Abstract
The nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase Sir2 regulates life-span in various species. Mammalian homologs of Sir2 are called sirtuins (SIRT1–SIRT7). In an effort to define the role of sirtuins in vascular homeostasis, we found that among the SIRT family, SIRT1 uniquely regulates angiogenesis signaling. We show that SIRT1 is highly expressed in the vasculature during blood vessel growth, where it controls the angiogenic activity of endothelial cells. Loss of SIRT1 function blocks sprouting angiogenesis and branching morphogenesis of endothelial cells with consequent down-regulation of genes involved in blood vessel development and vascular remodeling. Disruption of SIRT1 gene expression in zebrafish and mice results in defective blood vessel formation and blunts ischemia-induced neovascularization. Through gain- and loss-of-function approaches, we show that SIRT1 associates with and deacetylates the forkhead transcription factor Foxo1, an essential negative regulator of blood vessel development to restrain its anti-angiogenic activity. These findings uncover a novel and unexpected role for SIRT1 as a critical modulator of endothelial gene expression governing postnatal vascular growth.
Keywords: SIRT1, HDAC, endothelial cells, angiogenesis, Foxo
Chromatin dynamics control virtually all known biological processes governing mammalian development and the growth and survival of primary and cancer cells. Histones are the central structural elements of chromatin, and their N-terminal tails are subject to covalent modifications by the opposing actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Jenuwein and Allis 2001). HDACs act as cofactors that are recruited to promoters by specific DNA-binding factors resulting in the local modification of histones to promote chromatin compaction with subsequent inhibition of gene transcription (Cheung et al. 2000; Strahl and Allis 2000). In addition, an increasing number of nonhistone proteins, such as numerous transcription factors, are regulated by reversible acetylation and targeted by HDACs (Yang 2004).
HDACs are divided into three classes based on their homology with yeast proteins Rpd3 (class I), Hda1 (class II), and Sir2 (class III) (de Ruijter et al. 2003; North and Verdin 2004). While the class I and II HDACs are sensitive to trichostatin A (TSA), class III HDACs (SIRTs) are not, and their deacetylase activity uniquely relies on the cofactor nicotinamide adenine dinucleotide (NAD+). Class I and II HDACs play important roles in the transcriptional control of endothelial gene expression and vascular development (Kim et al. 2001; Deroanne et al. 2002; Rossig et al. 2002, 2005; Illi et al. 2003, 2005; Chang et al. 2006). Pharmacological inhibition of class I and II HDACs has been shown to block pathological angiogenesis during tumor growth (Kim et al. 2001) and to prevent adult progenitor cell differentiation toward the endothelial lineage (Rossig et al. 2005). Moreover, disruption of the HDAC7 gene, which is expressed specifically in endothelial cells during embryonic development, leads to embryonic lethality due to the loss of vascular integrity with subsequent blood vessel rupture (Chang et al. 2006). In contrast, the function of class III HDACs in endothelial cell biology and blood vessel development remains entirely unexplored.
The founding member of class III HDACs, Sir2, is evolutionarily conserved and regulates longevity and aging in multiple model organisms (e.g., Saccharomyces cerevisiae, Caenorhabditis elegans) (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000; Guarente and Picard 2005). Mammalian homologs of Sir2 are a family of seven proteins termed sirtuins (SIRT1–SIRT7) (Frye 2000), among which SIRT1 is the closest human homolog of the yeast Sir2 protein. SIRT1 has been shown to play a central role in regulating cellular differentiation and senescence and to control metabolic pathways in response to nutrient availability in a wide variety of tissues (Blander and Guarente 2004). SIRT1 interacts with a number of substrates—including transcription factors such as forkhead box o (Foxo), p53, PPAR-γ/NCoR/SMRT, Hey, or PGC1-α— that mediate specific functions of SIRT1 (Luo et al. 2001; Vaziri et al. 2001; Fulco et al. 2003; Takata and Ishikawa 2003; Brunet et al. 2004; Motta et al. 2004; Picard et al. 2004; Rodgers et al. 2005). SIRT1 binds and deacetylates Foxo transcription factors resulting in the differential regulation of Foxo target gene expression (Brunet et al. 2004; Daitoku et al. 2004; Motta et al. 2004; van der Horst et al. 2004; Frescas et al. 2005; Yang et al. 2005). Thus, SIRT1-dependent deacetylation of Foxo3a leads to the transcriptional repression of the proapoptotic target gene Bim, but up-regulation of the stress-resistance gene GADD45 (Brunet et al. 2004). Moreover, SIRT1-dependent deacetylation of Foxo1 represses its transcriptional and proapoptotic activity in prostate cancer cells (Yang et al. 2005). In addition, SIRT1 promotes fat mobilization by repressing PPAR-γ target genes (Picard et al. 2004) and controls glucose homeostasis through interaction with PGC1-α. In some cell types, SIRT1 promotes cell survival in response to cellular stress by deacetylating tumor suppressor protein p53, which down-regulates p53 stability and activity (Luo et al. 2001; Vaziri et al. 2001; Langley et al. 2002; Cheng et al. 2003). However, in fibroblasts, SIRT1 seems to increase total p53 levels upon conditions of chronic oxidative damage, causing cellular senescence (Chua et al. 2005).
Although SIRTs have been shown to play a critical role in the regulation of several essential physiological processes (Bordone and Guarente 2005), little information is available on a putative role of SIRTs in endothelial and vascular homeostasis. In the present study, we demonstrate that, uniquely among the SIRT proteins studied, SIRT1 plays a key role in angiogenesis signaling in vitro, in zebrafish, and in mice. We show that SIRT1 regulates the expression of multiple genes involved in vascular endothelial homeostasis and remodeling. These effects are mediated, at least in part, by regulating the forkhead transcription factor Foxo1. These findings reveal a key role for mammalian SIRT1 as a novel modulator of endothelial gene expression regulating postnatal angiogenesis.
Results
Sirtuins regulate the angiogenic activity of endothelial cells
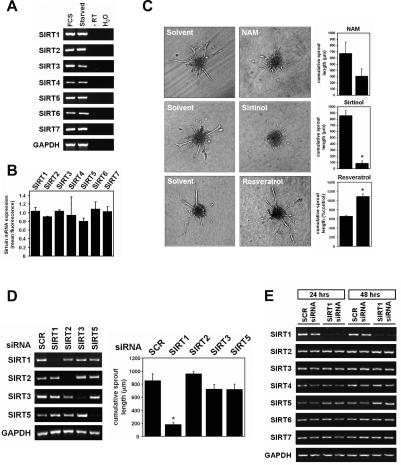
To investigate the role of SIRT proteins in the regulation of endothelial cell function, we first assessed the expression profile of the seven family members in endothelial cells. RT–PCR analysis showed that all seven sirtuin transcripts (SIRT1–SIRT7) are easily detectable in total RNA isolated from human umbilical vein endothelial cells (HUVECs) (Fig. 1A). Microarray analysis confirmed these results and revealed comparable expression levels among the various SIRT family members (Fig. 1B). To further test whether SIRTs regulate endothelial cell functions, we examined the role of SIRTs during endothelial differentiation and in vitro angiogenesis in a three-dimensional spheroidal assay. To this end, endothelial cell spheroids of defined size and cell number were treated with the general SIRT inhibitors nicotinamide and sirtinol. Both compounds severely impaired sprout-forming activity of endothelial cells (Fig. 1C). Consistent with these findings, treatment of spheroids with the sirtuin activator resveratrol increased the angiogenic response of endothelial cells (Fig. 1C), suggesting that some members of the SIRT family are important modulators of endothelial angiogenic functions.
Figure 1.
SIRT1 regulates the angiogenic activity of endothelial cells. (A) mRNA expression of SIRT1–SIRT7 in HUVECs as assessed by RT–PCR using the indicated primer pairs. GAPDH served as loading control. (B) Statistical summary of the SIRT expression profile as assessed in a microarray analysis of total RNA isolated from HUVECs. (C) Three-dimensional in vitro angiogenesis with collagen gel-embedded spheroids of solvent-treated, nicotinamide-treated (NAM, 5 mM), sirtinol-treated (100 μM), or resveratrol-treated (1 μM) endothelial cells. Cumulative length of all sprouts originating from an individual spheroid was quantified after 24 h. Representative micrographs and a statistical summary are shown. (D) HUVECs were transfected with different siRNAs targeting SIRT1, SIRT2, SIRT3, SIRT5, or a nonrelated scrambled control. A statistical summary of the cumulative sprout length after 24 h originating from individual spheroids in siRNA-transfected endothelial spheroids is shown. The mRNA expression of the SIRTs with deacetylase activity in siRNA-transfected HUVECs as assessed by RT–PCR using the indicated primer pairs is shown in the left panel. (E) The effects of SIRT1 gene silencing on the expression of mRNA transcripts for SIRT1–SIRT7 were assessed by RT–PCR using HUVEC RNA from SIRT1 siRNA-transfected endothelial cells.
Silencing of SIRT1 abolishes endothelial sprout formation
The above observations using pharmacological inhibitors indicate that SIRT deacetylase activity could be involved in endothelial angiogenic functions. To test this hypothesis, we used RNA interference (RNAi) to generate endothelial cells specifically deficient in SIRT1, SIRT2, SIRT3, and SIRT5, which are the only SIRT proteins with deacetylase activity reported so far (Fig. 1D; North et al. 2003). Interestingly, among the tested SIRT members, gene silencing of SIRT1 was uniquely associated with complete inhibition of in vitro endothelial sprout formation as measured in a three-dimensional spheroidal assay (Fig. 1D). Since small interfering RNA (siRNA)-mediated inhibition of SIRT1 did not alter the expression of SIRT2–SIRT7 (Fig. 1E), these experiments specifically implicate SIRT1 in the regulation of endothelial angiogenic signaling.
To confirm these results and exclude potential off-target effects of the siRNA duplex, we repeated the spheroidal assays using endothelial cells treated with a series of siRNAs targeting different sequences in the human SIRT1 mRNA. While the abilities of the various siRNA to inhibit SIRT1 expression were variable (Supplementary Fig. 1), they perfectly correlated with their capacity to inhibit sprout formation (Supplementary Fig. 1).
SIRT1 controls the angiogenic activity of endothelial cells
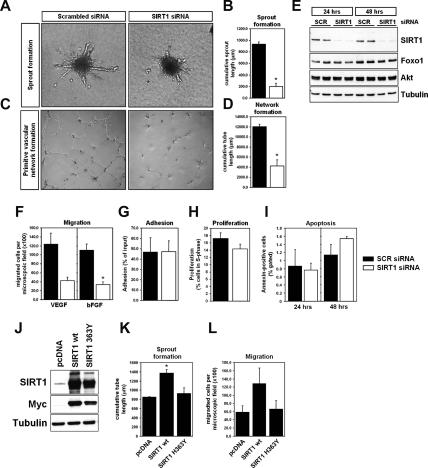
To unravel the cellular processes controlled by SIRT1 in endothelial cells, we further examined the properties of SIRT1-deficient endothelial cells. Consistent with the loss of sprout formation, inhibition of endogenous SIRT1 gene expression prevented the formation of a vascular-like network in vitro (Fig. 2A–E). In addition, knockdown of SIRT1 blocked endothelial cell migration toward different chemoattractants such as VEGF and bFGF, whereas the adhesion to different matrices was not significantly affected (Fig. 2F,G). The anti-angiogenic effect of SIRT1 gene silencing was unlikely to result from increased apoptosis or cell cycle arrest, since it had only minor effects on programmed cell death and cell cycle progression under basal conditions (Fig. 2H,I). Finally, overexpression of wild-type SIRT1, but not of a deacetylase-defective mutant of SIRT1 (SIRT1 H363Y) (Luo et al. 2001; Vaziri et al. 2001), increased the sprout-forming and migratory activity of endothelial cells (Fig. 2J–L). Taken together, these results demonstrate that the HDAC SIRT1 acts as a critical modulator of endothelial angiogenic functions and that the deacetylase activity is required for these effects.
Figure 2.
Effect of SIRT1 on cellular responses in endothelial cells. (A,B) Representative micrographs and statistical summary of three-dimensional in vitro angiogenesis assays with collagen gel-embedded spheroids generated from SIRT1 or scrambled siRNA-transfected endothelial cells. (C,D) Representative micrographs and statistical summary of in vitro Matrigel assays with SIRT1 or scrambled siRNA-transfected endothelial cells. (E) HUVECs were transfected with a SIRT1-specific siRNA or a nonrelated scrambled control. Cell lysates were subjected to Western blotting using antibodies against SIRT1. Antibodies recognizing Foxo1, Akt, or tubulin were used as a loading control. (F–I) Statistical summary of the effects of SIRT1 gene silencing (white bars) on migration, adhesion, proliferation, and apoptosis compared with scrambled siRNA-transfected controls (black bars). The VEGF and bFGF concentrations were 50 ng/mL and 30 ng/mL, respectively. (J) HUVECs were transfected with wild-type (wt) SIRT1, a deacetylation-defective SIRT1 mutant (H363Y), or mock control (pcDNA3). Cells were lysed after 24 h and subjected to Western blot analysis with antibodies against SIRT1 and Myc. (K) Statistical summary of the cumulative sprout length in pcDNA-, SIRT1 wild-type-, or SIRT1 H363Y-transfected endothelial spheroids. (L) Statistical summary of endothelial migration toward the chemoattractant VEGF (50 ng/mL) in pcDNA-, SIRT1 wild-type-, or SIRT1 363Y-transfected cells.
SIRT1 expression in the vascular endothelium
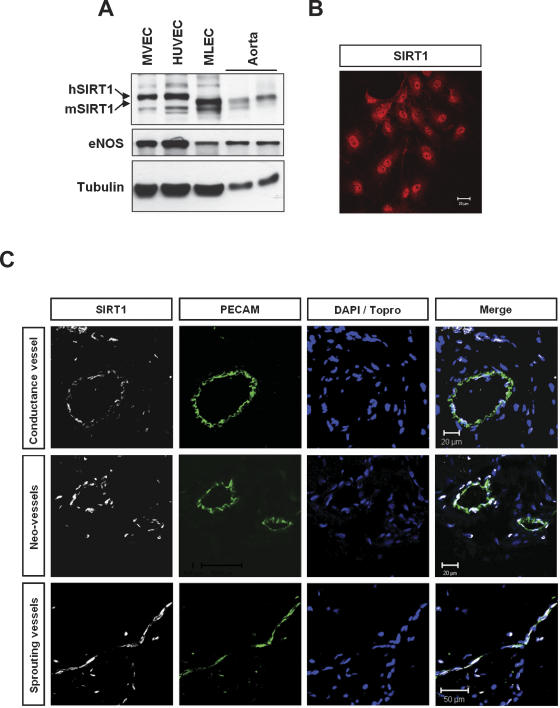
To examine the role of SIRT1 in the vasculature, we first assessed the expression and vascular tissue distribution of SIRT1 in cultured endothelial cells and in vivo in mice. Analysis of protein expression revealed that SIRT1 is widely expressed in different cultured endothelial cells such as HUVECs, human microvascular endothelial cells (HMVECs), and murine lung endothelial cells (MLECs), as well as in the intact aorta of mice (Fig. 3A). Immunofluorescence microscopy of cultured endothelial cells using a SIRT1-specific antibody showed a predominantly nuclear localization of SIRT1 in cultured endothelial cells (Fig. 3B).
Figure 3.
SIRT1 expression in the vascular endothelium. (A) Cell and tissue lysates were subjected to Western blot analysis with specific antibodies against SIRT1 and eNOS and tubulin. (HUVEC) Human umbilical vein endothelial cells; (HMVEC) human microvascular endothelial cells; (MLEC) murine lung endothelial cells; (Aorta) murine aorta of 129/SV mice. (B) Representative immunostaining is shown for the predominantly nuclear localization of SIRT1 in HUVEC. (C) Immunostainings of SIRT1 with a polyclonal antibody (white) in mouse vascular sections. No signal was detected with a rabbit IgG control antibody (data not shown). Endothelial cells were stained with an antibody recognizing CD31 (PECAM, green). Nuclei were stained in blue (Topro or DAPI).
Confocal microscopy of vascular sections from adult mice confirmed the robust expression of SIRT1 in the postnatal vascular endothelium (Fig. 3C). More importantly, SIRT1 expression was markedly induced in sprouting endothelial cells as well as in neo-vessels invading a Matrigel matrix (Fig. 3C). These data are consistent with an important role of SIRT1 during sprouting angiogenesis and suggest a signal-responsive regulation of SIRT1 during postnatal vessel formation.
Inhibition of SIRT1 expression results in defective vascular growth and patterning in zebrafish
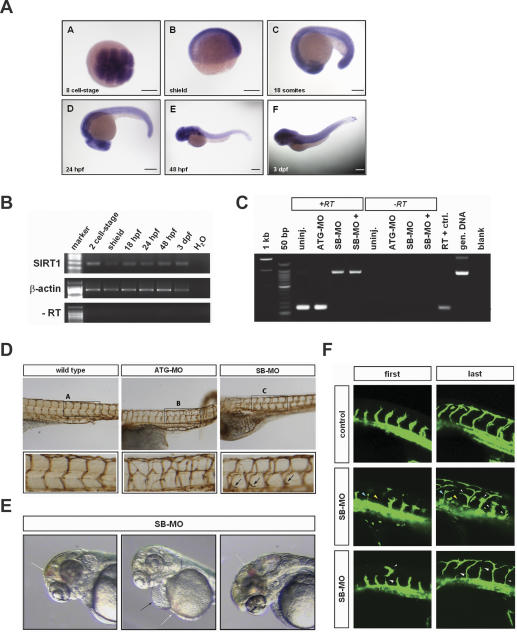
In order to determine the role of SIRT1 in an animal model, where the formation and patterning of the vasculature could be easily monitored, we perturbed SIRT1 expression in zebrafish embryos using antisense morpholino-modified oligonucleotides. In situ hybridization with SIRT1-specific cRNA probes and RT–PCR analysis showed that zebrafish SIRT1 is a maternal transcript (Fig. 4A,B, 8-cell stage) and that its expression persists in developing embryos in a diffuse pattern (Fig. 4A,B, from shield to 3 dpf [days post-fertilization]) because of the onset of zygotic expression at the mid-blastula transition stage. This pattern of expression prompted us to use two morpholinos: one targeting the start codon of zebrafish SIRT1 (ATG-MO) and, therefore, blocking translation of both maternal and zygotic SIRT1 mRNA, and one targeting the exon 2/intron 2 boundary (SB-MO) (Fig. 4C), which targets only the zygotic transcript. Embryos of the tg(fli1:eGFP) line (Lawson and Weinstein 2002) characterized by fluorescent endothelial cells were used to ease the analysis of the phenotype. Both morpholinos gave a similar phenotype, characterized by patterning defects of the trunk vasculature (Fig. 4D). Moreover, SIRT1 deficiency (SB-MO) resulted in spontaneous and induced hemorrhages throughout the zebrafish body (Fig. 4E, white arrows) indicating that SIRT1 is required for proper vascular growth and maturation.
Figure 4.
Cardiovascular defects in SIRT1-deficient zebrafish embryos. (A) SIRT1 expression at different stages of zebrafish development. (Panels A–F) SIRT1 antisense probe. (Panel A) Dorsal view. (Panels B–F) Lateral views. The embryonic stage is indicated at the bottom left corner. (8-cell stage) maternal expression; (shield) zygotic expression. Bars, 200 μm. (B) RT–PCR analysis performed on total RNA extracted from embryos at the indicated stages using primers specific for SIRT1. β-actin serves as loading control. (C) RT–PCR analysis of SIRT1 mRNA transcripts in control, ATG-MO-injected, and SB-MO-injected embryos. Specific and effective splice-blocking of the SIRT1 transcript in the SB-MO-injected embryos. SB-morpholinos were injected at two different concentrations (SB-MO+ indicates twice the amount of the morpholino as compared with SB-MO). The exon 2–exon 3 spliced form is 249 bp, and 701 bp is the fragment including the intron 2 sequence. (D) Lateral views of the vasculature from either control or SIRT1 knockdown (ATG-MO and SB-MO) zebrafish embryos. Pictures were taken from fixed embryos after immunohistochemistry with an anti-GFP antiserum using a stereomicroscope. (E) Hemorrhages (white arrows) and pericardial swelling (black arrow) in SIRT1 knockdown zebrafish (SB-MO). (F) First and last frame (GFP fluorescence) of time-lapse movies captured from control embryos (24 hpf) and embryos injected at one- to two-cell stage with a SIRT1 splice-blocking morpholino. The embryo’s head is to the left, dorsal is to the top. Arrowheads point to vessels defects.
To more closely characterize the cellular defects associated with SIRT1 deficiency during the angiogenic process, we assessed segmental artery formation in transgenic zebrafish embryos with fluorescent blood vessels [tg(fli1:eGFP) embryos] by time-lapse analysis focusing on intersomitic vessel (ISV) development at ∼24 h post-fertilization (hpf). Segmental arteries in tg(fli1:eGFP) embryos injected with a morpholino oligonucleotide that blocks splicing of SIRT1 display defects in the formation of ISVs (Fig. 4F; Supplementary Movies 1–3). Specifically, adjacent ISVs do extend asynchronously, with some growing no further than half way. The developing ISVs in the SIRT1 morphants extend and retract filopodia, but differently from the control embryos. They extend larger and thicker filopodia, which guide tip cells to follow an irregular path and to make extra branches or extra connections with adjacent vessels (Supplementary Movies 1–3). Thus, the phenotypes of the SIRT1-deficient zebrafish observed in the time-lapse analysis indicate that SIRT1 is required for endothelial cell migration during sprouting angiogenesis and that loss of SIRT1 leads to dysregulated tip cell activity in vivo.
SIRT1 is essential for postnatal blood vessel development
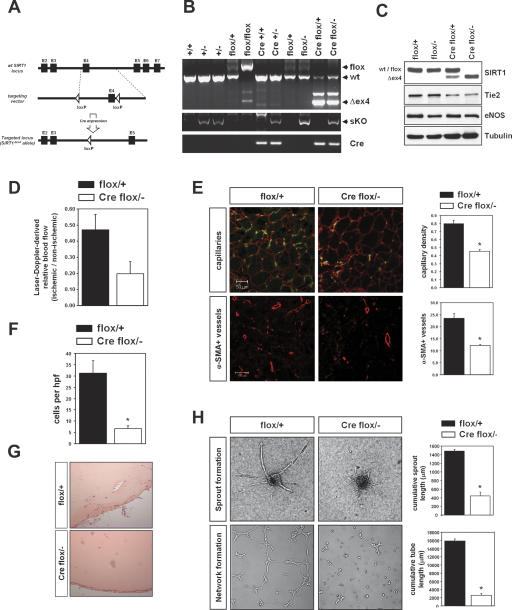
Since the early postnatal lethality of SIRT1-null mice precluded further analysis of potential vascular defects during postnatal angiogenesis (Cheng et al. 2003), we analyzed mice carrying a “floxed” SIRT1 allele (SIRT1flox), in which exon 4 encoding for 51 amino acids of the conserved SIRT1 catalytic domain is flanked by loxP sites (Fig. 5A). It has been shown previously that mice harboring homozygous deletion of this floxed exon 4 were phenotypically indistinguishable from SIRT1-null mice (SIRT1−/−), indicating that the resulting truncated SIRT1 protein is nonfunctional (Cheng et al. 2003). To achieve conditional deletion of SIRT1 specifically in the vascular endothelial cells, we crossed mice homozygous for the floxed SIRT1 allele (SIRT1flox/flox) allele with mice heterozygous for the SIRT1-null allele (SIRT1+/−), which carried at the same time the Cre-recombinase under the control of the Tie2-promoter enhancer (Tie2Cretg; SIRT1+/−). Introduction of the Cre recombinase driven by the endothelial-specific promoter of the Tie2 gene (Koni et al. 2001) led to the efficient Cre-mediated excision of exon 4 as evidenced by PCR analysis of genomic DNA (Fig. 5B). Western blot analysis from isolated MLECs of the Tie2Cretg;SIRT1flox/− mice showed that the floxed SIRT1 allele was completely deleted in the endothelial lineage (Fig. 5C).
Figure 5.
Endothelial cell-specific deletion of SIRT1. (A) Strategy to generate a conditional SIRT1 allele by inserting loxP sites flanking exon 4 of the mouse SIRT1 genomic locus. The structure of the genomic locus, the targeting vector, and the targeted allele are shown. Endothelial-specific deletion was achieved by breeding to transgenic mice harboring a Tie2-Cre transgene. (B) Genotyping of the endothelial-specific SIRT1 mutant mice (Tie2Cretg;SIRT1flox/−). (Cre) Tie2Cretg; (flox) SIRT1flox allele; (+/−) SIRT1+/−. (C) Western blot analysis with a specific antibody against SIRT1 showed that the floxed SIRT1 allele was completely deleted in endothelial cells isolated from the endothelial-restricted SIRT1 mutant mice. Cell lysates were also subjected to Western blot analysis with antibodies targeting eNOS and Tie2 to show the endothelial characteristics of cells; tubulin served as loading control. (D) Mice of the respective genotypes were subjected to hindlimb ischemia, and perfusion was assessed 14 d after onset of ischemia using laser Doppler imaging. Quantitative results are presented. (E) Capillary (CD31, green; laminin, red) and α-smooth muscle actin staining (red) in sections of mice after hindlimb ischemia. A statistical summary of capillary density and the number of α-smooth muscle actin-positive vessels is shown. (F) Mice of the respective genotypes were subcutaneously injected with Matrigel matrix. After 7 d, Matrigel plugs were explanted and the hemoglobin content and invading cell number of the plug were quantified. (G) Histological analysis of H&E stainings from Matrigel plugs isolated from SIRT1flox/+ and Tie2Cretg;SIRT1flox/− mice. (H) Murine endothelial cells of SIRT1flox/+ and Tie2Cretg;SIRT1flox/− mice were isolated by vascular endothelial-cadherin immunopurification. Representative micrographs and statistical summary of the three-dimensional spheroid and Matrigel assay of in vitro angiogenesis with endothelial cells generated from these mice.
Among the genotyped 3-wk-old pups born from crosses between young SIRT1flox/flox and Tie2Cretg; SIRT1flox/− parents, viable endothelial-restricted SIRT1 mutant mice (Tie2Cretg;SIRT1flox/−) were obtained at a frequency that approximated Mendelian ratios (Supplementary Table 1), suggesting that endothelial SIRT1 is dispensable for embryonic vascular development.
Despite the lack of an overt phenotype during embryonic and early postnatal development in the endothelial-restricted SIRT1 mutant mice, we wondered whether these mice might be sensitized to ischemia-induced neovascularization, as it would be predicted if SIRT1 is a mediator of stress-induced signaling pathways. Compared with the SIRT1flox/+ mice, limb perfusion after hindlimb ischemia was blunted in the endothelial-specific SIRT1 mutant mice after hindlimb ischemia (Fig. 5D). Histological analysis demonstrated that the blunted blood flow recovery in the SIRT1 mutant mice was a reflection of an impaired ability to form new vessels in the ischemic tissue. Thus, capillary density and the number of α-smooth muscle actin-positive vessels in the ischemic adductor muscle were significantly decreased in these mice (Fig. 5E). Moreover, the loss of SIRT1 in the endothelial lineage was associated with an increased incidence of foot and toe necrosis in response to tissue ischemia (1.43 ± 0.3 [Tie2Cretg;SIRT1flox/−] vs. 0.56 ± 0.28 [SIRT1flox/+]; P < 0.05). To further validate the in vivo relevance of SIRT1 for postnatal blood vessel development, we investigated the effect of SIRT1 deficiency in a growth factor-induced angiogenesis assay. Matrigel implants were administered subcutaneously to SIRT1flox/+ and the endothelial-restricted SIRT1 mutant mice, and blood vessel infiltration of the implants was quantified. Compared with wild-type mice, the number of lectin-positive endothelial cells (data not shown) and invading vessel-like structures (Fig. 5F,G) was markedly reduced in the SIRT1 mutant mice. Consistently, the hemoglobin content of the Matrigel plug was reduced in the endothelial cell-restricted SIRT1 mutant mice, indicating that SIRT1 activity is required for postnatal angiogenic responses.
In order to directly examine the cellular mechanisms that may explain the blunted angiogenic phenotype of the SIRT1 mutant mice, we isolated MLECs from SIRT1flox/+ and Tie2Cretg;SIRT1flox/− mice and assessed their angiogenic phenotype. As shown in Figure 5H, sprouting angiogenesis and branching morphogenesis were blocked in SIRT1 mutant MLECs consistent with an essential role of SIRT1 for the angiogenic activity of endothelial cells. In summary, these findings identify SIRT1 as a critical regulator of the signaling machinery within the endothelium that conveys an angiogenic/ischemic signal to the formation and growth of the postnatal vasculature.
Altered gene expression in SIRT1-silenced endothelial cells
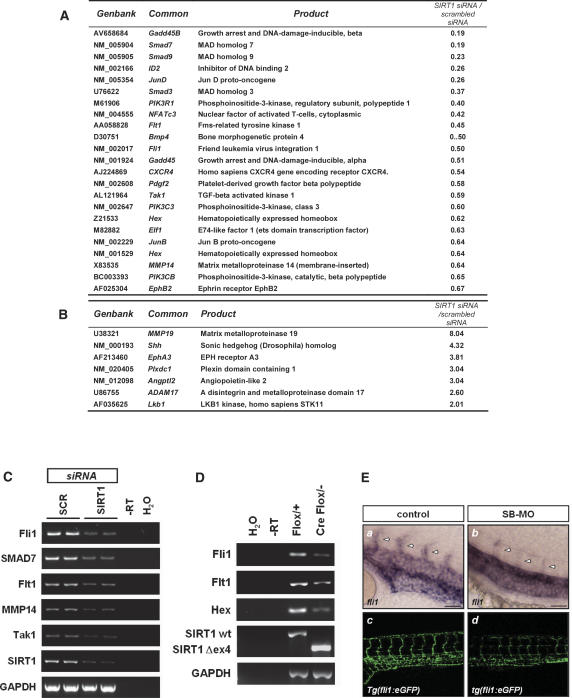
In an effort to identify related molecular mechanisms of the SIRT1-dependent regulation of angiogenesis, we compared gene expression profiles of scrambled and SIRT1 siRNA-transfected endothelial cells by microarray analysis. Numerous genes involved in vascular growth, maturation, and remodeling were dysregulated in SIRT1-silenced endothelial cells. These included genes encoding for Flt1, CXCR4, Pdgfß, angiopoietin-like 2, Mmp14, and EphB2 (Fig. 6A,B). Moreover, silencing of SIRT1 expression led to the down-regulation of genes essential for endothelial differentiation during vascular development, such as Hex and Fli1 (Fig. 6A). The down-regulation of gene expression associated with SIRT1 knockdown was confirmed by RT–PCR for several of these genes (Fig. 6C). Importantly, several of the angiogenesis- and vascular remodeling-related genes found in the microarray analysis, such as Fli1, Flt1, and Hex, were also down-regulated in endothelial cells freshly isolated from the endothelial-restricted SIRT1 mutant mice (Fig. 6D), suggesting that these genes are SIRT1-regulated genes in vivo. Consistent with these data, loss of SIRT1 expression in the zebrafish induced a marked reduction of Fli1 expression as determined by in situ hybridization as well as a reduction in the Fli1-driven GFP fluorescence signal using the transgenic tg(fli1:eGFP) zebrafish line. These data suggest that that Fli1 is a SIRT1-modulated gene in vivo (Fig. 6E).
Figure 6.
Dysregulated gene expression in SIRT1-silenced endothelial cells. HUVECs were transfected with a SIRT1-specific siRNA or a scrambled oligonucleotide siRNA (each n = 3). Total RNA was isolated after 24 h, and the gene expression profile was assessed with the Affymetrix gene chip expression assay. (A,B) Selected genes that were either down-regulated (A) or up-regulated (B) in SIRT1-silenced endothelial cells. (C) RT–PCR analysis of selected genes in scrambled and SIRT1 siRNA-transfected endothelial cells. (D) RT–PCR analysis of selected genes in lung endothelial cells freshly isolated from mice with the respective genotype. (E) Fli1 expression, as revealed by whole-mount in situ hybridization, is reduced in the ISVs (arrowheads) of SIRT1 morphants (SBmo-inj.) compared with that in the control embryos. The GFP signal visualized by fluorescence microscope in live embryos is weaker in SIRT1 morphants. (Panels a,b) Embryos 24 hpf, lateral view; Bar, 50 μm. (Panels c,d) Embryos 4 dpf, lateral view.
Interaction of SIRT1 with Foxo1 in endothelial cells
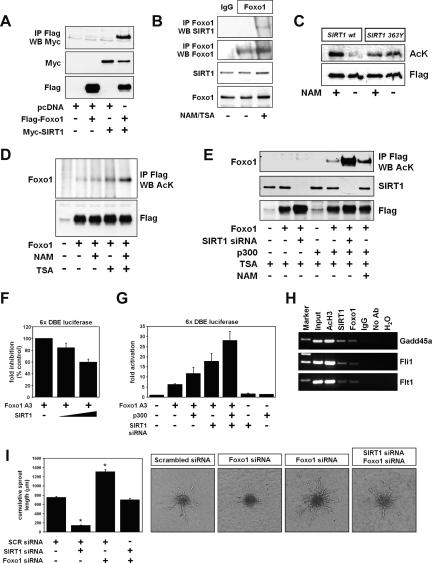
A close inspection of the results from the microarray analysis revealed that many of the genes modified in SIRT1-deficient endothelial cells are also regulated by members of the Foxo family of transcription factors. These are genes such as Gadd45, Id2, Bmp4, or CXCR4. Foxo transcription factors have been identified as molecular targets of SIRT1 (Giannakou and Partridge 2004). In addition, recent studies have identified Foxo1 as a crucial negative transcriptional regulator of blood vessel development (Potente et al. 2005; Paik et al. 2007). We thus tested the hypothesis that the role of SIRT1 during vascular growth and maturation could be mediated, at least in part, by a deacetylation-dependent regulation of Foxo1. To test whether SIRT1 could regulate Foxo acetylation in endothelial cells, we first performed coimmunoprecipitation experiments in HUVECs transfected with Myc-tagged SIRT1 and Flag-tagged Foxo1. An interaction was detected between SIRT1 and Foxo1 when the immunoprecipitation was performed with either an anti-Flag (Fig. 7A) or anti-Myc antibody (data not shown). More importantly, endogenous SIRT1 associated with endogenous Foxo1 under hyperacetylated conditions (Fig. 7B).
Figure 7.
SIRT1 interacts with Foxo1 in endothelial cells. (A) Interaction of SIRT1 with Foxo1. HUVECs were cotransfected with the Flag-tagged Foxo1 and the Myc-tagged SIRT1. Twenty-four hours after transfection, cells were lysed and Flag-tagged Foxo1 was immunoprecipitated. The immune complexes were assessed for the presence of SIRT1 (Myc) by Western blot analysis with antibodies to Myc. The amounts of Flag-Foxo1 and Myc-SIRT1 present in the cell extracts were analyzed with the respective antibodies. (B) Interaction of endogenous SIRT1 with Foxo1. HUVECs were left untreated or incubated with the SIRT1 inhibitor nicotinamide (NAM) and the class I/II HDAC inhibitor TSA for 2 h. Endogenous Foxo1 was immunoprecipitated with a goat polyclonal antibody to Foxo1 or a control preimmune goat serum (IgG). The immune complexes were assessed for the presence of SIRT1 by Western blot with an antibody to SIRT1. Amounts of endogenous Foxo1 and SIRT1 in the cell extracts were determined with antibodies to SIRT1 and Foxo1. (C) In vitro SIRT1 deacetylation assay. Acetylated Flag-tagged Foxo1 was immunoprecipitated using an antibody targeting the Flag epitope. The acetylated Foxo1 was incubated with immunoprecipitated wild-type SIRT1 (wt) or the deacetylase-defective SIRT1 mutant (H363Y) in the presence or absence of nicotinamide (NAM). Acetylation of Foxo1 was assessed by Western blot with antibodies to acetylated lysine. (D) HUVECs expressing Flag-tagged Foxo1 were incubated for 6 h in the absence or presence of the SIRT1 inhibitor nicotinamide (NAM), the class I/II HDAC inhibitor TSA, or combinations thereof. Flag-tagged Foxo1 was immunoprecipitated, and the acetylation of Foxo1 was assessed by Western blot with antibodies to acetylated lysine. (E) HUVECs were transfected with a SIRT1-specific siRNA or a scrambled control oligonucleotide and 24 h later with pcDNA, Flag-tagged Foxo1, p300, or combinations thereof. Endothelial cells were then incubated with TSA for 2 h. Flag-tagged Foxo1 was immunoprecipitated with antibodies targeting the Flag epitope. The acetylation of Foxo1 was assessed by Western blot analysis. (F) HUVECs were transiently transfected with expression plasmids encoding Foxo1 A3, SIRT1, and a forkhead-responsive element-driven luciferase reporter. Twenty-four hours after transfection, cells were lysed and cell extracts were assayed for luciferase expression. (G) HUVECs were transfected with a SIRT1-specific siRNA or a scrambled control siRNA and 24 h later with expression plasmids encoding pcDNA3, Foxo1 A3, p300, and a forkhead-responsive element-driven luciferase reporter. Twenty-four hours after transfection, cells were lysed and cell extracts were assayed for luciferase expression. (H) ChIP assays were performed with chromatin prepared from HUVECs incubated with nicotinamide (NAM) and TSA for 3 h. Chromatin was immunoprecipitated with mouse IgG or antibodies against SIRT1, Foxo1, or acetylated histone H3, and precipitated genomic DNA was analyzed by PCR using primers flanking conserved Foxo-binding sites in the promoter regions of the Gadd45a, Fli1, and Flt1 genes. (I) Representative images and statistical summary of a three-dimensional in vitro angiogenesis assay with collagen gel-embedded endothelial spheroids transfected with combinations of scrambled, SIRT1, and Foxo1 siRNAs. Cumulative sprout length was quantified after 24 h.
We next tested the possibility that SIRT1 might interact with Foxo1 to regulate its deacetylation. For this purpose, we next determined whether SIRT1 could deacetylate Foxo1 directly. Flag-tagged Foxo1 was immunopurified from HUVECs that had been treated with nicotinamide and TSA. Purified Foxo1 was then incubated with SIRT1 in the presence of NAD. As shown in Figure 7C, wild-type SIRT1, but not the deacetylase-defective SIRT1 H363Y mutant, shows the ability to deacetylate Foxo1 in vitro, which was inhibited by nicotinamide.
To confirm these observations in intact cells, we treated HUVECs with nicotinamide or/and TSA. Flag-Foxo1 was immunoprecipitated from cells treated with various combinations of HDAC inhibitors and analyzed with an anti-acetylated lysine antibody. Treatment with TSA alone or nicotinamide alone had only a minor effect on Foxo1 acetylation in cells. Consistent with previous reports (Brunet et al. 2004), incubation of cells with a combination of both inhibitors led to a marked increase in Foxo1 acetylation (Fig. 7D), suggesting that both SIRT1 and some unidentified class I/II HDAC regulate the acetylation levels of Foxo1 in cells. Moreover, loss of SIRT1 induced an enhanced acetylation of Foxo1 either under basal conditions or in the presence of the HAT p300, demonstrating that SIRT1 is a crucial Foxo1 deacetylase in endothelial cells (Fig. 7E).
To gain insight into the regulation of Foxo1 by SIRT1, we next determined the effect of SIRT1 on Foxo-mediated transcription. HUVECs were transfected with a Foxo expression vector and increasing amounts of a SIRT1 expression vector. The transcriptional activity of Foxo1 was determined by cotransfection of a forkhead-responsive element-driven reporter gene. Increasing amounts of SIRT1 correlated with decreased luciferase activity, indicating that SIRT1 negatively regulates Foxo1 transcriptional activity (Fig. 7F). Consistent with this hypothesis, hyperacetylation of Foxo1 by the HAT p300 (Fig. 7E; data not shown) correlated with an increase in Foxo1-dependent transcriptional activity (Fig. 7G). More importantly, knockdown of SIRT1 induced a marked induction of the Foxo1-dependent transcriptional activity, which was further enhanced by cotransfection of p300 (Fig. 7G). Thus, SIRT1-mediated deacetylation of Foxo1 modulates its transcriptional activity and represses its biological activity.
To further validate the biological significance of the SIRT1–Foxo1 interaction, we performed chromatin immunoprecipitation (ChIP) assays and assessed the association of endogenous SIRT1 and Foxo1 with conserved Foxo DNA-binding elements in the promoters of the SIRT1-modulated genes. As shown in Figure 7H, SIRT1 and Foxo1 were detected on the promoters of the SIRT1-regulated genes Gadd45a, Fli1, and Flt1 in the context of native chromatin in HUVECs. We conclude that SIRT1 and Foxo1 associate with promoters of SIRT1-targeted genes.
Knockdown of Foxo1 partially rescues the inhibitory effect of SIRT1 gene silencing on the angiogenic activity of endothelial cells
In order to address whether SIRT1-mediated negative regulation of Foxo1 is capable of restraining the anti-angiogenic activity of Foxo1, we transfected endothelial cells with siRNAs targeting SIRT1 and Foxo1, and assessed the sprout-forming activity of the transfected cells in a spheroid assay. Silencing of Foxo1 gene expression led to an enhanced basal angiogenic activity, as previously reported (Fig. 7I; Potente et al. 2005). More importantly, however, knockdown of Foxo1 partially rescued the inhibitory effects of SIRT1 gene silencing on the sprout-forming activity of endothelial cells (Fig. 7I), suggesting that SIRT1 regulates endothelial angiogenic functions, at least in part, by modulating the transcriptional activity of Foxo1.
Discussion
The transcriptional regulation of vascular homeostasis and blood vessel development is governed by a complex set of several transcription factors and their association with cofactors, which allow for a time- and signal-dependent regulation of gene expression. HDACs act as important cofactors that are recruited by transcription factors to regulate chromatin remodeling, transcription factor activity, and gene transcription (for review, see Cheung et al. 2000; Strahl and Allis 2000). The results of the present study reveal a key role of the class III HDAC SIRT1 in the regulation of endothelial angiogenic functions during blood vessel formation. We show that SIRT1 is highly expressed in the vascular endothelium during sprouting angiogenesis, and that blocking the function of SIRT1 abolishes endothelial sprout formation and migration and the assembly of a primitive vascular network in vitro. Consistent with these results, disruption of SIRT1 gene expression in zebrafish and mice results in dysregulated vascular growth, thus providing evidence for a critical role of SIRT1 in the control of vascular homeostasis.
SIRT1 is the closest mammalian homolog of the yeast protein Sir2 known to be involved in cell and organismal aging. While it is not clear whether SIRT1 similarly regulates ageing and longevity in mammals, SIRT1 has been shown to play a central role in regulating cellular differentiation and senescence and to control metabolic pathways in response to nutrient availability in a wide variety of tissues (Blander and Guarente 2004). The results of the present study expand the role of SIRT1 as an important regulator of tissue homeostasis and point to SIRT1 as a specific modulator of the angiogenic activity of endothelial cells. The physiological significance of SIRT1 functions in the vascular endothelium is underscored by our finding that endothelial-restricted SIRT1 mutant mice display a severe impairment in postnatal neovascularization due to the lack of appropriate vessel growth. The phenotype of these SIRT1 mutant mice demonstrates that SIRT1 acts in the postnatal vascular endothelium to maintain neovascularization capacity in response to angiogenic cues. In the absence of ischemic stress, these mice developed normally, consistent with a role of SIRT1 as a mediator of stress-induced signaling as it has been shown for other cells types and tissues (Luo et al. 2001; Vaziri et al. 2001; Brunet et al. 2004). These findings imply that the angiogenic processes during postnatal vascular growth are sensitive to the proper function of SIRT1 and suggest that none of the other sirtuin family members, which are also expressed in the postnatal endothelium, can fully compensate for a reduction in SIRT1 function.
Moreover, the regulatory role of SIRT1 was not conserved among the sirtuin family members with deacetylase activity in a three-dimensional assay of in vitro angiogenesis. Although we did not explore the functions of SIRT4, SIRT6, and SIRT7, which do not possess deacetylase activity, we hypothesize that SIRT1 plays a unique role among the different family members in the regulation of endothelial angiogenic functions. The ability of wild-type SIRT1, but not of a catalytically inactive mutant, to enhance endothelial angiogenic functions demonstrates that the deacetylase activity of SIRT1 is required for the regulation of endothelial angiogenic functions. The impaired ischemia-induced angiogenesis in the endothelial-restricted SIRT1 mutant mice, in which the SIRT1 protein is nonfunctional due to Cre-mediated excision of the catalytic domain, suggests that this mechanism is also operational during angiogenic processes in vivo.
The development and maturation of the vasculature is a complex process involving multiple gene products all contributing to an integrated sequence of events. Consistent with an important role of SIRT1 in the tightly regulated process of vessel formation, our study identified a number of novel SIRT1-modulated genes involved at different stages of angiogenesis. For example, our data show that loss of SIRT1 down-regulates the expression of genes important for early embryonic vascular development such as the transcription factors Hex and Fli1. Moreover, knockdown of SIRT1 was associated with a reduction of MMP14 (MT1-MMP) expression, a membrane-anchored matrix metalloproteinase critical for the regulation of matrix remodeling and tip cell activity during sprouting angiogenesis (Hiraoka et al. 1998; Yana et al. 2007). Given the defective sprout formation in response to SIRT1 knockdown observed in our study, MMP14 is a likely downstream effector in the SIRT1-dependent signaling cascade controlling endothelial angiogenic functions. In addition, we found several other SIRT1-modulated genes with important functions in the nascent vasculature, such as Flt1, CXCR4, Pdgfβ, and EphB2. Notably, inhibition of SIRT1 expression in zebrafish also induced a marked reduction in the Fli1-driven GFP signal, indicating that Fli1 is a SIRT1-regulated gene in vivo, which might contribute to the defective vascular patterning observed in the SIRT1-deficient zebrafish embryos.
In light of the dysregulated expression of several genes involved in endothelial differentiation and embryonic vascular development, such as Fli1 and Hex, it is indeed tempting to speculate that SIRT1 controls early endothelial differentiation processes. However, the viable phenotype of the endothelial-restricted SIRT1 mutant mice, which does not display obvious abnormalities during embryonic vascular development, indicates that embryonic endothelial differentiation is unaffected by disruption of the SIRT1 gene. Instead, our data support the model that SIRT1 is essentially required for postnatal neovascularization by controlling the angiogenic activity rather than the differentiation of endothelial cells. Consistent with this model, SIRT1-deficient zebrafish have normal numbers of endothelial cells but are characterized by dysregulated endothelial migration and tip cell activity with aberrantly fused blood vessels.
Unlike yeast Sir2, which exclusively deacetylates histones, SIRT1 interacts with several substrates, including transcription factors, which mediate specific functions of SIRT1 (Luo et al. 2001; Vaziri et al. 2001; Fulco et al. 2003; Takata and Ishikawa 2003; Brunet et al. 2004; Motta et al. 2004; Picard et al. 2004; Rodgers et al. 2005). Foxo transcription factors are important targets for SIRT1-dependent deacetylation (Greer and Brunet 2005), and SIRT1 has been shown to tip the balance of Foxo functions away from cell death toward stress resistance (Brunet et al. 2004; Motta et al. 2004). Although we did not observe a dramatic increase in programmed cell death under basal conditions in SIRT1-silenced cells, Foxos are likely molecular targets of SIRT1 in endothelial cells. Foxo1 is highly expressed in the vascular endothelium and has been shown to be an essential negative transcriptional regulator of embryonic and postnatal vessel formation (Daly et al. 2004; Furuyama et al. 2004; Hosaka et al. 2004; Potente et al. 2005; Paik et al. 2007). Indeed, our results demonstrate that SIRT1 binds, deacetylates, and represses the transcriptional activity of Foxo1 and, thus, point to this transcription factor as an effector in the SIRT1-dependent angiogenic signaling pathway. However, the mode of Foxo regulation by SIRT1 remains controversial, with some data suggesting that deacetylation decreases (Motta et al. 2004) and others that it increases Foxo activity (Brunet et al. 2004; Daitoku et al. 2004; van der Horst et al. 2004). We hypothesize that the effects of SIRT1 on Foxo function vary depending on cell type and Foxo target genes. Using a forkhead-responsive element-driven reporter gene, our data show that SIRT1 can act as a repressor of Foxo1-dependent transcriptional activity in endothelial cells. More importantly, knockdown of Foxo1 partially rescued the inhibitory effects of SIRT1 gene silencing on the angiogenic activity of endothelial cells indicating that SIRT1 regulates endothelial angiogenic functions, at least in part, by modulating the transcriptional activity of Foxo1. Nevertheless, the inhibitory effects of SIRT1 on Foxo1 activity were incomplete, suggesting that additional cofactors contribute to the regulation of Foxo1 activity. Given the experimental observation that Foxo1 is strongly acetylated in the presence of a combined inhibition of all three HDAC classes, it is tempting to speculate that an unidentified class I/II HDAC acts as additional Foxo1 deacetylase/corepressor to fully block its transcriptional and biological activity.
In addition to Foxo transcription factors, we speculate that SIRT1 may have other transcriptional partners in vascular endothelial cells to mediate its specific effects. For example, SIRT1 has been shown to interact with Hairy and Enhancer-of-split basic helix–loop–helix (bHLH) transcriptional repressors such as Hey2 to mediate transcriptional repression (Rosenberg and Parkhurst 2002; Takata and Ishikawa 2003). Indeed, Hey2, which is the human homolog of the zebrafish gene gridlock, is an important regulator of endothelial gene expression and differentiation (Chi et al. 2003), and mice homozygous for a combined loss of Hey1 and Hey2 are embryonically lethal due to defects in vascular development (Fischer et al. 2004). In the future, it will be interesting to identify novel SIRT1-regulated transcription factors and to investigate their involvement in vascular development and disease.
Taken together, the findings of the present study identify SIRT1 as a critical regulator of postnatal angiogenic responses and vascular growth by functioning as a transcriptional cofactor regulating the expression of genes involved in angiogenesis and endothelial homeostasis. Since SIRT1 has been linked to mammalian ageing, the regulation of SIRT1 activity and/or expression might provide an opportunity for counteracting vascular rarefaction associated with organismal ageing. Moreover, it will be interesting to study the involvement of SIRT1 in metabolic syndrome- and atherosclerosis-related vascular disease, given its role as a metabolic sensor regulating glucose and fatty acid metabolism. Thus, these findings point to SIRT1 as a potential therapeutic target for vascular-related diseases in the adult.
Materials and methods
Generation of SIRT1 mutant mice
SIRT1flox/flox, SIRT1−/−, and Tie2-Cre transgenic mouse lines have been described (Koni et al. 2001; Cheng et al. 2003; Chua et al. 2005). Endothelium-specific deletion of the floxed exon 4 of the SIRT1 gene was achieved by crossing mice heterozygous for the SIRT1-null allele with Tie2-Cre transgenic mice, which were subsequently crossed to SIRT1flox/flox mice. Mice were on a mixed genetic background (129/Sv × C57BL/6) and received standard rodent chow.
Morpholino inhibition in zebrafish
Fish of the tg(fli1:EGFP) line (Lawson and Weinstein 2002) were maintained/raised according to EU regulations on laboratory animals. cDNA sequences corresponding to Danio rerio SIRT1 were identified through Blast search. In situ hybridization was performed with probes transcribed from two EST clones: BI563162 (fr89c08.y1) and BM316937 (fw77b01.y1); both gave similar results. To identify genomic sequences corresponding to SIRT1 we used the University of California at Santa Cruz genomic browser and the Blat tool. Embryos were injected at the one-cell stage with the morpholinos ATGmo (5′-CCGTTTA TTTTCGCCGTCCGCCATC-3′; 4 nL of a 0.3 mM, 0.2 mM, or 0.15 mM solution) and SBmo (5′-CGAACCAAACTCACCAAT CTGTGGC-3′; 4 nL of a 0.6 mM or 1.2 mM solution). Immunohistochemistry was performed as described previously (Costagli et al. 2002). “Standard control morpholinos” (GeneTools) were injected at the same concentrations and gave no phenotype. Pictures were taken from living embryos with a confocal microscope to visualize the GFP fluorescent signal or from fixed embryos after immunohistochemistry with anti-GFP antiserum (Upstate Biotechnology) using a stereoscope microscope.
Isolation of murine endothelial cells
Murine endothelial cells were isolated and purified as described (Fleming et al. 2005). Mice of the respective genotypes were sacrificed, and the lungs were removed and incubated with dispase. Thereafter, the homogenate was filtered through a cell strainer, collected by centrifugation, and washed with phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA). The resulting cell suspension was incubated with anti-mouse vascular endothelial-cadherin antibody-coated (Pharmingen, Becton Dickinson) magnetic beads (Dynal Biotech). The magnetic beads were washed with PBS/BSA; resuspended in D-MEM/F12 (Invitrogen) supplemented with 20% fetal calf serum, endothelial cell growth factor (Promocell), penicillin, and streptomycin; and seeded on gelatin-coated culture dishes. For the first three passages the cells were repurified with vascular endothelial-cadherin antibody-coated magnetic beads.
RNAi
To silence SIRT gene expression, transfection of siRNA duplexes was performed using GeneTrans II (MoBiTec) as described previously (Potente et al. 2005). SIRT1, SIRT2, SIRT3, SIRT5, and Foxo1 siRNAs were synthesized by Eurogentec. A nonrelated scrambled siRNA without any other match in the human genomic sequence was used as a control (Supplemental Material).
ChIP assay
A ChIP kit was used for the assays according to manufacturer’s instructions (Upstate Biotechnology). Acetyl-histone3 antibody and SIRT1 antibodies were purchased from Upstate Biotechnology. Foxo1 antibodies were purchased from Santa Cruz Biotechnology (H-128) and Cell Signaling Technology. Primer sequences are available on request.
Spheroid-based angiogenesis assay
Endothelial cell spheroids of defined cell number were generated as described previously (Potente et al. 2005). Spheroids were generated overnight, after which they were embedded into collagen gels. Angiogenic activity was quantified by measuring the cumulative length of the sprouts that had grown out of each spheroid using a digital imaging software (Axioplan, Zeiss) analyzing five to 10 spheroids per experimental group and experiment.
Murine model of hindlimb ischemia
The role of endothelial SIRT1 in ischemia-induced vessel formation was assessed in a murine model of hindlimb ischemia using SIRT1flox/+ and Tie2Cretg;SIRT1flox/− mice. The present study was performed with the permission of the State of Hesse, Regierungspräsidium Darmstadt, according to section 8 of the German Law for the Protection of Animals, and conforms to the German Guide for the Care and Use of Laboratory Animals. The model and histological analysis was performed as described previously (Potente et al. 2005).
Acknowledgments
We are indebted to B. Zimmermann, A. Fischer, M. Muhly-Reinholz, and T. Röxe for expert technical assistance. This work is supported by the DFG (PO1306/1-1 and FOR 503 Di 600/2-3) and by the European Vascular Genomics Network (http://www.evgn.org), a Network of Excellence supported by the European Community’s sixth Framework Programme for Research Priority 1 “Life sciences, genomics and biotechnology for health” (contract no. LSHM-CT-2003-503254).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.435107
References
- Blander G., Guarente L., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bordone L., Guarente L., Guarente L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Ross S.E., Mostoslavsky R., Cohen H.Y., Mostoslavsky R., Cohen H.Y., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Chang S., Young B.D., Li S., Qi X., Richardson J.A., Olson E.N., Young B.D., Li S., Qi X., Richardson J.A., Olson E.N., Li S., Qi X., Richardson J.A., Olson E.N., Qi X., Richardson J.A., Olson E.N., Richardson J.A., Olson E.N., Olson E.N. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F., Bronson R., Appella E., Alt F.W., Chua K.F., Appella E., Alt F.W., Chua K.F., Alt F.W., Chua K.F., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Allis C.D., Sassone-Corsi P., Allis C.D., Sassone-Corsi P., Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Chi J.T., Chang H.Y., Haraldsen G., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Chang H.Y., Haraldsen G., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Haraldsen G., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Chang D.S., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Wang Z., Rockson S.G., de van Rijn M., Botstein D., Rockson S.G., de van Rijn M., Botstein D., de van Rijn M., Botstein D., Botstein D., et al. Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.F., Mostoslavsky R., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Mostoslavsky R., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Cheng H.L., Fischer M.R., Stokes N., Fischer M.R., Stokes N., Stokes N., et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Costagli A., Kapsimali M., Wilson S.W., Mione M., Kapsimali M., Wilson S.W., Mione M., Wilson S.W., Mione M., Mione M. Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J. Comp. Neurol. 2002;450:73–93. doi: 10.1002/cne.10292. [DOI] [PubMed] [Google Scholar]
- Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A., Miyagishi M., Nakajima T., Fukamizu A., Nakajima T., Fukamizu A., Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly C., Wong V., Burova E., Wei Y., Zabski S., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Wong V., Burova E., Wei Y., Zabski S., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Burova E., Wei Y., Zabski S., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Wei Y., Zabski S., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Zabski S., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Griffiths J., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Lai K.M., Lin H.C., Ioffe E., Yancopoulos G.D., Lin H.C., Ioffe E., Yancopoulos G.D., Ioffe E., Yancopoulos G.D., Yancopoulos G.D., et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1) Genes & Dev. 2004;18:1060–1071. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter A.J., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B., Caron H.N., Kemp S., van Kuilenburg A.B., Kemp S., van Kuilenburg A.B., van Kuilenburg A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroanne C.F., Bonjean K., Servotte S., Devy L., Colige A., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Bonjean K., Servotte S., Devy L., Colige A., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Servotte S., Devy L., Colige A., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Devy L., Colige A., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Colige A., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Clausse N., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Blacher S., Verdin E., Foidart J.M., Nusgens B.V., Verdin E., Foidart J.M., Nusgens B.V., Foidart J.M., Nusgens B.V., Nusgens B.V., et al. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M., Schumacher N., Maier M., Sendtner M., Gessler M., Maier M., Sendtner M., Gessler M., Sendtner M., Gessler M., Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes & Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming I., Fisslthaler B., Dixit M., Busse R., Fisslthaler B., Dixit M., Busse R., Dixit M., Busse R., Busse R. Role of PECAM-1 in the shear-stress-induced activation of Akt and the endothelial nitric oxide synthase (eNOS) in endothelial cells. J. Cell Sci. 2005;118:4103–4111. doi: 10.1242/jcs.02541. [DOI] [PubMed] [Google Scholar]
- Frescas D., Valenti L., Accili D., Valenti L., Accili D., Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fulco M., Schiltz R.L., Iezzi S., King M.T., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., Schiltz R.L., Iezzi S., King M.T., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., Iezzi S., King M.T., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., King M.T., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., Zhao P., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., Kashiwaya Y., Hoffman E., Veech R.L., Sartorelli V., Hoffman E., Veech R.L., Sartorelli V., Veech R.L., Sartorelli V., Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Furuyama T., Kitayama K., Shimoda Y., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Kitayama K., Shimoda Y., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Shimoda Y., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Hisatsune H., Nishikawa S., Nakayama K., Ikeda K., Nishikawa S., Nakayama K., Ikeda K., Nakayama K., Ikeda K., Ikeda K., et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J. Biol. Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Giannakou M.E., Partridge L., Partridge L. The interaction between FOXO and SIRT1: Tipping the balance towards survival. Trends Cell Biol. 2004;14:408–412. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Greer E.L., Brunet A., Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Guarente L., Picard F., Picard F. Calorie restriction—The SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Hiraoka N., Allen E., Apel I.J., Gyetko M.R., Weiss S.J., Allen E., Apel I.J., Gyetko M.R., Weiss S.J., Apel I.J., Gyetko M.R., Weiss S.J., Gyetko M.R., Weiss S.J., Weiss S.J. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 1998;95:365–377. doi: 10.1016/s0092-8674(00)81768-7. [DOI] [PubMed] [Google Scholar]
- Hosaka T., Biggs W.H., Tieu D., Boyer A.D., Varki N.M., Cavenee W.K., Arden K.C., Biggs W.H., Tieu D., Boyer A.D., Varki N.M., Cavenee W.K., Arden K.C., Tieu D., Boyer A.D., Varki N.M., Cavenee W.K., Arden K.C., Boyer A.D., Varki N.M., Cavenee W.K., Arden K.C., Varki N.M., Cavenee W.K., Arden K.C., Cavenee W.K., Arden K.C., Arden K.C. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illi B., Nanni S., Scopece A., Farsetti A., Biglioli P., Capogrossi M.C., Gaetano C., Nanni S., Scopece A., Farsetti A., Biglioli P., Capogrossi M.C., Gaetano C., Scopece A., Farsetti A., Biglioli P., Capogrossi M.C., Gaetano C., Farsetti A., Biglioli P., Capogrossi M.C., Gaetano C., Biglioli P., Capogrossi M.C., Gaetano C., Capogrossi M.C., Gaetano C., Gaetano C. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ. Res. 2003;93:155–161. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- Illi B., Scopece A., Nanni S., Farsetti A., Morgante L., Biglioli P., Capogrossi M.C., Gaetano C., Scopece A., Nanni S., Farsetti A., Morgante L., Biglioli P., Capogrossi M.C., Gaetano C., Nanni S., Farsetti A., Morgante L., Biglioli P., Capogrossi M.C., Gaetano C., Farsetti A., Morgante L., Biglioli P., Capogrossi M.C., Gaetano C., Morgante L., Biglioli P., Capogrossi M.C., Gaetano C., Biglioli P., Capogrossi M.C., Gaetano C., Capogrossi M.C., Gaetano C., Gaetano C. Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circ. Res. 2005;96:501–508. doi: 10.1161/01.RES.0000159181.06379.63. [DOI] [PubMed] [Google Scholar]
- Imai S., Armstrong C.M., Kaeberlein M., Guarente L., Armstrong C.M., Kaeberlein M., Guarente L., Kaeberlein M., Guarente L., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kim M.S., Kwon H.J., Lee Y.M., Baek J.H., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Kwon H.J., Lee Y.M., Baek J.H., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Lee Y.M., Baek J.H., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Baek J.H., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., Kim H.S., Lee S.K., Chung H.Y., Lee S.K., Chung H.Y., Chung H.Y., et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat. Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- Koni P.A., Joshi S.K., Temann U.A., Olson D., Burkly L., Flavell R.A., Joshi S.K., Temann U.A., Olson D., Burkly L., Flavell R.A., Temann U.A., Olson D., Burkly L., Flavell R.A., Olson D., Burkly L., Flavell R.A., Burkly L., Flavell R.A., Flavell R.A. Conditional vascular cell adhesion molecule 1 deletion in mice: Impaired lymphocyte migration to bone marrow. J. Exp. Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J., Slama J.T., Sternglanz R., Slama J.T., Sternglanz R., Sternglanz R. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Langley E., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T., Minucci S., Pelicci P.G., Kouzarides T., Pelicci P.G., Kouzarides T., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N.D., Weinstein B.M., Weinstein B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W., Chen D., Su F., Shiloh A., Guarente L., Gu W., Su F., Shiloh A., Guarente L., Gu W., Shiloh A., Guarente L., Gu W., Guarente L., Gu W., Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Motta M.C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L., Gu W., Bultsma Y., McBurney M., Guarente L., Bultsma Y., McBurney M., Guarente L., McBurney M., Guarente L., Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- North B.J., Verdin E., Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E., Marshall B.L., Borra M.T., Denu J.M., Verdin E., Borra M.T., Denu J.M., Verdin E., Denu J.M., Verdin E., Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Miao L., Tothova Z., Horner J.W., Carrasco D.R., Tothova Z., Horner J.W., Carrasco D.R., Horner J.W., Carrasco D.R., Carrasco D.R., et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., Chung N., Topark-Ngarm A., Senawong T., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., Topark-Ngarm A., Senawong T., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., Senawong T., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., De Machado Oliveira R., Leid M., McBurney M.W., Guarente L., Leid M., McBurney M.W., Guarente L., McBurney M.W., Guarente L., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M., Urbich C., Sasaki K., Hofmann W.K., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Urbich C., Sasaki K., Hofmann W.K., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Sasaki K., Hofmann W.K., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Hofmann W.K., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Heeschen C., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Aicher A., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., Kollipara R., DePinho R.A., Zeiher A.M., Dimmeler S., DePinho R.A., Zeiher A.M., Dimmeler S., Zeiher A.M., Dimmeler S., Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P., Gygi S.P., Spiegelman B.M., Puigserver P., Spiegelman B.M., Puigserver P., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rosenberg M.I., Parkhurst S.M., Parkhurst S.M. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- Rossig L., Li H., Fisslthaler B., Urbich C., Fleming I., Forstermann U., Zeiher A.M., Dimmeler S., Li H., Fisslthaler B., Urbich C., Fleming I., Forstermann U., Zeiher A.M., Dimmeler S., Fisslthaler B., Urbich C., Fleming I., Forstermann U., Zeiher A.M., Dimmeler S., Urbich C., Fleming I., Forstermann U., Zeiher A.M., Dimmeler S., Fleming I., Forstermann U., Zeiher A.M., Dimmeler S., Forstermann U., Zeiher A.M., Dimmeler S., Zeiher A.M., Dimmeler S., Dimmeler S. Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ. Res. 2002;91:837–844. doi: 10.1161/01.res.0000037983.07158.b1. [DOI] [PubMed] [Google Scholar]
- Rossig L., Urbich C., Bruhl T., Dernbach E., Heeschen C., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Urbich C., Bruhl T., Dernbach E., Heeschen C., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Bruhl T., Dernbach E., Heeschen C., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Dernbach E., Heeschen C., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Heeschen C., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Chavakis E., Sasaki K., Aicher D., Diehl F., Seeger F., Sasaki K., Aicher D., Diehl F., Seeger F., Aicher D., Diehl F., Seeger F., Diehl F., Seeger F., Seeger F., et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J. Exp. Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Brachmann C.B., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Brachmann C.B., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Grubmeyer C., Wolberger C., Wolberger C., et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Takata T., Ishikawa F., Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- van der Horst A., Tertoolen L.G., de Vries-Smits L.M., Frye R.A., Medema R.H., Burgering B.M., Tertoolen L.G., de Vries-Smits L.M., Frye R.A., Medema R.H., Burgering B.M., de Vries-Smits L.M., Frye R.A., Medema R.H., Burgering B.M., Frye R.A., Medema R.H., Burgering B.M., Medema R.H., Burgering B.M., Burgering B.M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J. Biol. Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A., Pandita T.K., Guarente L., Weinberg R.A., Guarente L., Weinberg R.A., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Yana I., Sagara H., Takaki S., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Sagara H., Takaki S., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Takaki S., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Katsuki M., Taniguchi S., Aoki T., Sato H., Taniguchi S., Aoki T., Sato H., Aoki T., Sato H., Sato H., et al. Crosstalk between neovessels and mural cells directs the site-specific expression of MT1-MMP to endothelial tip cells. J. Cell Sci. 2007;120:1607–1614. doi: 10.1242/jcs.000679. [DOI] [PubMed] [Google Scholar]
- Yang X.J. Lysine acetylation and the bromodomain: A new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hou H., Haller E.M., Nicosia S.V., Bai W., Hou H., Haller E.M., Nicosia S.V., Bai W., Haller E.M., Nicosia S.V., Bai W., Nicosia S.V., Bai W., Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]