studies in zebrafish embryos, D. melanogaster, and human cells have shown that miRNAs accelerate the deadenylation and decapping of their targets by recruiting components of the general mRNA degradation machinery (Bagga et al. 2005; Behm-Ansmant et al. 2006a, b; Giraldez et al. 2006; Wu et al. 2006). miRNA-mediated mRNA decay requires the Argonaute proteins, the P-body component GW182, the CAF1:CCR4:NOT deadenylase complex, the decapping enzyme DCP2, and the decapping activator DCP1 (Jakymiw et al. 2005; Liu et al. 2005a; Meister et al. 2005; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a, b; Chu and Rana 2006). Moreover, in human cells, the decapping activator RCK/p54 was implicated in silencing by miRNAs (Chu and Rana 2006).
miRNAs not only promote mRNA degradation, but they can also repress protein production, often without detectable changes in the abundance of target mRNAs (for review, see Pillai et al. 2006; Jackson and Standart 2007; Nilsen 2007). Several mechanisms for the miRNA-mediated down-regulation of protein expression have been proposed. These include (1) cotranslational protein degradation, (2) inhibition of translation elongation, (3) premature termination of translation (ribosome drop-off), or (4) inhibition of translation initiation (for review, see Pillai et al. 2006; Jackson and Standart 2007; Nilsen 2007).
Two recent reports demonstrated that miRNAs inhibit translation; surprisingly, each proposed a different underlying mechanism (Chendrimada et al. 2007; Kiriakidou et al. 2007). Kiriakidou et al. (2007) showed that the human Argonaute-2 (AGO2) polypeptide sequence exhibits sequence similarities with the cytoplasmic cap-binding protein eIF4E (eukaryotic initiation factor 4E) and binds the mRNA cap structure. This supports the argument that miRNAs (in complex with human AGO2) can inhibit translation at the initiation step by competing with eIF4E for the cap structure.
Chendrimada et al. (2007) identified the eukaryotic initiation factor 6 (eIF6) as a binding partner of AGO2 in human cells (Chendrimada et al. 2007). eIF6 interacts with the large ribosomal subunit and prevents its premature association with the small subunit (Ceci et al. 2003). If AGO2 recruits eIF6 to miRNA targets, then it may repress translation at an early step by blocking association of the large ribosomal subunit (Chendrimada et al. 2007). Even though eIF6 is a translation factor, its depletion suppressed the silencing of targets regulated mainly at the mRNA level. The most straightforward interpretation of these results is that miRNAs silence genes primarily by inhibiting protein synthesis, and mRNA degradation is only a consequence of this primary event (Chendrimada et al. 2007).
To shed light on the mechanisms that allow miRNAs to repress expression of their targets, we screened an RNA interference (RNAi) library for suppressors of silencing mediated by miR-12. This screen identified genes already known to be required for miRNA biogenesis and function (Drosha, Pasha, Dicer-1, AGO1, and GW182), as well as the decapping activator Ge-1. We show that depleting Ge-1 alone and/or in combination with other decapping activators relieves silencing of targets by several miRNAs, indicating that miRNAs promote decapping and the subsequent mRNA degradation. We show further that some miRNA targets are stabilized in the absence of active translation, whereas others are nevertheless degraded. These findings suggest the existence of more than one mechanism of miRNA function and may explain conflicting reports regarding the mechanisms of silencing by miRNAs.
Results
A genome-wide RNAi screen identified the decapping activator Ge-1 as an effector of the miRNA pathway
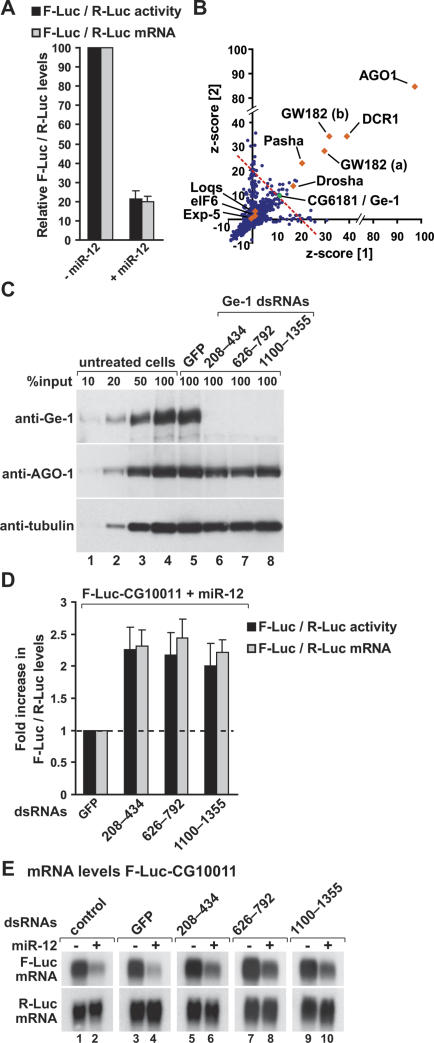
We screened for suppressors of miRNA-mediated gene silencing using a reporter expressing the firefly luciferase (F-Luc) coding region with a 3′ UTR from the D. melanogaster CG10011 transcript, an mRNA silenced by miR-12 (Rehwinkel et al. 2005, 2006; Behm-Ansmant et al. 2006a, b). In the presence of miR-12, expression of both F-Luc protein and mRNA is reduced to similar extents (Fig. 1A), indicating that miR-12 directs this reporter for degradation (Behm-Ansmant et al. 2006a, b).
Figure 1.
A genome-wide RNAi screen reveals a role for Ge-1 in miRNA-mediated gene silencing. (A) S2 cells were transfected with a mixture of plasmids containing the F-Luc-CG10011 reporter, a plasmid expressing miR-12 (+miR-12) or the corresponding empty vector (−miR-12), and a plasmid expressing Renilla luciferase (R-Luc). Firefly luciferase activity (black bars) and mRNA levels (gray bars) are normalized to that of the Renilla luciferase and set to 100 in cells transfected with the empty vector. Mean values ± standard deviations from three independent experiments are shown. (B) The results of the RNAi screen are represented as z-scores of two independent screens (see Materials and Methods). (Blue dots) Individual dsRNAs in the library; (red diamonds) components of the miRNA pathway; (green dot) Ge-1 (CG6181); (dashed red line) cutoff used (average z-score > 10). (C–E) S2 cells were treated with the indicated dsRNAs on days 0 and 4. On day 6, cells were transfected with the mixture of plasmids described in A. (C) The effectiveness of the depletions was analyzed by Western blotting, with the antibodies indicated on the left. In lanes 1–4, dilutions of untreated cells are loaded. Anti-tubulin antibodies were used as a loading control. Numbers above the lanes indicate the regions targeted by the dsRNAs and correspond to amino acids in the Ge-1 protein sequence. (D) Firefly luciferase activity (black bars) and mRNA levels (gray bars) are normalized to that of the Renilla luciferase. Furthermore, for each knockdown, the normalized values of F-Luc activity and mRNA levels in the presence of the miRNA are divided by those obtained in the absence of the miRNA. These ratios are set to unity in cells treated with GFP dsRNA. Mean values ± standard deviations from three independent experiments are shown. (E) Northern blot analysis of representative RNA samples isolated from S2 cells shown in D.
The screen was performed in duplicate with a double-stranded RNA (dsRNA) library that targets ∼90% of all annotated genes in the D. melanogaster genome (Boutros et al. 2004; Gesellchen et al. 2005). We identified 47 dsRNAs that relieved the silencing by miR-12, including six previously identified components of the D. melanogaster miRNA pathway; i.e., Drosha, Pasha, Dicer-1, AGO1, and GW182 (Fig. 1B; Table 1). Exportin-5, Loquacious, and eIF6, also implicated in miRNA biogenesis and function (Förstemann et al. 2005; Saito et al. 2005; Lund and Dahlberg 2006; Chendrimada et al. 2007), did not score positive in our screen (Fig. 1B; Table 1).
Table 1.
z-Scores of the dsRNAs targeting known effectors of the miRNA pathway and Ge-1 in two independent screens
GW182 is represented in the library by two nonoverlapping dsRNAs (a and b).
We also identified several new candidates. To determine the specificity of the new candidates, we resynthesized and tested the dsRNAs using the same reporter either in the absence or presence of miR-12. When we took into account nonspecific effects of the dsRNAs on F-Luc-CG10011 expression (i.e., those observed in the absence of miR-12), only the dsRNA targeting D. melanogaster Ge-1 (CG6181) interfered specifically with silencing of the reporter (data not shown).
To confirm that suppression of silencing by Ge-1 dsRNA was specific, and not due to off-target effects, we designed two additional dsRNAs that target nonoverlapping regions of the Ge-1 mRNA. These dsRNAs also depleted endogenous Ge-1 efficiently, as judged by Western blot (Fig. 1C). Moreover, all three dsRNAs partially relieved the inhibition of F-Luc expression caused by miR-12 (Fig. 1D), and led to an approximately twofold increase of F-Luc-CG10011 mRNA levels (Fig. 1D,E). The partial restoration of luciferase activity and mRNA levels observed in these experiments (i.e., full restoration would require a fivefold increase of F-luc activity and mRNA levels) (Fig. 1A) is consistent with the effects observed in the screen (z-score ≈ 10 for Ge-1 vs. z-score ≈ 90 for AGO1) (Table 1) and is probably due to the redundancy of decapping activators (see Fig. 4, below). Importantly, depleting Ge-1 does not alter the expression levels of AGO1 or GW182 (Fig. 1C; data not shown). Together these results indicate that Ge-1 is required for miR-12 to efficiently destabilize the F-Luc-CG10011 reporter mRNA.
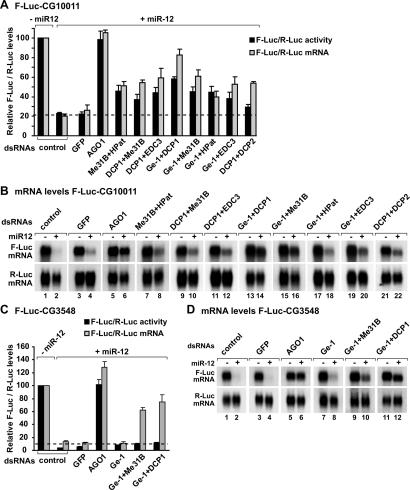
Figure 4.
Decapping activators have partially redundant functions in silencing. (A–D) S2 cells were treated with the indicated dsRNAs on days 0 and 4. On day 6, cells were cotransfected with a mixture of plasmids expressing firefly luciferase (F-Luc) followed by the indicated 3′ UTRs, plasmids expressing miRNA primary transcripts (+miR-12) or the corresponding empty vector (−miR-12), and a plasmid expressing Renilla luciferase (R-Luc). (A,C) Firefly luciferase activity and mRNA levels are normalized to that of the Renilla luciferase. For each knockdown, the normalized values of F-Luc activity and mRNA levels are set to 100 in cells transfected with the empty vector (not shown except for control cells). Mean values ± standard deviations from three independent experiments are shown. (B,D) Northern blot analysis of representative RNA samples isolated from S2 cells shown in A and C, respectively.
Ge-1 is a P-body component that belongs to the β-propeller family of proteins
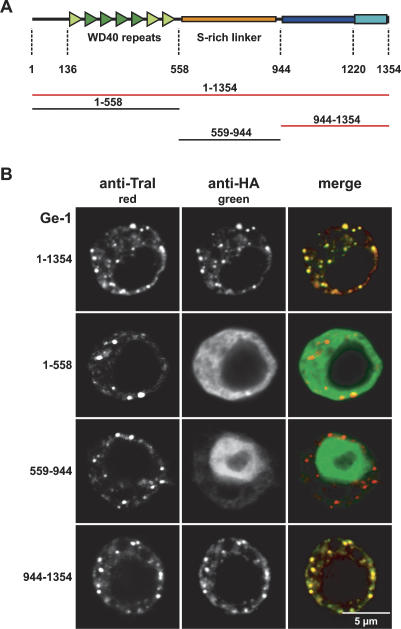
The Ge-1 protein family is characterized by four canonical N-terminal WD40 repeats (Fig. 2A, dark-green triangles; Deyholos et al. 2003; Fenger-Grøn et al. 2005; Yu et al. 2005; Xu et al. 2006). WD40 repeats are protein: protein interaction modules present in proteins with unrelated functions (Li and Roberts 2001). These repeats adopt a six- to seven-blade propeller fold and usually require six to seven repeats for structural stability (Li and Roberts 2001). Sequence alignments and secondary structure predictions of the region encompassing amino acids 136–558 of D. melanogaster Ge-1 revealed three additional WD40 repeats (Fig. 2A, light-green triangles), probably adapted to a specialized function in this protein family; thus standard computer programs might not detect statistically significant sequence similarities to known WD40s. The Ge-1 protein family is also characterized by a conserved C-terminal domain containing a highly conserved motif (Fig. 2A, blue and cyan boxes, respectively). The N-terminal β-propeller and the C-terminal domains are separated by a low-complexity region rich in serines (S-rich linker) (Fig. 2A, orange box) that probably provides a flexible linker between these two protein domains.
Figure 2.
Ge-1 is a conserved component of metazoan P-bodies. (A) Domain architecture of Ge-1. (Dark-green triangles) Canonical WD40 repeats; (light-green triangles) divergent WD40 repeats; (yellow box) flexible linker rich in serines; (dark-blue box) C-terminal conserved domain; (cyan box) highly conserved motif. Numbers below the protein outline represent amino acid positions at fragment boundaries for the D. melanogaster protein. The protein domains sufficient for the localization to P-bodies are shown in red. (B) Confocal fluorescent micrograph of S2 cells expressing HA fusions of the Ge-1 domains indicated on the left. Cells were stained with anti-HA and affinity-purified anti-Tral antibodies. Bar, 5 μm.
The Ge-1 proteins in Homo sapiens, D. melanogaster, and Arabidopsis thaliana localize to P-bodies and are required for P-body integrity (Fenger-Grøn et al. 2005; Yu et al. 2005; Xu et al. 2006; Eulalio et al. 2007a, b). Analysis of the subcellular localization of HA-tagged Ge-1 protein domains (the N-terminal β-propeller domain, the S-rich linker, and the C-terminal domain) indicated that the C-terminal conserved domain (amino acids 944– 1354) localizes to cytoplasmic foci (Fig. 2B), as with the full-length protein. These foci corresponded to endogenous P-bodies, because they were labeled with antibodies recognizing the protein Trailer hitch (Tral) (Fig. 2B), an endogenous P-body marker in D. melanogaster (Eulalio et al. 2007a, b). The N-terminal domain encompassing the WD40 repeats spread diffusely throughout the cytoplasm, whereas the S-rich linker domain accumulated within the nucleus (Fig. 2B). Overexpressing these protein fragments did not affect the integrity of endogenous P-bodies, as evidenced by the presence of foci stained with anti-Tral antibodies (Fig. 2B). Thus, Ge-1 is a conserved component of P-bodies.
Decapping activators regulate a subset of AGO1 targets
To investigate whether in addition to Ge-1, other decapping activators participate in miRNA-mediated gene silencing, and to determine how generally silencing required decapping activators, we compared mRNA expression profiles in cells depleted of AGO1, DCP1, Ge-1, HPat, EDC3, or LSm1, using high-density oligonucleotide microarrays. The profiles of DCP2- and Me31B-depleted cells could not be analyzed for technical reasons (see Materials and Methods). For each protein, we obtained expression profiles from two or three independent RNA samples. As a reference, RNA samples were isolated from mock-treated cells. We also examined the mRNA profiles in cells treated with GFP (green fluorescent protein) dsRNA to identify mRNAs that are likely to be regulated nonspecifically in response to the dsRNA treatment.
We grouped detectable transcripts to three classes according to their relative expression levels. These were underrepresented transcripts (at least 1.5-fold underrepresented compared with the reference sample) (Fig. 3, blue), transcripts that did not significantly change (<1.5-fold different from the reference) (Fig. 3, yellow), and overrepresented transcripts (at least 1.5-fold overrepresented) (Fig. 3, red). We only considered transcripts that were assigned to the same class in the three and two independent profiles obtained for AGO1 and decapping activators, respectively. We validated changes in mRNA levels for selected mRNAs by Northern blotting (data not shown).
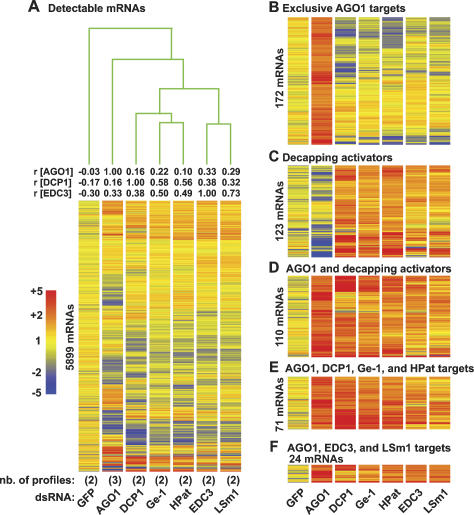
Figure 3.
Expression profiles of D. melanogaster S2 cells depleted of AGO1 or decapping activators. (A) S2 cells were treated with the dsRNAs indicated below the lanes. The number (nb) of independent expression profiles obtained per depleted protein is indicated in brackets. Average expression levels of transcripts detectable in all profiles (5899 mRNAs) are shown. RNAs are represented as lines colored relative to their expression levels, as indicated on the left. Numbers above the lanes indicate rank correlation coefficients relative to AGO1, DCP1, or EDC3 as indicated. The experiment tree was calculated using the distance option in the GeneSpring software (Euclidean distance). (B) RNAs at least 1.5-fold overrepresented in the three independent profiles obtained for AGO1, and <1.5-fold up-regulated in at least nine of 10 profiles of decapping activators. (C) RNAs at least 1.5-fold overrepresented in the two profiles obtained for two or more decapping activators, and <1.5 up-regulated in the AGO1 knockdown. (D) mRNAs at least 1.5-fold overrepresented in the three independent profiles obtained for AGO1, and in the two profiles obtained for at least two decapping activators. (E) RNAs at least 1.5-fold overrepresented in the three independent profiles obtained for AGO1, and in five of six profiles obtained for DCP-1, Ge-1, and HPat. (F) RNAs at least 1.5-fold overrepresented in the three independent profiles obtained for AGO1 and in all profiles obtained for EDC3 and LSm1. Note that the lines representing individual mRNAs are compressed in A. The number of mRNAs displayed per panel is indicated on the left.
In contrast to the profiles of cells depleted of the P-body component GW182, which are highly correlated with those depleted of AGO1 (Behm-Ansmant et al. 2006a), the profiles of cells depleted of decapping activators were not correlated with those depleted of AGO1, although they were correlated with each other (Fig. 3A). The rank correlation coefficients between DCP1 and Ge-1 or DCP1 and HPat profiles are r = 0.58 or r = 0.56, respectively, and between Ge-1 and HPat r = 0.68. These results indicate that DCP1, Ge-1, and HPat regulate common targets (for proteins acting in the same pathway—e.g., AGO1 and Drosha—the rank correlation coefficient is r ≥ 0.7) (Rehwinkel et al. 2006). A subset of transcripts commonly regulated by DCP1, Ge-1, and HPat was also regulated by EDC3 and LSm1 (Fig. 3A,C–F). However, the profiles of cells depleted of EDC3 and LSm1 were more similar to each other (rank correlation coefficient r = 0.73) than to those from cells depleted of DCP1, Ge-1, or HPat (Fig. 3A). Thus, EDC3 and LSm1–7 may form a subclass of decapping activators distinct from DCP1, HPat, and Ge-1. Because transcripts up-regulated in cells depleted of AGO1 are enriched in predicted and validated miRNA targets (Behm-Ansmant et al. 2006a; Rehwinkel et al. 2006), we next focused on these transcripts.
The expression profiles in cells depleted of AGO1 differ from expression profiles in cells depleted of decapping activators. In the three independent AGO1 profiles, 498 transcripts were at least 1.5-fold up-regulated. Of those, 34.5% (172 transcripts) were unchanged or down-regulated in nine of 10 profiles obtained for decapping activators (Fig. 3B; Supplementary Table 1). Conversely, depleting at least two decapping activators, up-regulated 396 mRNAs, of those 31.1% (123 mRNAs) remained unchanged or were down-regulated in the AGO1 profiles (Fig. 3C; Supplementary Table 1).
Nevertheless, decapping activators do regulate a subset of AGO1 targets. Indeed, out of 498 transcripts overrepresented in the AGO1 knockdown, 110 (22.1%) were also overrepresented in knockdowns of at least two decapping activators (Fig. 3D; Supplementary Table 1). The probability that this would happen by chance is P = 1.6E−32. A subset of these (71 mRNAs) was also up-regulated in five of six profiles obtained for DCP1, Ge-1, and HPat (Fig. 3E) (the likelihood to have 71 or more common targets is P = 6E−23). Furthermore, of the 498 mRNAs overrepresented in the AGO1 knockdown, 24 also changed levels concordantly in all profiles obtained for EDC3 and LSm1 (Fig. 3F) (the likelihood to have 24 or more common targets is P = 1.5E−06). Thus, by comparing gene expression profiles in cells depleted of AGO1 or decapping activators, we estimate that 15% of AGO1-targets are also regulated by Ge-1, DCP1, and HPat, whereas ∼5% are dependent on EDC3 and LSm1–7.
Transcripts commonly regulated by AGO1 and decapping activators have longer 3′ UTRs on average (Supplementary Fig. 1) and are significantly enriched for predicted targets of K-Box miRNAs and miR-34 (P ∼ 1E−04–1E−03) (Supplementary Table 2), indicating that these transcripts represent predicted targets of a limited number of miRNAs.
Decapping activators have partially redundant functions in miRNA-mediated mRNA decay
The similarity of decapping activator profiles prompted us to investigate whether, in addition to Ge-1, other decapping activators could relieve silencing of the F-Luc-CG10011 reporter by miR-12 (Fig. 4). In this analysis, we also tested Me31B (the D. melanogaster homolog of human RCK/p54), a protein that interacts with HPat (data not shown) and is part of the decapping complex in human cells (Fenger-Grøn et al. 2005). Depleted cells were transiently cotransfected with plasmids expressing F-Luc-CG10011, miR-12, or the corresponding vector without an insert (empty vector), and the Renilla luciferase (R-Luc) transfection control. In these experiments, we compensated for any differences in transfection efficiencies, by normalizing the firefly luciferase activity and mRNA levels to the Renilla luciferase control. To compensate for nonspecific effects that the depletions might have on mRNA levels, the normalized values of firefly luciferase activity or mRNA levels in the presence of the miRNA were divided by those obtained in the absence of the miRNA for each knockdown.
Depleting single decapping activators (DCP1, EDC3, HPat1, or Me31B, but not LSm1 or LSm3), slightly relieved silencing of F-Luc-CG10011 by miR-12 and led to a 1.5- to 1.8-fold increase in F-Luc activity (data not shown). These effects are below the twofold threshold that we define as significant, and may explain why, apart from Ge-1, these genes were not identified in the screen.
We showed before that codepleting DCP1 with DCP2 suppresses F-Luc-CG10011 mRNA decay by miR-12. Nevertheless, F-Luc activity is only partially restored, most likely because the accumulated transcripts are deadenylated (see change in mobility in Fig. 4B, lanes 21,22; Behm-Ansmant et al. 2006a, b), and consequently, are translated less efficiently. On the basis of these observations, we next tested the effect of codepleting decapping activators in pairwise combinations.
Strikingly, codepletions of two decapping activators (with the exception of LSm1 or LSm3) (data not shown) restored F-Luc CG10011 mRNA levels more than single depletions, with the best results obtained when Ge-1 and DCP1 were codepleted (Fig. 4A,B, lanes 13,14). As a positive control, the previously reported depletion of AGO1 was included; and as expected, this depletion restored both reporter mRNA level and F-Luc activity (Fig. 4A,B; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a, b). The results obtained by codepleting decapping activators indicate that these proteins have partially redundant functions in silencing.
We next tested whether another target of miR-12 similarly depended on decapping activators. In particular, we tested a reporter in which the F-Luc ORF was fused to the 3′ UTR of the D. melanogaster CG3548 mRNA. miR-12 destabilized this reporter significantly (Fig. 4C,D). Depleting Ge-1 alone did not prevent miR-12-mediated decay of F-Luc-CG3548 (Fig. 4C,D), in agreement with the observation that the level of endogenous CG3548 mRNA did not change in cells depleted of Ge-1. However, when Ge-1 was codepleted with Me31B or DCP1, the levels of reporter mRNA were significantly restored (Fig. 4C,D). The accumulated transcripts were deadenylated, as reflected by the increased electrophoretic mobility of the mRNAs in the presence of the miRNA (Fig. 4D, lanes 10,12). This observation may explain why restoring mRNA levels did not correspondingly increase firefly luciferase activity. Alternatively, this reporter may still be silenced at the translational level (see Fig. 8, below). The observations described above, together with previous published results (Behm-Ansmant et al. 2006a, b; Giraldez et al. 2006; Wu et al. 2006), indicate that miRNAs destabilize target mRNAs via deadenylation and subsequent decapping.
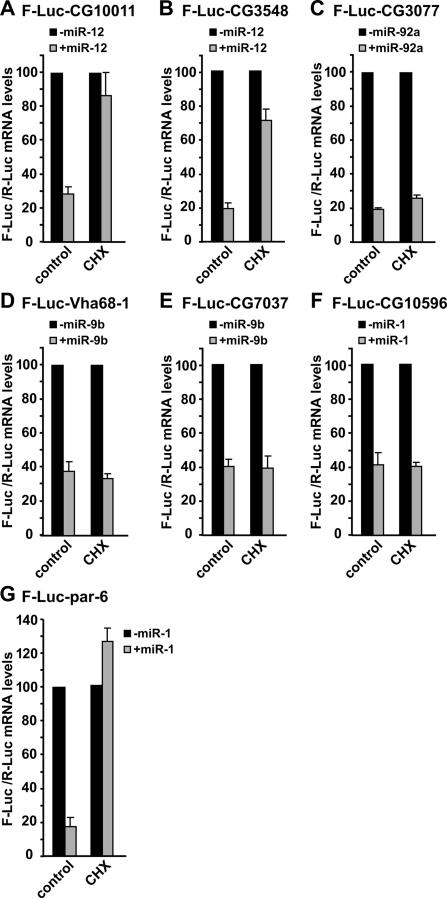
Figure 8.
miRNAs elicit mRNA decay via translation-dependent and translation-independent mechanisms. (A–G) S2 cells were cotransfected with a mixture of plasmids as described in Figure 4. Cells were treated with cycloheximide (CHX) as indicated. Firefly luciferase mRNA levels are normalized to that of the Renilla luciferase. The normalized values of F-Luc mRNA are set to 100 in cells transfected with the empty vector (i.e., in the absence of the miRNA; black bars). Mean values ± standard deviations from three independent experiments are shown.
Depleting decapping activators suppress mRNA decay mediated by several miRNAs
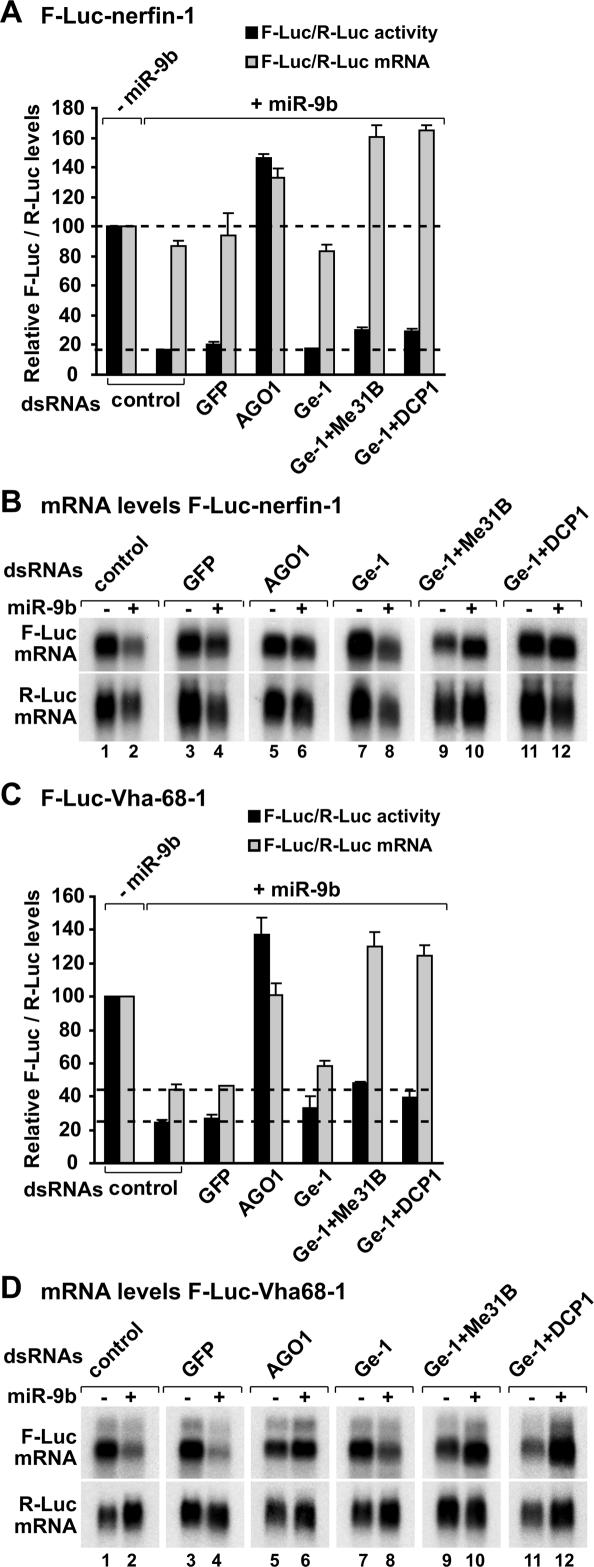
We also tested how depleting decapping activators affects silencing of additional miRNA targets (Figs. 5, 6, 7). In particular, we analyzed targets of miR-9b (nerfin-1, Vha68-1, CG7037), miR-1 (par-6 and CG10596), and miR-92a (CG3077). The nerfin-1 reporter is regulated mainly at the translational level. Indeed, in the presence of miR-9b, F-Luc expression was reduced sixfold, whereas the corresponding mRNA levels were only reduced 1.2-fold (Fig. 5A,B; Behm-Ansmant et al. 2006a, b). Depleting Ge-1 in combination with Me31B or DCP1 did not restore luciferase expression from the F-Luc-nerfin-1 reporter (Fig. 5A,B). Although cells depleted of decapping activators accumulated deadenylated F-Luc-nerfin-1 transcripts (Fig. 5B; see change in the mRNA mobility in lanes 8,10,12; data not shown), it is unlikely that deadenylation causes the translational repression, because inhibiting deadenylation by depleting NOT1 does not suppress silencing of this reporter by miR-9b (Behm-Ansmant et al. 2006a, b).
Figure 5.
Decapping activators suppress silencing of miRNA targets regulated at the mRNA level. (A–D) S2 cells were treated with the indicated dsRNAs on days 0 and 4. On day 6, cells were transfected with a mixture of plasmids as described in Figure 4. Firefly luciferase activity and mRNA levels are analyzed as described in Figure 4. (B,D) Northern blot analysis of representative RNA samples isolated from S2 cells shown in A and C, respectively.
Figure 6.
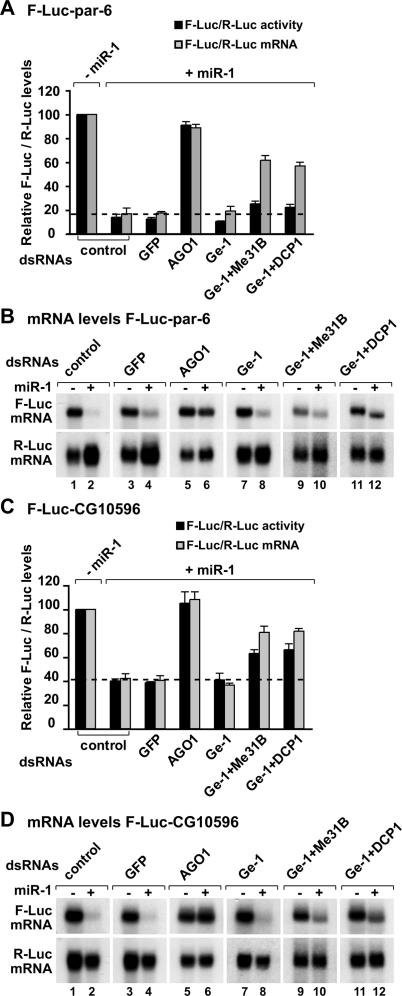
Decapping activators suppress silencing of miR-1 targets. (A–D) S2 cells were treated with the indicated dsRNAs on days 0 and 4. On day 6, cells were transfected with a mixture of plasmids as described in Figure 4. Firefly luciferase activity and mRNA levels are analyzed as described in Figure 4. (B,D) Northern blot analysis of representative RNA samples isolated from S2 cells shown in A and C, respectively.
Figure 7.
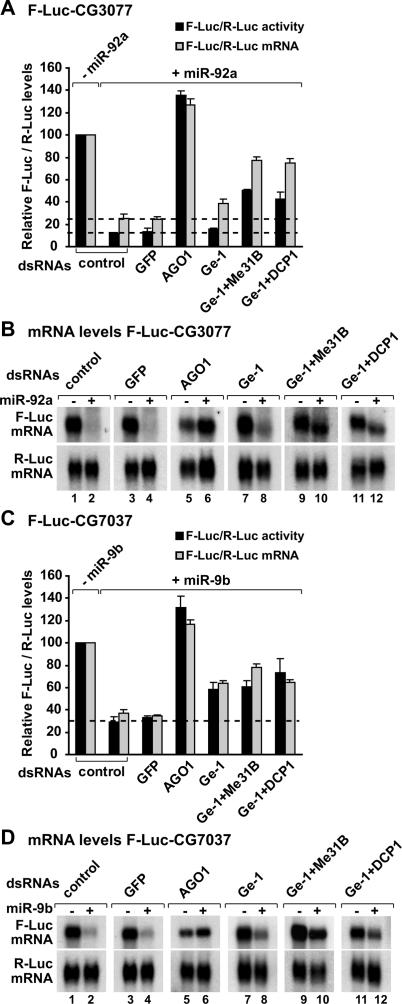
Decapping activators suppress silencing of miRNA targets regulated at the mRNA levels. (A–D) S2 cells were treated with the indicated dsRNAs on days 0 and 4. On day 6, cells were transfected with a mixture of plasmids as described in Figure 4. Samples are analyzed as described in Figure 4. (B,D) Northern blot analysis of representative RNA samples isolated from S2 cells shown in A and C, respectively.
In contrast to the nerfin-1 reporter, Vha68-1 is mainly regulated at the mRNA level (Fig. 5C,D). Targets of miR-1 and miR-92a are also regulated predominantly at the mRNA level (Figs. 6 7A,B). Depleting Ge-1 alone did not restore F-Luc activities or mRNA levels of these reporters (Figs. 5C,D, 6A–D 7A,B), consistent with the observation that the level of endogenous transcripts did not change in cells depleted of Ge-1. However, mRNA levels were restored significantly in cells codepleted of Ge-1 and Me31B or Ge-1 and DCP1 (Figs. 5C,D, 6A–D 7A,B). Again, the accumulated transcripts were deadenylated (see change in the mobility of the mRNAs in the presence of the miRNA) (Figs. 5D, 7B, lanes 10,12).
Finally, CG7037 is a predicted target of miR-9b and is also regulated at the mRNA level. Depleting Ge-1 alone or in combination with other decapping activators led to an approximately twofold increase of F-Luc-CG7037 mRNA levels (Fig. 7C). This is in agreement with the regulation of the corresponding endogenous mRNA in cells depleted of Ge-1 or additional decapping activators (Supplementary Table 1).
In summary, for the reporters regulated mainly at the mRNA level, depleting Ge-1 in combination with Me31B or DCP1 suppresses miRNA-mediated mRNA degradation. However, firefly luciferase expression is not similarly restored. One possible explanation is that the accumulated mRNAs are deadenylated and are therefore translated less efficiently. Another possibility is that the depletions do not suppress silencing at the translational level. Together, these results indicate that miRNAs mediate down-regulation by at least two modes: They can repress protein production without significantly changing mRNA levels (as typified by the nerfin-1 reporter); or increase mRNA degradation via decapping (as observed for the additional reporters analyzed in this study).
miRNAs can destabilize target mRNAs in the absence of active translation
Having established that miRNAs elicit the decapping and subsequent degradation of target mRNAs, it was important to determine whether the decay is linked to the inhibition of protein expression. If mRNA decay by miRNAs were a consequence of inhibiting translation, miRNA targets should be stabilized in cells in which translation is inhibited. We therefore tested the effect of translational inhibitors such as cycloheximide, homoharringtonine, and hippuristanol on the reporters described above in the presence or absence of the cognate miRNAs. The inhibitors tested repress protein synthesis, but whereas cycloheximide and homoharringtonine block translation elongation, hippuristanol is a specific inhibitor of eIF4A and blocks eIF4A-dependent translation initiation (Chan et al. 2004; Bordeleau et al. 2006).
As shown above, for all the reporters tested, the miRNAs triggered significant mRNA decay in control cells (Fig. 8; Supplementary Fig. 2). Strikingly, in cells treated with cycloheximide, homoharringtonine, or hippuristanol, the levels of F-Luc-CG10011, F-Luc-CG3548, and F-Luc-par-6 mRNAs were restored, despite the presence of the cognate miRNAs (Fig. 8A,B,G; Supplementary Fig. 2; data not shown). In contrast, the translational inhibitors did not prevent degradation of F-Luc-CG3077 by miR-92a, of F-Luc-Vha68-1 or F-Luc-CG7037 by miR-9b, and of F-Luc-CG10596 by miR-1 (Fig. 8C–F; Supplementary Fig. 2). The negative effect of the inhibitors on translation was demonstrated by the reduction of F-Luc and R-Luc activities, as well as by the analysis of polysome profiles in treated cells (data not shown). Similar results were obtained following 3 h incubation with cycloheximide (data not shown).
The differential effects of the translational inhibitors on miRNA-mediated mRNA decay are likely to reflect the influence of 3′ UTRs on the final outcome of silencing for the following reasons. First, suppression of mRNA decay by the inhibitors is not miRNA specific, as demonstrated by the observation that miR-1-mediated decay of par-6, but not of CG10596, is suppressed in the absence of active translation (Fig. 8F,G). Secondly, the differential effect of hippuristanol treatment on reporter levels is unlikely to reflect differences in eIF4A dependency, because all the reporters tested have the same 5′ UTR, and hippuristanol inhibits F-Luc expression from these reporters to similar extents in the absence of miRNAs (data not shown). Finally, the half-lives of the reporters shown in Figure 8 are in the same range (i.e., 2–3 h in the absence of the miRNA, and 15–30 min in the presence of the miRNA) (data not shown), excluding the possibility that the effects observed are a consequence of differences in turnover rates.
In summary, the destabilization of CG10011 and CG3548 reporters by miR-12, or of par-6 by miR-1, requires active translation. Thus, it is possible that these reporters are silenced after translation has been initiated as suggested before (Olsen and Ambros 1999; Kim et al. 2004; Nelson et al. 2004; Maroney et al. 2006; Nottrott et al. 2006; Petersen et al. 2006). Nevertheless, the possibility that the suppression of miRNA-mediated mRNA decay is an indirect effect of the general inhibition of protein synthesis cannot be excluded (e.g., a labile factor might be specifically required for the degradation of these mRNAs).
In contrast, miRNA-mediated degradation of CG3077, Vha68-1, CG7037, and CG10596 mRNAs is not affected by the inhibitors of translation. Thus, for these reporters, mRNA degradation either represents an independent mechanism through which miRNAs down-regulate their expression or is a consequence of a primary inhibitory effect exerted by the miRNAs on a translation step upstream of the action of the inhibitors, as suggested by previous studies (Humphreys et al. 2005; Pillai et al. 2005; Kiriakidou et al. 2007; Mathonnet et al. 2007; Thermann and Hentze 2007; Wakiyama et al. 2007). Together, these results are consistent with the hypothesis that there may be more than one mechanism by which miRNAs mediate repression of gene expression (Behm-Ansmant et al. 2006a.b; for review, see Pillai et al. 2006; Jackson and Standart 2007; Nilsen 2007).
Discussion
miRNAs silence gene expression by interfering with protein expression directly and/or by promoting mRNA degradation. mRNA degradation by miRNAs requires the Argonaute proteins, the P-body component GW182, the CAF1:CCR4:NOT deadenylase complex, and the decapping DCP1:DCP2 complex (Jakymiw et al. 2005; Liu et al. 2005b; Meister et al. 2005; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a, b). Moreover, a role for the decapping activator RCK/p54 in miRNA-mediated silencing was reported in human cells (Chu and Rana 2006). All of these proteins colocalize to cytoplasmic foci or mRNA processing bodies (P-bodies) (for review, see Eulalio et al. 2007a). In addition, the Argonaute proteins, miRNAs, and miRNA targets are also detected in P-bodies, which strongly suggests that P-body components participate in miRNA-mediated gene silencing (Jakymiw et al. 2005; Liu et al. 2005a, b; Meister et al. 2005; Pillai et al. 2005; Sen and Blau 2005; Behm-Ansmant et al. 2006a, b; Bhattacharyya et al. 2006; Leung et al. 2006; Pauley et al. 2006).
The results presented here indicate that additional P-body components are required for silencing of endogenous miRNA targets. These include Ge-1, Me31B, HPat, EDC3, and the LSm1–7 complex, all shown to enhance decapping (Parker and Song 2004; Fenger-Grøn et al. 2005; Xu et al. 2006).
The decapping activator Ge-1 is required for silencing a subset of miRNA targets
We set out to identify novel components of the miRNA pathway by screening a dsRNA library targeting nearly all annotated D. melanogaster genes for suppressors of silencing. In addition to known components of the miRNA pathway (i.e., Drosha, Pasha, Dicer-1, and AGO1), the screen identified two P-body components: GW182 and Ge-1 (Fig. 1B). A role for GW182 in the miRNA pathway was reported before (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005b; Meister et al. 2005; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a, b; Chu and Rana 2006; Wakiyama et al. 2007).
AGO1- and GW182-depleted cells exhibit strikingly similar expression profiles, indicating that GW182 functions in the miRNA pathway and is unlikely to have additional roles in general mRNA turnover. Indeed, GW182 regulates nearly all the AGO1-targets that are detectable by microarray (i.e., that change levels in AGO1-depleted cells) (Behm-Ansmant et al. 2006a). In contrast, comparing the gene expression profiles of AGO1- and Ge-1-depleted cells revealed that Ge-1 is required for silencing ∼15% of AGO1-targets (76 of 498 mRNAs) in D. melanogaster (Fig. 3).
Ge-1 belongs to a protein family characterized by N-terminal WD40-repeats, a central S-rich linker, and a conserved C-terminal domain (Fig. 2A). Multiple sequence alignment of all proteins possessing these domains revealed two paralogs in Oryza sativa and A. thaliana (Varicose and Varicose-related, VCS and VCR, respectively), and a single ortholog in animals and fungi, with the exception of S. cerevisiae (Deyholos et al. 2003; Fenger-Grøn et al. 2005; Yu et al. 2005; Xu et al. 2006).
The N-terminal WD40-repeat region of VCS is required for its interaction with DCP1, whereas the C-terminal domain is required for its oligomerization and interaction with DCP2 (Xu et al. 2006). These interactions stimulate the catalytic activity of DCP2 (Fenger-Grøn et al. 2005; Xu et al. 2006). Apart from the interaction with DCP2, the C-terminal domain of Ge-1 is necessary and sufficient to localize the protein to P-bodies (Fig. 2B), which is in turn required for P-body integrity (Yu et al. 2005). Since in human cells, Ge-1 interacts with DCP2, DCP1, EDC3, and RCK/p54 (Fenger-Grøn et al. 2005), this suggests that Ge-1 may act as a molecular scaffold bringing together DCP2 and decapping activators, and possibly nucleating the assembly of P-bodies. Understanding how various Ge-1 domains interact with mRNA decay enzymes and influence P-body integrity awaits the further biochemical characterization of these domains.
Role of decapping activators in the miRNA pathway
The presence of Ge-1 in a multiprotein complex consisting of the decapping enzyme DCP2 and additional decapping activators including DCP1, EDC3, and RCK/p54 in human cells (Fenger-Grøn et al. 2005) prompted us to investigate the role of additional enhancers of decapping in the miRNA pathway. Comparing gene expression profiles in cells depleted of AGO1, DCP1, Ge-1, HPat, EDC3, or LSm1 showed that 15% of the transcripts that are up-regulated in cells depleted of AGO1 are also up-regulated in cells depleted of DCP1, Ge-1, or HPat; whereas in cells depleted of EDC3 or LSm1, only 5% show concordant changes (Fig. 3). The fraction of AGO1 targets that decapping activators also regulated is underestimated when comparing expression profiles of cells depleted of the proteins individually, because these proteins have partially redundant functions in silencing. This is evidenced by the observation that the depletion of decapping activators, with the exception of Ge-1, suppresses silencing (above the twofold threshold that we defined as significant) only when they are codepleted with Ge-1 or additional activators (Fig. 4). This observation also provides an explanation for why these proteins were not identified in this or previous screens.
An important finding from our experiments is that even in cases in which the codepletion of decapping activators restored reporter mRNA levels, luciferase expression was not always restored, because the accumulated transcripts were deadenylated (Figs. 5, 6, 7). Consequently, screens based on protein expression (luciferase or GFP) are very likely to overlook factors required for silencing.
Transcripts up-regulated by both AGO1 and decapping activators are enriched in predicted and validated targets of a subset of miRNAs (Supplementary Table 2). This enrichment is relevant because transcripts exclusively regulated by decapping activators but not by AGO1 are not significantly enriched in miRNA-binding sites. A feature that distinguishes transcripts regulated by both AGO1 and decapping activators from transcripts exclusively regulated by AGO1 is that the former have longer 3′ UTRs on average (721 nucleotides [nt] vs. 629 nt) (Supplementary Fig. 1). Nevertheless, the specific features of miRNA targets that lead to the dependence of decapping activators for their silencing are unknown.
Mechanisms of miRNA-mediated gene silencing
Our screen was performed with a reporter that is mainly regulated at the mRNA level. Analyzing of miRNA activity in cells treated with cycloheximide showed that degradation of this reporter by miR-12 requires ongoing translation (Fig. 8A). Therefore, one might expect to identify translation factors in the screen. However, there are many reasons why these factors may escape detection, including inefficient depletions or a general inhibition of translation affecting firefly and/or Renilla luciferase expression in such a way that the suppression of silencing is masked by nonspecific effects.
In particular, eIF6 did not score positively in the screen (Fig. 1B; Table 1). We tested whether two consecutive eIF6 knockdowns (instead of a single depletion as in the screen) and prolonged depletion times (9 d instead of 7 d) restored F-Luc-CG10011 expression. Under these conditions (i.e., 90% of the endogenous protein was depleted), we observed a partial suppression of silencing (an approximate twofold to 2.5-fold increase in firefly luciferase expression) for F-Luc-CG10011 and several other reporters (Supplementary Fig. 3). Nevertheless, further studies are needed to elucidate the role of eIF6 in the miRNA pathway in D. melanogaster.
The inhibition of translation by miRNAs is not always followed by mRNA decay. Conversely, we show that some targets are degraded in the absence of active translation in D. melanogaster cells (Fig. 8C–F). Similarly, in human cells and zebrafish embryos, it was reported that miRNA targets that cannot be translated (e.g., because of the presence of a strong stem–loop in the 5′ UTR) are nevertheless subject to deadenylation and subsequent decay (Mishima et al. 2006; Wu et al. 2006).
Our results, together with previous studies, indicate that miRNAs use at least three mechanisms to down-regulate their targets: (1) inhibition of protein production without a significant change in mRNA level (which can be achieved in different ways; see introduction), (2) translation-dependent mRNA decay, or (3) translation-independent mRNA decay. Because at least two modes of regulation have been observed for targets of the same miRNA (i.e., miR-9b and miR-1), it is unlikely that these different modes are specified by the miRNA itself. Moreover, because the extent of miRNA regulation occurs over a wide range of magnitudes and differs for different miRNA:target pairs, we favor a model whereby the specific features and/or composition of the mRNP itself influence the outcome of miRNA regulation. This may explain the conflicting reports regarding the mechanisms of silencing by miRNAs. A major challenge for future studies will be to elucidate how mRNP features and/or composition influence miRNA regulation.
Materials and methods
Luciferase reporter assays
Luciferase reporters were described before (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a, b). Additional wild-type 3′ UTRs of predicted miRNA targets were amplified by PCR from a D. melanogaster S2 cell cDNA library and cloned downstream from the firefly luciferase coding region, between the EcoRI and SalI sites of plasmid pAc5.1A-F-Luc (Rehwinkel et al. 2005). All 3′ UTRs contained the natural 3′ polyadenylation signals. Renilla luciferase cloned between the EcoRI and XhoI sites of vector pAc5.1A (Invitrogen) served as a transfection control. For expression of miRNAs, a genomic fragment of ∼400 nt encompassing the miRNA gene was amplified from genomic DNA and cloned in vector pAc5.1A downstream from the actin 5C promoter. When transfections were performed in six-well plates, the transfection mixtures contained 0.1 μg of firefly luciferase reporter plasmid, 0.4 μg of the Renilla transfection control, and 1 μg of plasmids expressing miRNA primary transcripts. Cells were collected 3 d after transfection. All transfections were performed with Effectene (Qiagen). Luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega). Cycloheximide (10 μg/mL; Sigma), homoharringtonine (5.5 μg/mL; Sigma), and hippuristanol (4 μM) were added to the cells 3 or 7 h before isolating the RNA samples.
RNAi screens
RNAi screens were performed as described before (Boutros et al. 2004). Complete primer and amplicon sequences can be found at http://rnai.dkfz.de. The screen was performed in duplicate using 384-well plates. D. melanogaster Schneider cells (S2 cells) were added to the dsRNAs on day 0. On day 4, cells were transfected with the F-Luc-CG10011 reporter, a plasmid encoding the primary miR-12 transcript, and a plasmid constitutively expressing Renilla luciferase (R-Luc) that served as a transfection control. Firefly and Renilla luciferase activities were measured on day 7. Firefly luciferase activity was normalized to that of the Renilla luciferase and quantified by a significance factor, or z-score. z-Scores were calculated plate-by-plate and represent the number of median absolute deviations an individual measurement differs from the median. An average z-score of the two measurements for each dsRNA was calculated, and an arbitrary cutoff of z > 10 was set.
RNAi, Northern and Western blotting, and immunofluorescence
RNAi, Northern blotting, and RT–PCR were performed as described before (Rehwinkel et al. 2005). For Western blots, the polyclonal antibodies were diluted 1:1000. Bound primary antibodies were detected with alkaline-phosphatase-coupled secondary antibodies (Western-Star kit from Tropix). Immunofluorescence stainings using monoclonal anti-HA antibodies (Covance) and affinity-purified anti-Tral antibodies were performed essentially as described by Eulalio et al. (2007b).
RNA isolation and genome-wide expression analysis
High-density oligonucleotide microarrays (Affymetrix D. melanogaster array 2) covering >18,500 transcripts from D. melanogaster were used. The microarray data have been submitted to the ArrayExpress database at EBI under accession number E-MEXP-1012.
Total RNA was isolated using Trifast Reagent (PeqLab). To reduce potential variations in RNA preparation, two RNA preparations were isolated from a single knockdown experiment. These preparations were pooled with the equivalent preparations isolated from an independent knockdown, to minimize differences in knockdown efficiencies. These pools of four RNA preparations from two independent knockdowns are referred to as RNA samples.
Biotinylated targets were prepared from 5 μg of total RNA following standard Affymetrix procedures. These include an oligo-dT-primed amplification step during sample preparation. This step introduces a bias in samples isolated from DCP2- or Me31B-depleted cells because a significant fraction of up-regulated transcripts in these cells is deadenylated (as expected for transcripts degraded via a deadenylation-dependent decapping mechanism). Indeed, the hybridization signals of the poly(A) control transcripts (Affymetrix GeneChip Eukaryotic Poly-A RNA Control Kit) spiked in samples isolated from DCP2- or Me31B-depleted cells were stronger compared with control samples. This indicates that total mRNA was either less abundant or deadenylated in samples isolated from these knockdowns. Thus, these samples could not be analyzed. This was not observed in cells depleted of additional decapping activators, although we cannot rule out that some transcripts are deadenylated.
Standard Affymetrix protocols were used for hybridization, washing, and data acquisition (Fluidics station 400, GeneArray 2500 scanner, Affymetrix Microarray suite version 5.1). Control parameters were within recommended limits. Data were imported into GeneSpring 6 (Silicon Genetics) (mock-treated cells = control channel, knockdown experiment = signal channel). All experiments were normalized using an intensity-dependent normalization scheme (Lowess). The enrichment of miRNA targets among regulated genes was assessed by the probability (P) that an equally high or higher enrichment could be obtained by chance, given the frequency of the targets among detectable genes as described by Stark et al. (2005) and Rehwinkel et al. (2006).
Acknowledgments
We are grateful to members of the EMBL Gene Core Facility for performing the array hybridizations, S.M. Cohen for miRNA expression vectors and the nerfin-1 reporter, J. Pelletier for his generous gift of hippuristanol, and D.J. Thomas for comments on the manuscript. This study was supported by the Max Planck Society, the European Molecular Biology Organization (EMBO), the Human Frontier Science Program Organization (HFSPO), and by grants from the Deutsche Forschungsgemeinschaft (to M.B. and E.I.). A.E. and E.H. are recipients of fellowships from the Portuguese Foundation for Science and Technology and EMBO, respectively.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.443107
References
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E., Doerks T., Stark A., Bork P., Izaurralde E., Stark A., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Izaurralde E., Rehwinkel J., Izaurralde E., Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W., Habermacher R., Martine U., Closs E.I., Filipowicz W., Martine U., Closs E.I., Filipowicz W., Closs E.I., Filipowicz W., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Belsham G.J., Wagner G., Tanaka J., Pelletier J., Wagner G., Tanaka J., Pelletier J., Tanaka J., Pelletier J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Boutros M., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N., Hild M., Koch B., Haas S.A., Paro R., Perrimon N., Koch B., Haas S.A., Paro R., Perrimon N., Haas S.A., Paro R., Perrimon N., Paro R., Perrimon N., Perrimon N., Heidelberg Fly Array Consortium Genome-wide RNAi analysis of growth and viability in D. melanogastercells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Ceci M., Gaviraghi C., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S., Gaviraghi C., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S., Offenhauser N., Marchisio P.C., Biffo S., Marchisio P.C., Biffo S., Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chan J., Khan S.N., Harvey I., Merrick W., Pelletier J., Khan S.N., Harvey I., Merrick W., Pelletier J., Harvey I., Merrick W., Pelletier J., Merrick W., Pelletier J., Pelletier J. Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA. 2004;10:528–543. doi: 10.1261/rna.5200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R., Pasquinelli A.E., Shiekhattar R., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Chu C.Y., Rana T.M., Rana T.M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos M.K., Cavaness G.F., Hall B., King E., Punwani J., Van Norman J., Sieburth L.E., Cavaness G.F., Hall B., King E., Punwani J., Van Norman J., Sieburth L.E., Hall B., King E., Punwani J., Van Norman J., Sieburth L.E., King E., Punwani J., Van Norman J., Sieburth L.E., Punwani J., Van Norman J., Sieburth L.E., Van Norman J., Sieburth L.E., Sieburth L.E. VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130:6577–6588. doi: 10.1242/dev.00909. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., Spencer A., Morita K., Han M., Morita K., Han M., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E., Behm-Ansmant I., Izaurralde E., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E., Behm-Ansmant I., Schweizer D., Izaurralde E., Schweizer D., Izaurralde E., Izaurralde E. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol. Cell. Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J., Fillman C., Norrild B., Lykke-Andersen J., Norrild B., Lykke-Andersen J., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Förstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Klattenhoff C., Theurkauf W.E., Zamore P.D., Theurkauf W.E., Zamore P.D., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesellchen V., Kuttenkeuler D., Steckel M., Pelte N., Boutros M., Kuttenkeuler D., Steckel M., Pelte N., Boutros M., Steckel M., Pelte N., Boutros M., Pelte N., Boutros M., Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T., Westman B.J., Martin D.I., Preiss T., Martin D.I., Preiss T., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Standart N., Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Hamel J.C., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kim J., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G., Grad Y., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G., Hayes G.D., Kosik K.S., Church G.M., Ruvkun G., Kosik K.S., Church G.M., Ruvkun G., Church G.M., Ruvkun G., Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z., De Planell-Saguer M., Nelson P.T., Mourelatos Z., Nelson P.T., Mourelatos Z., Mourelatos Z. An mRNA m(7)G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A., Calabrese J.M., Sharp P.A., Sharp P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Roberts R., Roberts R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J., Yates J.R., Parker R., Hannon G.J., Parker R., Hannon G.J., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R., Valencia-Sanchez M.A., Hannon G.J., Parker R., Hannon G.J., Parker R., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J.E., Dahlberg J.E. Substrate selectivity of Exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- Maroney P.A., Yu Y., Fisher J., Nilsen T.W., Yu Y., Fisher J., Nilsen T.W., Fisher J., Nilsen T.W., Nilsen T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Merrick W.C., Darzynkiewicz E., Pillai R.S., Darzynkiewicz E., Pillai R.S., Pillai R.S., et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007 doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T., Urlaub H., Luhrmann R., Tuschl T., Luhrmann R., Tuschl T., Tuschl T. Identification of novel Argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A.J., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K., Giraldez A.J., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K., Fujiwara T., Sakamoto H., Schier A.F., Inoue K., Sakamoto H., Schier A.F., Inoue K., Schier A.F., Inoue K., Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P.T., Hatzigeorgiou A.G., Mourelatos Z., Hatzigeorgiou A.G., Mourelatos Z., Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D., Simard M.J., Richter J.D., Richter J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pauley K.M., Eystathioy T., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Eystathioy T., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Hamel J.C., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A., Bordeleau M.E., Pelletier J., Sharp P.A., Pelletier J., Sharp P.A., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Basyuk E., Bertrand E., Filipowicz W., Bertrand E., Filipowicz W., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Filipowicz W., Bhattacharyya S.N., Filipowicz W., Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2006;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E., Behm-Ansmant I., Gatfield D., Izaurralde E., Gatfield D., Izaurralde E., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Natalin P., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Stark A., Brennecke J., Cohen S.M., Izaurralde E., Brennecke J., Cohen S.M., Izaurralde E., Cohen S.M., Izaurralde E., Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Ishizuka A., Siomi H., Siomi M.C., Ishizuka A., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Thermann R., Hentze M.W., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Takimoto K., Ohara O., Yokoyama S., Takimoto K., Ohara O., Yokoyama S., Ohara O., Yokoyama S., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C.J., Wilusz J., Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G., Fan J., Belasco J.G., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J.Y., Niu Q.W., Chua N.H., Yang J.Y., Niu Q.W., Chua N.H., Niu Q.W., Chua N.H., Chua N.H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Yang W.H., Gulick T., Bloch K.D., Bloch D.B., Yang W.H., Gulick T., Bloch K.D., Bloch D.B., Gulick T., Bloch K.D., Bloch D.B., Bloch K.D., Bloch D.B., Bloch D.B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]