Abstract
One activity that controls mRNA translation in vertebrate oocytes, embryos, and neurons is cytoplasmic polyadenylation. In Xenopus oocytes, where much of the biochemistry of this process has been elucidated, nuclear pre-mRNAs containing a cytoplasmic polyadenylation element (CPE) in their 3′ untranslated regions (UTRs) have long poly(A) tails; once the RNAs are spliced and transported to the cytoplasm, the tails are shortened. Following the resumption of meiosis, the poly(A) tails are lengthened and translation ensues. CPEB is a sequence-specific RNA-binding protein that coordinates these events and does so by binding to the CPE as well as several factors including Gld2, a poly(A) polymerase, and PARN [poly(A)-specific ribonuclease], a deadenylase. Here, we show that ePAB, embryonic poly(A)-binding protein, transiently associates with the polyadenylation complex; it initially interacts with CPEB, but after polyadenylation, it binds the poly(A) tail. ePAB dissociation from CPEB is regulated by RINGO (Rapid Inducer of G2/M progression in Oocytes), a cyclin B1-like cofactor that activates cdk1, a protein kinase that phosphorylates CPEB. Subsequent ePAB binding to the poly(A) tail is necessary to protect the homopolymer from degradation by deadenylating enzymes. Poly(A)-bound ePAB also interacts with eIF4G, which instigates translation initiation of CPEB-bound mRNAs.
Keywords: RINGO, CPEB, cytoplasmic polyadenylation, poly(A), translation, ePAB
Cytoplasmic polyadenylation is one process that controls the translation of several key mRNAs in vertebrate germ cells, embryos, and neurons (Klann and Richter 2007; Richter 2007; Thompson et al. 2007). Much of the biochemistry of polyadenylation-induced translation has been elucidated in Xenopus oocytes: Here, several RNAs containing a cytoplasmic polyadenylation element (CPE) and the hexanucleotide AAUAAA have short poly(A) tails; in response to progesterone stimulation of a surface-associated receptor, the tails are elongated and translation ensues. Cytoplasmic polyadenylation and translation during this oocyte maturation period are controlled by several factors including CPEB, an RNA-binding protein whose recognition of the CPE dictates which mRNAs undergo this 3′ end processing. While CPE-lacking and CPE-containing pre-RNAs both acquire long poly(A) tails in the nucleus, following splicing and export to the cytoplasm, only the CPE-containing RNAs undergo deadenylation and translational silencing. The deadenylation is controlled by two CPEB-associated factors: Gld2 (germline development 2) and PARN [poly(A)-specific ribonuclease]. Gld2 is a poly(A) polymerase (Wang et al. 2002; Kwak et al. 2004) and PARN is a deadenylase (Korner et al. 1998; Copeland and Wormington 2001); in oocytes, both of these proteins are active, but because PARN activity is particularly robust, the poly(A) tails are shortened and maintained in that manner (Kim and Richter 2006). Progesterone-triggered signaling activates the kinase Aurora A, which phosphorylates CPEB Ser 174 (Mendez et al. 2000) (note that mitogen-activated protein [MAP] kinase has also been reported to phosphorylate CPEB [Keady et al. 2007]). This event causes the expulsion of PARN from the RNP complex, thereby allowing Gld2 to catalyze poly(A) addition by default (Kim and Richter 2006). Two additional factors required for polyadenylation are symplekin (Barnard et al. 2004), which may act as a scaffold for RNP assembly (Takagaki and Manley 2000), and cleavage and polyadenylation specificity factor (CPSF), a group of proteins that bind the hexanucleotide AAUAAA (Dickson et al. 1999). Translation is controlled most proximally by maskin, which interacts with both CPEB and the cap-binding factor eIF4E. The maskin–eIF4E association precludes the binding of eIF4E with eIF4G, which is required for cap-dependent initiation. Following polyadenylation, maskin dissociates from eIF4E, thereby allowing eIF4G to bind eIF4E and recruit the 40S ribosomal subunit to the 5′ end of the mRNA (Richter and Sonenberg 2005; Cao et al. 2006).
In addition to Aurora A phosphorylation of CPEB, two other upstream events are necessary for polyadenylation. First, RINGO (Rapid Inducer of G2/M progression in Oocytes), a cyclin B1-like factor that activates the kinase cdk1, must be synthesized (Ferby et al. 1999). While oocytes have little RINGO protein, they do contain dormant RINGO mRNA that is translated soon after the oocytes are stimulated by progesterone; this translational control event is mediated by Pumilio-2, a sequence-specific RNA-binding protein (Padmanabhan and Richter 2006). A second essential upstream event involves the activation of Aurora A; the control of this kinase in various cell types is complex (Marumoto et al. 2005), but in oocytes, it is at least partly regulated by phosphorylation catalyzed glycogen synthase kinase 3 (GSK-3) (Sarkissian et al. 2004). Finally, CPEB undergoes additional phosphorylation events subsequent to that catalyzed by Aurora A; these “late-round” phosphorylations are catalyzed by cdk1 and cause partial destruction of CPEB at the very end of meiotic maturation (Mendez et al. 2002).
In the cytoplasm, following CPEB stimulation by Aurora A, the number of adenosine residues that are polymerized on the mRNA 3′ end is tightly regulated; poly(A) tails rarely exceed ∼200 bases. However, when the cytoplasmic polyadenylation complex is first immunoselected by symplekin coimmunoprecipitation (co-IP), polyadenylation surpasses 1000 bases (Barnard et al. 2004). These data suggest that a factor(s) that regulates poly(A) tail length is lost during the RNP selection. We have sought to determine how poly(A) length is regulated and the reason for this regulation. We demonstrate that ePAB [embryonic poly(A)-binding protein] (Voeltz et al. 2001) is initially tethered to CPEB in oocytes, but dissociates from this factor during maturation and binds newly elongated poly(A) tails. The stimulus for the CPEB–ePAB dissociation is RINGO activation of the kinase cdk1 (Ferby et al. 1999; Padmanabhan and Richter 2006), which phosphorylates CPEB on six residues (Mendez et al. 2002). The ePAB liberated by CPEB phosphorylation then binds the newly elongated poly(A) tail, where it not only protects the homopolymer from nuclease attack, but also binds eIF4G to help promote translation initiation.
Results
Transient association of ePAB with the cytoplasmic polyadenylation complex
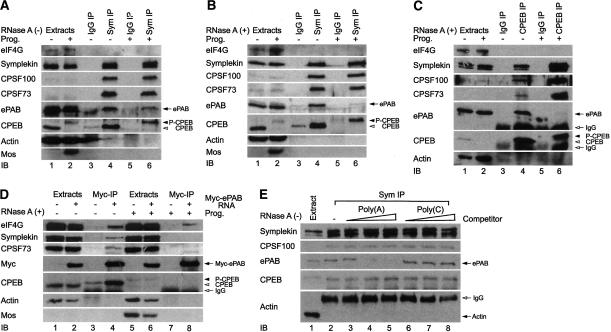
One protein we thought might be involved in maintaining poly(A) tail length is poly(A)-binding protein (PABP). However, oocytes contain very little PABP, although they do have a related protein, ePAB (Voeltz et al. 2001). To examine the relationship between ePAB and cytoplasmic polyadenylation, symplekin was used to coimmunoprecipitate the cytoplasmic polyadenylation complex from oocytes (Kim and Richter 2006). Symplekin is particularly useful for these co-IP experiments because it is relatively abundant, an antibody directed against it is strongly precipitating, and the antibody is mouse monoclonal, thereby facilitating immunoblotting of the proteins collected in this manner with rabbit polyclonal antibodies. In the absence or presence of progesterone (but in the presence of RNase inhibitors), CPSF100, CPSF73, CPEB, and ePAB, but not eIF4G, were all coprecipitated; actin, a very abundant protein, served as the negative control and was not precipitated. Mos also served as a control; it is synthesized only upon the induction of maturation and thus indicates that progesterone effectively stimulated the oocytes. As expected, mos was not coprecipitated with symplekin from progesterone-treated oocytes (Fig. 1A). When the same experiment was performed with extracts treated with RNase A, CPSF, and CPEB, but again not eIF4G, were coprecipitated with symplekin from untreated or progesterone-treated oocytes; however, ePAB was coprecipitated only from untreated oocytes (Fig. 1B). A similar result was obtained when CPEB was used for the IP in the presence of RNase A; ePAB was coprecipitated with CPEB from oocytes, but not those treated with progesterone (Fig. 1C). Finally, oocytes were injected with mRNA encoding myc-ePAB; symplekin, CPSF, and CPEB were coprecipitated with ePAB from oocyte extracts treated with RNAse A, but not from mature oocyte extracts also treated with RNase A. However, eIF4G was coimmunoprecipitated with ePAB before and after progesterone treatment, probably indicating that ePAB is present on the poly(A) tails of many non-CPE-containing mRNAs when it interacts with eIF4G (Fig. 1D). Thus, ePAB association with the polyadenylation complex changes from one that is RNase insensitive in oocytes to one that is RNase sensitive following maturation.
Figure 1.
ePAB interacts with the cytoplasmic polyadenylation machinery. Extracts from oocytes, some of which were incubated with progesterone to induce maturation, were immunoprecipitated (IP) with symplekin antibody or control mouse IgG in the presence of RNase inhibitor (100 U/mL) (A) or RNase A (100 μg/mL) (B). Immunoblots of the precipitates were probed for eIF4G, symplekin, CPSF100, CPSF73, ePAB, CPEB, and, as negative controls, actin and mos. (P-CPEB) Phospho-CPEB. (C) Extracts from untreated or progesterone-treated oocytes were subjected to IP in the presence of RNase A with CPEB antibody or control rabbit IgG, which was followed by immunoblotting. (D) mRNA encoding myc-ePAB was injected into oocytes; following 12 h incubation, some of them were incubated with progesterone to induce maturation, and the oocytes were lysed and subjected to IP with myc-specific antibody (9E10). The coprecipitating proteins were analyzed on immunoblots. (E) Extracts from eggs (no RNase A) were subjected to a symplekin IP in the absence or presence of increasing amounts (0.1, 0.5, and 2.5 μg) of poly(A) or poly(C). The coprecipitating proteins were analyzed on immunoblots.
To determine whether ePAB dissociates from direct contact with the polyadenylation complex to an interaction with elongated poly(A) tails, a symplekin co-IP experiment was performed using egg extracts (no RNase A) supplemented with poly(A) or, as a control, poly(C). Figure 1E shows that ePAB was coprecipitated with symplekin only in the absence of competitor poly(A), indicating that ePAB binds newly elongated poly(A); this conclusion is corroborated by in vitro binding analysis demonstrating that ePAB has a strong avidity for poly(A) (Supplementary Fig. 1).
RINGO/cdk1-mediated phosphorylation of CPEB controls ePAB association with the polyadenylation complex
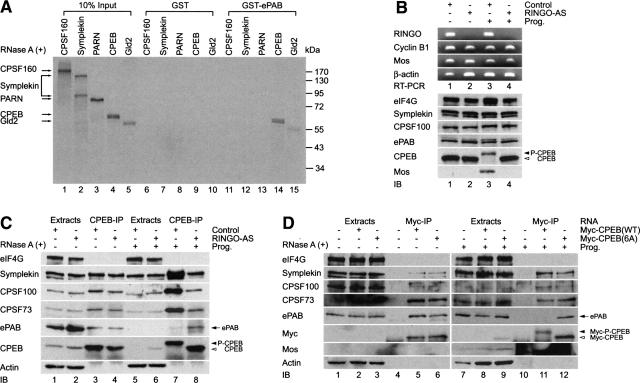
The cytoplasmic polyadenylation complex is (minimally) composed of CPEB, symplekin, CPSF, PARN, and Gld2. To determine which of these proteins directly contacts ePAB, GST–ePAB beads were mixed with each of them following their metabolic labeling with [35S]methionine in rabbit reticulocyte lysates. Figure 2A shows that CPEB, and to a lesser extent Gld2, interacted with ePAB. Because CPEB becomes phosphorylated on Ser 174 during maturation and this event is necessary for the expulsion of PARN from the polyadenylation complex (Kim and Richter 2006), we surmised that CPEB S174 phosphorylation would also expel ePAB from the complex; however, S174 phosphorylation had no effect on the interaction between CPEB and ePAB (data not shown).
Figure 2.
CPEB phosphorylation abrogates binding of ePAB to the cytoplasmic polyadenylation machinery. (A) mRNAs encoding CPSF160, symplekin, PARN, CPEB, and Gld2 were translated in a rabbit reticulocyte lysate in the presence of [35S]methionine and applied to glutathione columns containing GST or GST-ePAB. The columns were washed and the bound material was analyzed by SDS-10% PAGE. (B) RINGO is required for ePAB dissociation from the cytoplasmic polyadenylation machinery. Oocytes were injected with AS ODN against RINGO mRNA or a control-nonspecific ODN and incubated for 5 h to overnight. Following a further incubation for 9 h in the absence or presence of progesterone activation, total RNA was extracted and the efficiency of destruction of RINGO mRNA by RINGO-AS ODN and control ODN was assessed by RT–PCR (RT–PCR panel). Cyclin B1, mos, and β-actin were amplified as negative controls. (C) Extracts from 75 oocytes were immunoprecipitated with CPEB antibody and the coprecipitating proteins were analyzed on immunoblots. (D) Six cdk1 phosphorylation sites in CPEB that may be important for the dissociation of ePAB from the cytoplasmic polyadenylation machinery were examined. mRNAs encoding myc-CPEB(WT) or myc-CPEB(6A) were injected into oocytes; 12 h later, the oocytes were incubated further in the absence (lanes 1–6) or presence (lanes 7–12) of progesterone. Seventy-five oocytes were collected and subjected to the IP with myc-specific antibody. The coprecipitating proteins were analyzed on immunoblots.
In addition to S174, CPEB undergoes six additional phosphorylation events (on serine residues 138, 144, 184, 210, 248, and 423) that are important for its partial destruction late in maturation (Mendez et al. 2002). These events are catalyzed by cdk1, which presumably is activated by its cofactor cyclin B1. cdk1 can also be activated by RINGO, a cyclin B1-like cofactor (Ferby et al. 1999). Oocytes contain little RINGO protein, but do contain dormant RINGO mRNA that is activated soon after the initiation of maturation. RINGO mRNA translation is necessary for CPEB phosphorylation, cytoplasmic polyadenylation, and oocyte maturation (Padmanabhan and Richter 2006). These observations suggest that RINGO/cdk1 might phosphorylate at least some CPEB early during maturation, and that these modifications could potentially mediate CPEB association with ePAB. To investigate this possibility, we injected oocytes with an oligodeoxynucleotide (ODN) complementary to RINGO mRNA (RINGO-AS) or, as a control, an ODN with an irrelevant sequence. The RINGO-AS ODN induced the destruction of RINGO mRNA as determined by RT–PCR but had no effect on three other mRNAs (Fig. 2B, RT–PCR panel). The RINGO-AS ODN also prevented the hyperphosphorylation of CPEB as well as mos synthesis (Fig. 2B, IB panel; note that partial destruction of CPEB is caused by its hyperphosphorylation; cf. lanes 3 and 4). In untreated oocytes, the RINGO-AS ODN had no effect on CPEB co-IP of ePAB or other proteins, while in progesterone-treated oocytes, the RINGO-AS ODN prevented the dissociation of ePAB from CPEB (Fig. 2C). These data suggest that RINGO mRNA, and by extension RINGO activation of cdk1, is necessary for ePAB dissociation from CPEB.
To investigate whether an early RINGO/cdk1-catalyzed phosphorylation of CPEB could mediate ePAB binding, mRNA encoding myc-tagged wild-type (WT) or a mutant CPEB with alanine substitutions for the phospho-serines (6A) was injected into oocytes followed by myc co-IP; symplekin, CPSF, and ePAB, but not eIF4G, were coprecipitated with both wild-type and 6A CPEB proteins (Fig. 2D, left). While symplekin and CPSF were coprecipitated with both wild-type and 6A CPEB after progesterone treatment, ePAB was not precipitated with wild-type CPEB (Fig. 2D, right). Thus, ePAB association with CPEB is mediated by the six cdk1-catalyzed phosphorylated residues of CPEB.
ePAB promotes long poly(A) tails
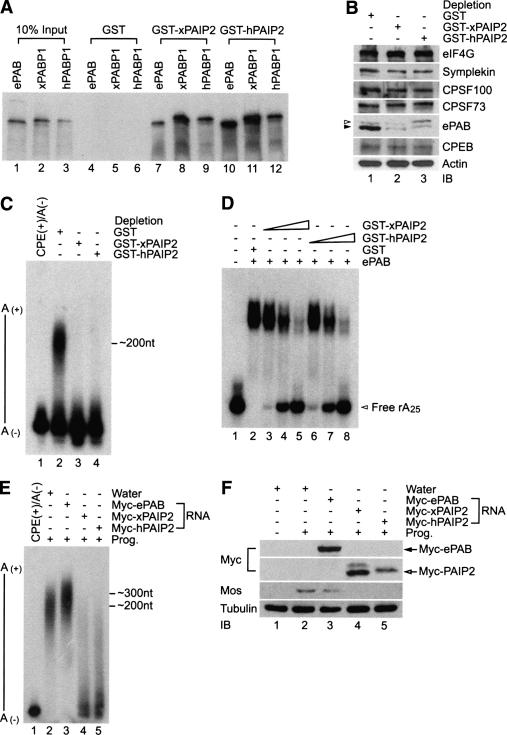
To next examine the influence of ePAB on cytoplasmic polyadenylation, we attempted to immunodeplete this protein from egg extracts; however, the antibody was not adequate for this purpose (data not shown). Consequently, we used an alternative approach first noted by Svitkin and Sonenberg (2004), who used poly(A)-binding protein-interacting protein, PAIP, to remove PABP from the poly(A) tail (Khaleghpour et al. 2001). In vitro, both Xenopus and human PAIP2 have a strong affinity for Xenopus and human PABP as well as ePAB (Fig. 3A). We next determined whether Xenopus and human PAIP2 could deplete ePAB from an egg extract. Figure 3B demonstrates that while PAIP2 did not detectably deplete eIF4G, symplekin, CPSF100, CPSF73, or CPEB, it efficiently depleted ePAB. To examine the effect of this ePAB depletion on polyadenylation, the extracts were primed with CPE-containing RNA. Figure 3C shows that while the mock GST depletion had no effect on poly(A) tail growth, ePAB depletion appeared to completely disrupt this process. As expected, PAIP2 inhibited ePAB from binding poly(A) in vitro (Fig. 3D). In addition, ePAB mRNA injection into oocytes resulted in a hyperextended poly(A) tail (by an average of ∼100 nucleotides [nt]); injection of mRNA Xenopus or human PAIP2 inhibited poly(A) tail extension (Fig. 3E). The injection of PAIP2 mRNA into oocytes also inhibited mos synthesis, whose encoding mRNA requires cytoplasmic polyadenylation to be translated (Fig. 3F; Sheets et al. 1994). These data show that ePAB controls poly(A) tail length and resulting translational activation.
Figure 3.
ePAB is required for cytoplasmic polyadenylation. (A) mRNAs encoding ePAB, Xenopus PABP (xPABP1), and human PABP (hPABP1) were translated in a rabbit reticulocyte lysate in the presence of [35S]methionine and applied to glutathione columns containing GST and GST-PAIP2. The columns were washed and the bound material was analyzed by SDS-10% PAGE. (B) Depletion of ePAB with GST-PAIP2 in the egg extracts. Egg extracts were subjected to depletion of ePAB with GST-PAIP2 (Xenopus and human) or control GST columns. Immunoblots of depleted extracts were probed for eIF4G, symplekin, CPSF100, CPSF73, ePAB, CPEB, and actin. The closed arrowhead represents ePAB; the open arrowhead refers to a nonspecific band. (C) ePAB-depleted egg extracts were primed with 32P-labeled cyclin B1 RNA. The RNA was isolated and resolved on a 5% denaturing gel. (D) Binding of ePAB to A25 RNA in the presence of GST-PAIP2 was analyzed by EMSA. 6xhistidine-tagged recombinant ePAB (70 kDa, 0.35 μg; 5 pmol) was incubated in a reaction buffer for 10 min at 23°C in the presence of increasing concentrations (0.25, 0.5, and 1.0 μg; 5, 10, and 20 pmol, respectively) of GST-PAIP2 (50 kDa) or 0.5 μg (20 pmol) of GST (25 kDa) and further incubated with 25,000 counts per minute (cpm) (0.04 μg; ∼5 pmol) of 32P end-labeled rA25 for 10 min at 30°C. The reaction mixtures (20 μL final) containing RNA–protein complexes were applied to an 8% nondenaturing PAGE, and samples were analyzed by autoradiography. (E) Oocytes were injected with mRNA encoding myc-ePAB or myc-PAIP2; after 12 h incubation, 32P-labeled cyclin B1 RNA was injected and oocytes were incubated with progesterone to induce maturation. RNA from the oocytes was extracted and resolved by 5% denaturing PAGE. Note that myc-PAIP2-injected oocytes did not show any germinal vesicle breakdown (GVBD); therefore, these oocytes were harvested at the time point of GVBD of control water-injected and myc-ePAB mRNA. (F) Expression levels of myc-ePAB and PAIP2 were analyzed with anti-myc antibody. The expression level of mos and tubulin were also assessed by immunoblots.
ePAB prevents poly(A) tail destruction
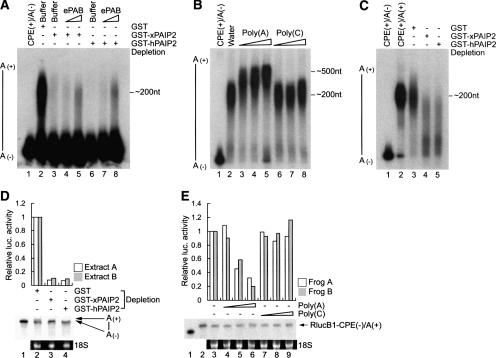
We surmised that ePAB could control polyadenylation in two ways: by potentiating Gld2-catalyzed polyadenylation or by inhibiting the activity of PARN, which would presumably be present and active in the extract after its expulsion from the polyadenylation complex following CPEB S174 phosphorylation (Kim and Richter 2006). To distinguish between these possibilities, ePAB was depleted from an egg extract with Xenopus or human PAIP2; this procedure inhibited polyadenylation. However, the addition of recombinant ePAB to the depleted extract restored polyadenylation, demonstrating that ePAB is the key factor regulating poly(A) tail length (Fig. 4A). Next, an egg extract was supplemented with 32P-labeled CPE(+) RNA in the absence or presence of poly(A) or poly(C). While poly(C) had little effect on poly(A) tail length, the poly(A) caused a substantial increase in polyadenylation, ∼300 nt (Fig. 4B). Finally, egg extracts depleted of ePAB by treatment with Xenopus or human PAIP2 were supplemented CPE(+) RNA preadenylated in vitro; while the poly(A) tail was retained in the nondepleted extract, it was removed in the ePAB-depleted extract (Fig. 4C). These data show clearly that ePAB stabilizes newly elongated poly(A) tails by protecting them from deadenylase activity.
Figure 4.
ePAB stabilizes newly elongated poly(A) tails. (A) Egg extracts depleted of ePAB with GST-PAIP2 or control GST column were supplemented with increasing amounts of recombinant ePAB; the extracts were then primed with 32P-labeled cyclin B1 RNA and examined for polyadenylation. (B) Nondepleted and precleared egg extracts were mixed with increasing amounts (20, 100, and 500 ng/μL) of poly(A) or poly(C) and then primed with 32P-labeled cyclin B1 RNA. (C) 32P-labeled cyclin B1 RNA was preadenylated with ∼200 nt in vitro with E. coli poly(A) polymerase. ePAB-depleted egg extracts with GST-PAIP2 or control GST column were supplemented with this RNA; polyadenylation was then examined. (D) ePAB-depleted egg extracts from two different frogs were in vitro translated with 32P-labeled RlucB1-CPE(+) mRNA. In the histogram of activity, Renilla luciferase activity (Rluc) was normalized to the RlucB1 mRNA. The relative activity (Rluc/RlucB1 mRNA) of RlucB1-CPE(+) mRNA plus egg extract depleted with GST was arbitrarily set to 1. 32P-labeled RlucB1 mRNAs were resolved on a 3.5% denaturing polyacrylamide gel and analyzed by a PhosphorImager; the mRNAs were examined by electrophoresis on a 1% denaturing agarose gel followed by 18S rRNA detection by ethidium bromide staining. (Bottom panel) In the RlucB1 mRNAs analysis, a representative experiment is shown; the radiolabeled probe is shown in lane 1. (E) Oocytes from two different frogs were injected with increasing amounts (10, 20, and 50 ng per oocyte) of poly(A) or poly(C), followed by a second injection of 32P-labeled RlucB1-CPE(−) mRNA (0.5 fmol/oocyte) that was preadenylated around ∼200 nt in vitro [RlucB1-CPE(−)/A(+)]. After 4 h, 25 oocytes were homogenized and prepared for either luciferase assay or recovery of RNA. (Lane 3) The relative activity (Rluc/RlucB1 mRNA) of RlucB1 mRNA without any homopolymer injection was arbitrarily set to 1. The radiolabeled probes without or with preadenylation are shown in lanes 1 and 2, respectively.
Finally, we examined mRNA translation in the ePAB-depleted extracts. Treatment of egg extracts with Xenopus or human PAIP2 elicited an ∼90% reduction in the translation of CPE-containing luciferase reporter RNA (Fig. 4D). The panel below the histogram in Figure 4D shows that when ePAB was depleted from extracts by Xenopus or human PAIP2, radioactive luciferase mRNA had a short poly(A) tail. The ethidium bromide-stained gel depicting 18S rRNA indicates that the RNA was substantially intact. Extracts A and B were derived from eggs from two different frogs. Moreover, the injection of poly(A) but not poly(C) to oocytes reduced by ∼80% the translation of an RNA preadenylated in vitro (Fig. 4E). The panels below the histogram in Figure 4E show that luciferase mRNA was polyadenylated and was intact, as was 18S rRNA. Frogs and B refer to experiments performed with oocytes from two different animals. These results point to the importance of ePAB for translation as well as maintenance of the poly(A) tail after maturation.
Discussion
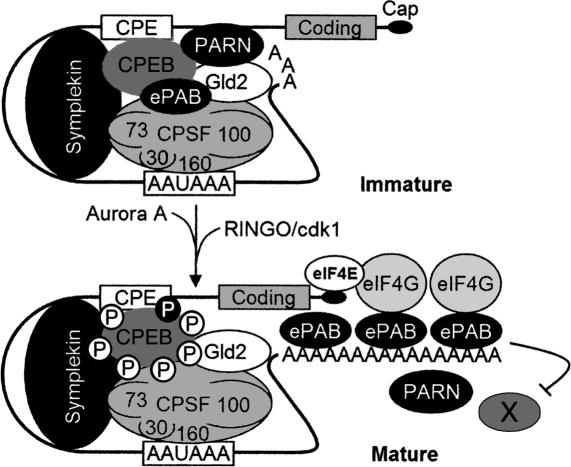
Our current model for cytoplasmic polyadenylation is summarized in Figure 5. As noted by Kim and Richter (2006), probably most nuclear pre-mRNAs have long poly(A) tails; following splicing and export to the cytoplasm, those containing a CPE are shortened by CPEB-bound PARN, whose robust activity overrides that of active CPEB-bound Gld2. During maturation, Aurora A phosphorylates CPEB S174, causing the expulsion of PARN from the RNP complex and consequent default Gld2-catalyzed polyadenylation. At a similar time, RINGO mRNA translation begins; RINGO binds and activates cdk1, which phosphorylates CPEB on six residues. The cdk1-catalyzed phosphorylations induce ePAB dissociation from CPEB and subsequent binding to the newly elongated poly(A) tail. Here, ePAB protects poly(A) from degradation by PARN and other nucleases. Poly(A)-bound ePAB also interacts with eIF4G, which helps it initiate translation of CPE-containing mRNAs.
Figure 5.
Model of poly(A) tail dynamics of CPE-containing mRNA in the cytoplasm. In the cytoplasm, CPE-containing mRNA associates with CPEB, symplekin, the four subunits of CPSF (sizes ∼30, 73, 100, and 160 kDa), Gld2, PARN, and ePAB. PARN activity is robust, and overrides the polyadenylating activity of the Gld2, which results in a short poly(A) tail and translation repression. Progesterone stimulates the activation of Aurora A, which phosphorylates CPEB S174 (black circled P), an event that causes the expulsion of PARN and Gld2-catalyzed default polyadenylation. At a similar time, RINGO mRNA is translated; RINGO associates with and activates cdk1, which phosphorylates CPEB on six residues (empty circled P). These phosphorylations cause ePAB dissociation from the CPEB-containing complex and subsequent binding to the elongated poly(A) tail. Here, ePAB protects poly(A) from hydrolysis by PARN and other nucleases (designated with an “X”). ePAB also binds eIF4G, which helps initiate translation. This model was modified from Kim and Richter (2006) with permission from Elsevier (© 2006).
Mendez et al. (2002) showed that the first prominent CPEB phosphorylation is catalyzed by Aurora A and occurs on S174, which stimulates polyadenylation, and that subsequently six additional cyclin B1/cdk1-catalyzed serine phosphorylations induce partial CPEB destruction later during maturation. Based on the data presented here, we propose that at least some CPEB undergoes early RINGO/cdk1-catalyzed phosphorylation, which was indicated initially by RINGO mRNA depletion with an antisense ODN (AS ODN); in the absence of RINGO, ePAB remained associated with CPEB. Moreover, ePAB did not dissociate from a CPEB protein containing alanine substitutions for the six serine residues that are phosphorylated by cdk1. Finally, roscovitin, a specific inhibitor of the kinase cdk1, also prevented ePAB dissociation from CPEB (data not shown).
Several additional points require comment. CPEB and symplekin co-IP experiments indicate that while ePAB is associated with the CPEB-containing complex in oocytes, eIF4G is not present in the same complex. However, because eIF4G was coimmunoprecipitated with ePAB from control and progesterone-treated oocytes, we surmise that ePAB is probably associated with eIF4G on the poly(A) tails of non-CPE-containing mRNAs at this time (Fig. 1A-D). Indeed, eIF4G co-IP experiments indicate that it interacts with a substantial amount of ePAB (Supplementary Fig. 3). These results might suggest that the number of CPE-containing RNAs that undergo polyadenylation in oocytes is relatively few compared with the total amount of mRNA in the cell. While this parameter has not been determined in oocytes, it has been estimated that in cultured hippocampal neurons following synaptic activation, ∼7% of RNAs undergo cytoplasmic polyadenylation (Du and Richter 2005). In these cells, CPEB promotes polyadenylation and translation following synaptic stimulation of N-methyl-D-aspartate receptors (Wu et al. 1998; Wells et al. 2001; Huang et al. 2002). If the same approximate value holds true for oocytes as well, then differences in the amount of eIF4G–ePAB interaction before and after maturation would be difficult to detect. It should also be noted that the seemingly simple CPE sequence (UUUUUAU) can be found in many 3′ untranslated regions (UTRs), but structural constraints or distance from the AAUAAA might prevent them from promoting polyadenylation (McGrew and Richter 1990; see also Huang et al. 2006). Thus, the presence of a CPE in a 3′UTR may not indicate, a priori, that the mRNA will undergo cytoplasmic polyadenylation.
Following CPEB-dependent polyadenylation, poly(A)-bound ePAB recruits eIF4G, presumably to stimulate translation by displacing maskin from eIF4E. We attempted to identify the enzyme that, in the absence of ePAB, deadenylates RNA after maturation. PARN was immunodepleted to >90% without loss of deadenylase activity (data not shown). We also attempted to immunodeplete CCR4, but were not successful. We suspect that ePAB protects the poly(A) tails from PARN as well as other deadenylases—possibly including CCR4—that may be active in the oocyte cytoplasm. Irrespective of which deadenylase(s) are involved at this time, the results presented here as well as in Barnard et al. (2004) and Kim and Richter (2006) demonstrate the complex regulation of poly(A) tail length and consequent translational control during early development.
We do not know the stoichiometry of ePAB binding to CPEB in oocytes; if it is one to one, then it would seem that ePAB is probably not sequestered on the CPEB complex prior to polyadenylation. That is, PABP, which is slightly larger than ePAB, binds poly(A) about every 25 bases (Sachs et al. 1987); a newly elongated poly(A) tail of 200 bases would then be expected to associate with eight ePAB molecules. We think it unlikely that this many ePAB molecules are released from one CPEB. However, it is possible that ePAB binding to poly(A) is cooperative, and that a single ePAB released from CPEB following RINGO/cdk1 phosphorylation is the first to bind poly(A), which then facilitates continued ePAB binding from a store of these molecules.
Because ePAB is replaced by “conventional” PABP during Xenopus development and in at least one cell line (Voeltz et al. 2001), is the mechanism of ePAB protection of poly(A) and ePAB recruitment of eIF4G restricted to oocytes and not, for example, neurons? To begin to assess this, we have attempted to deplete ePAB from oocyte extracts with PAIP2 and then to replace it with PABP. However, CPEB was codepleted with ePAB, thus preventing an analysis of poly(A) tail length with PABP (data not shown). However, myc-tagged PABP derived from injected mRNA interacts with eIF4G, symplekin, CPEB, and CPSF to the same extent as ePAB in parallel experiments (Supplementary Fig. 2). These results indicate that PABP is likely to functionally substitute for ePAB in somatic cells such as neurons that support cytoplasmic polyadenylation.
Materials and methods
Plasmids
For the generation of pMyc-ePAB, pGEX-KG-ePAB, and pRSETA-ePAB, plasmid containing full-length cDNA of Xenopus laevis ePAB (Open Biosystems) was PCR-amplified with ePAB-specific oligomers and then cloned into pMyc (Kim and Richter 2006), pGEX-KG, and pRSETA (Invitrogen), respectively. Plasmids pMyc-xPAIP2 and pGEX-KG-xPAIP2 were constructed with amplified products by RT–PCR with total RNA from Stage VI oocytes. X. laevis PABP (xPABP1) was amplified from a X. laevis cDNA library (Clontech) and cloned into pMyc. Myc-hPAIP2, pGEX-KG-hPAIP2, and pMyc-hPABP1 were constructed with amplified products by PCR with human fetal liver cDNA library (Clontech). Plasmids pB1(WT), pRluc-B1(WT), pRluc-B1(MT), pRSETA-CPSF160, pBS-symplekin, pMyc-PARN(WT), pBS-Myc-CPEB(WT), pBS-Myc-CPEB(6A), and pBS-Gld2(WT) have been described previously (Mendez et al. 2002; Barnard et al. 2004; Kim and Richter 2006). All constructs were verified by DNA sequencing.
Egg extract preparation, depletion of ePAB, in vitro polyadenylation, and in vitro translation
Extracts from unfertilized X. laevis eggs were prepared by the method of Murray and Kirschner (1989) with slight modifications. For the ePAB depletion, 2.5 μg of GST or GST-PAIP2 conjugated with 10 μL of MagneGST (Promega) bead particles were incubated with 25 μL of egg extract (containing 7.5 mM creatine phosphate [Roche], 1 mM ATP, and 1 mM MgCl2) plus 1 U/μL RNaseOUT (Invitrogen) three times followed by one time with unconjugated beads for 30 min each at 4°C. Polyadenylation assays were carried out for 1.5 h at 23°C with 0.4 vol of egg extract, 25,000 cpm of 32P-lableled RNA, and 0.6 vol of reaction mixture containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 2.5 mM MnCl2, 50 μg/mL bovine serum albumin, and 10% glycerol (v/v) at final concentration, followed by phenol/chloroform extraction. RNA probes were analyzed on 3.5% or 5% denaturing polyacrylamide gels and polyadenylation was monitored by phosphorimaging or autoradiography. For the in vitro translation experiments, a previous method (Patrick et al. 1989) was adapted and modified. Egg extract (0.6 vol) was mixed with 0.4 vol of reaction mixture containing 0.1 fmol/μL 32P-labeled RlucB1-CPE(+) mRNA, 10 mM Tris-HCl (pH 7.5), 20 mM creatine phosphate, 0.2 μg/μL creatine phosphokinase (Roche), 0.1 μg/μL calf liver tRNA (Novagen), 20 μM complete amino acid mix (Promega), 0.5 mM spermidine, 40 mM KCl, and 1 mM MnCl2 at final concentration. The final reaction mixture was incubated for 1.5 h at 23°C, diluted appropriately with passive lysis buffer (Promega), and subjected to the luciferase assay.
Recombinant ePAB purification and electromobility shift assay
Escherichia coli strain BL21(DE3)pLysS (Promega) was used to produce recombinant ePAB from plasmid pRSETA-ePAB. Isopropyl-β-D-thiogalactopyranoside (final concentration, 0.5 mM) was added to induce ePAB protein expression at OD600 0.5. After incubation for 5 h at 25°C, cells were harvested, resuspended in lysis buffer (20 mM Na-phosphate at pH 7.6, 300 mM NaCl, 0.5 mM phenylmethanesulphonylfluoride, 10 mM imidazole, 1 mM β-mercaptoethanol, 10% glycerol [v/v]), and sonicated. The resulting cell extracts were loaded onto a Ni-nitrilotriacetic acid agarose column (Qiagen) and washed with lysis buffer containing 20 mM imidazole, and the bound ePAB was eluted with 200 mM imidazole. Purified recombinant ePAB was dialyzed with buffer containing 20 mM HEPES-KOH (pH 7.5), 50 mM KCl, 5 mM MgCl2, and 10% glycerol (v/v). For the electromobility shift assay, reactions were assessed in 20-μL vol containing 25,000 cpm of 32P end-labeled 25 stretch oligo(A)-ribonucleotide (rA25), 10 mM HEPES-KOH (pH 7.5), 70 mM KCl, 1 mM EDTA, 1 mM MgCl2, 5% glycerol (v/v), 0.5 mM dithiothreitol, 1 U/μL RNaseOUT, and an appropriate amount of recombinant proteins or dialysis buffer. After incubation 10–20 min at 30°C, RNA–protein complexes were loaded on 8% nondenaturing gel and samples were analyzed by autoradiography.
Knockdown of RINGO mRNA, RT–PCR, and IP
Knockdown of X. laevis RINGO mRNA was described elsewhere (Ferby et al. 1999; Padmanabhan and Richter 2006). Briefly, oocytes injected with 100 ng of control (5′-TAGAGAA GATAATCGTCATCTTA-3′) or RINGO mRNA-specific (5′-ATATGCTAGAACCATTGCTATGAGA-3′) AS ODN were incubated 5 h to overnight and then incubated further with or without progesterone treatment. After 9 h (50% of oocytes showed germinal vesicle breakdown by 6–7 h after injection with control AS ODN), the oocytes were lysed and used for RT–PCR or CPEB-IP. For the RT–PCR, RNA pellets were dissolved in nuclease-free water and 4 μg of sample were used for the reaction. Twenty-five cycles of PCR amplification used the following primers for RINGO mRNA: 5′-ATGAGGCATATG CAGAGTGCAACC-3′ and 5′-GCCATGAATCTCCTAGTGC CCAGG3′. Oligomers for cyclin B1, mos, and β-actin were described in Kim and Richter (2006). For the CPEB co-IP, lysates from 75 oocytes were precleared with protein A-Sepharose 4B (Invitrogen) and incubated with CPEB antibody-conjugated protein A beads for 3 h to overnight at 4°C. The collected beads were then washed four to six times and the coprecipitating proteins were analyzed on immunoblots. As a secondary antibody, horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG (Vector Laboratories) or protein A-HRP (Invitrogen) was used.
Details of the oocyte manipulation and microinjection, recombinant GST fusion protein purification, and in vitro binding analysis were described elsewhere (Kim and Richter 2006; Padmanabhan and Richter 2006).
Acknowledgments
We thank D. Bentley (University of Colorado), I. Daar (National Cancer Institute-Frederick), B. Keiper (East Carolina University), J. Lykke-Andersen (University of Colorado), J. Manley (Columbia University), J. Ruderman (Harvard Medical School), J. Steitz (Yale University), A. Virtanen (Uppsala University, Sweden), E. Wahle (University of Halle, Germany), and M. Wormington (University of Virginia) for gifts of antibodies or clones. We also thank D. Schoenberg (Ohio State University) for suggestions of experimental protocols. Finally, we thank D. Barnard, D. Burns, Q. Cao, C-L. Lin, and K. Padmanabhan for sharing material, critical comments, and technical assistance. We are also grateful to other members of laboratory for helpful comments. This work was supported by a grant from the NIH (GM46779).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1593007
References
- Barnard D.C., Ryan K., Manley J.L., Richter J.D., Ryan K., Manley J.L., Richter J.D., Manley J.L., Richter J.D., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Cao Q., Kim J.H., Richter J.D., Kim J.H., Richter J.D., Richter J.D. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Wormington M., Wormington M. The mechanism and regulation of deadenylation: Identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson K.S., Bilger A., Ballantyne S., Wickens M.P., Bilger A., Ballantyne S., Wickens M.P., Ballantyne S., Wickens M.P., Wickens M.P. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol. Cell. Biol. 1999;19:5707–5717. doi: 10.1128/mcb.19.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Richter J.D., Richter J.D. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I., Blazquez M., Palmer A., Eritja R., Nebreda A.R., Blazquez M., Palmer A., Eritja R., Nebreda A.R., Palmer A., Eritja R., Nebreda A.R., Eritja R., Nebreda A.R., Nebreda A.R. A novel p34cdc2-binding and activating protein that is necessary and sufficient to trigger G2/M progression in Xenopus oocytes. Genes & Dev. 1999;13:2177–2189. doi: 10.1101/gad.13.16.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Jung M.Y., Sarkissian M., Richter J.D., Jung M.Y., Sarkissian M., Richter J.D., Sarkissian M., Richter J.D., Richter J.D. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Kan M.C., Lin C.L., Richter J.D., Kan M.C., Lin C.L., Richter J.D., Lin C.L., Richter J.D., Richter J.D. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady B.T., Kuo P., Martinez S.E., Yuan L., Hake L.E., Kuo P., Martinez S.E., Yuan L., Hake L.E., Martinez S.E., Yuan L., Hake L.E., Yuan L., Hake L.E., Hake L.E. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J. Cell Sci. 2007;120:1093–1103. doi: 10.1242/jcs.03416. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N., Deo R.C., Burley S.K., Sonenberg N., Burley S.K., Sonenberg N., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Richter J.D., Richter J.D. Opposing polymerase–deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Klann E., Richter J.D., Richter J.D. Translation control of synaptic plasticity and learning and memory. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 485–506. [Google Scholar]
- Korner C.G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E., Muckenthaler M., Schneider S., Dehlin E., Wahle E., Schneider S., Dehlin E., Wahle E., Dehlin E., Wahle E., Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Wang L., Ballantyne S., Kimble J., Wickens M., Wang L., Ballantyne S., Kimble J., Wickens M., Ballantyne S., Kimble J., Wickens M., Kimble J., Wickens M., Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., Zhang D., Saya H., Zhang D., Saya H., Saya H. Aurora-A—A guardian of poles. Nat. Rev. Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- McGrew L.L., Richter J.D., Richter J.D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: Characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990;9:3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R., Hake L.E., Andresson T., Littlepage L.E., Ruderman J.V., Richter J.D., Hake L.E., Andresson T., Littlepage L.E., Ruderman J.V., Richter J.D., Andresson T., Littlepage L.E., Ruderman J.V., Richter J.D., Littlepage L.E., Ruderman J.V., Richter J.D., Ruderman J.V., Richter J.D., Richter J.D. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez R., Barnard D., Richter J.D., Barnard D., Richter J.D., Richter J.D. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W., Kirschner M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K., Richter J.D., Richter J.D. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes & Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick T.D., Lewer C.E., Pain V.M., Lewer C.E., Pain V.M., Pain V.M. Preparation and characterization of cell-free protein synthesis systems from oocytes and eggs of Xenopus laevis. Development. 1989;106:1–9. doi: 10.1242/dev.106.1.1. [DOI] [PubMed] [Google Scholar]
- Richter J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Richter J.D., Sonenberg N., Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Davis R.W., Kornberg R.D., Davis R.W., Kornberg R.D., Kornberg R.D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkissian M., Mendez R., Richter J.D., Mendez R., Richter J.D., Richter J.D. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes & Dev. 2004;18:48–61. doi: 10.1101/gad.1136004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M.D., Fox C.A., Hunt T., Vande Woude G., Wickens M., Fox C.A., Hunt T., Vande Woude G., Wickens M., Hunt T., Vande Woude G., Wickens M., Vande Woude G., Wickens M., Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes & Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Sonenberg N., Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Manley J.L., Manley J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B., Wickens M., Kimble J., Wickens M., Kimble J., Kimble J. Translation control in development. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 507–544. [Google Scholar]
- Voeltz G.K., Ongkasuwan J., Standart N., Steitz J.A., Ongkasuwan J., Standart N., Steitz J.A., Standart N., Steitz J.A., Steitz J.A. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes & Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J., Kadyk L.C., Wickens M., Kimble J., Wickens M., Kimble J., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wells D.G., Dong X., Quinlan E.M., Huang Y.S., Bear M.F., Richter J.D., Fallon J.R., Dong X., Quinlan E.M., Huang Y.S., Bear M.F., Richter J.D., Fallon J.R., Quinlan E.M., Huang Y.S., Bear M.F., Richter J.D., Fallon J.R., Huang Y.S., Bear M.F., Richter J.D., Fallon J.R., Bear M.F., Richter J.D., Fallon J.R., Richter J.D., Fallon J.R., Fallon J.R. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wells D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Wells D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Tay J., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Mendis D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Abbott M.A., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Barnitt A., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Quinlan E., Heynen A., Fallon J.R., Richter J.D., Heynen A., Fallon J.R., Richter J.D., Fallon J.R., Richter J.D., Richter J.D. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]