Abstract
Adenosine receptor ligands have anti-inflammatory effects and modulate immune responses by up-regulating IL-10 production by immunostimulated macrophages. The adenosine receptor family comprises G protein-coupled heptahelical transmembrane receptors classified into four types: A1, A2A, A2B, and A3. Our understanding of the signaling mechanisms leading to enhanced IL-10 production following adenosine receptor occupancy on macrophages is limited. In this study, we demonstrate that adenosine receptor occupancy increases IL-10 production by LPS-stimulated macrophages without affecting IL-10 promoter activity and IL-10 mRNA levels, indicating a posttranscriptional mechanism. Transfection experiments with reporter constructs containing sequences corresponding to the AU-rich 3′-untranslated region (UTR) of IL-10 mRNA confirmed that adenosine receptor activation acts by relieving the translational repressive effect of the IL-10 3′-UTR. By contrast, adenosine receptor activation failed to liberate the translational arrest conferred by the 3′-UTR of TNF-α mRNA. The IL-10 3′-UTR formed specific complexes with proteins present in cytoplasmic extracts of RAW 264.7 cells. Adenosine enhanced binding of proteins to a region of the IL-10 3′-UTR containing the GUAUUUAUU nonamer. The stimulatory effect of adenosine on IL-10 production was mediated through the A2B receptor, because the order of potency of selective agonists was 5′-N-ethylcarboxamidoadenosine (NECA) > N6-(3-iodobenzyl)-adenosine-5′-N-methyluronamide (IB-MECA) > 2-chloro-N6-cyclopentyladenosine (CCPA) = 2-p-(2-carboxyethyl)-phenethylamino-5′-N-ethyl-carboxamidoadenosine (CGS-21680). Also, the selective A2B antagonist, alloxazine, prevented the effect of adenosine. Collectively, these studies identify a novel pathway in which activation of a G protein-coupled receptor augments translation of an anti-inflammatory gene.
Interleukin-10 is an important immunomodulatory cytokine which has attracted much attention because of its anti-inflammatory and immunosuppressive properties. IL-10 was initially described as a Th2 product that restrained the secretion of cytokines by Th1 T cell clones (1). Subsequently, it has become clear that IL-10 is also produce by cells of the monocyte/macrophage lineage (2) and monocytes/macrophages appear to be the predominant cell type secreting IL-10 following LPS administration in vivo (3). Macrophages exposed to LPS secrete IL-10 with a later onset when compared with proinflammatory cytokines. This belated production of IL-10 represents an essential autoregulatory mechanism that limits excessive production of the proinflammatory cytokines TNF-α, IL-1, and IL-12 (4), thereby contributing to both the limitation and resolution of inflammation (5).
In recent years, progress has been made toward delineating the cellular mechanisms by which macrophage IL-10 expression is regulated, and it has been shown that this regulation occurs both at the transcriptional and posttranscriptional levels. Induction of IL-10 gene expression following LPS administration depends on a transcriptional process initiated by binding of the transcription factors Sp1 (6–9), Sp3 (9), Stat3 (10), C/EBPβ and γ (9), and c-Maf (11) to the IL-10 promoter. cAMP-elevating agents trigger IL-10 promoter activity and IL-10 secretion in human monocytic cells, secondary to an enhanced binding of CREB, as well as C/EBPα and β, to the IL-10 promoter (12). Much less is known about the posttranscriptional regulation of IL-10. A recent study reported that transfer of sequences in the 3′-untranslated region (3′-UTR)3 of the mouse IL-10 gene to a luciferase reporter construct decreased luciferase activity in transfected mouse macrophages when compared with the empty vector (13). This observation indicates that the 3′-UTR of the IL-10 mRNA may regulate the translation and/or stability of the IL-10 mRNA.
Adenosine, an endogenous purine nucleoside, is a biologically active extracellular signaling molecule that is formed at sites of metabolic stress associated with hypoxia, ischemia, trauma, or inflammation. Adenosine mediates many of its effects through one or more of four receptors, the adenosine A1, A2A, A2B, and A3 receptors (14, 15). These receptors are seven membrane-spanning proteins that couple to heterotrimeric G proteins to trigger a variety of intracellular signaling pathways (14, 15). Cells of the monocyte/macrophage lineage have been documented to express all four adenosine receptors (16). Stimulation of adenosine receptors has been shown to result in an anti-inflammatory, deactivated macrophage phenotype (16, 17). In LPS-stimulated monocytes/macrophages, adenosine receptor activation attenuates the production of several proinflammatory cytokines, including TNF-α (18–24) and IL-12 (22, 25, 26). In contrast to the suppressive effect of adenosine on the production of these proinflammatory mediators, adenosine up-regulates IL-10 production by LPS-stimulated monocytes/macrophages (22, 26). The mode of action of adenosine in up-regulating IL-10 production is unclear and so is the identity of receptors responsible for the adenosine enhancement of IL-10 production in LPS-activated macrophages.
In this study, we demonstrate that although LPS induces both IL-10 promoter activity and IL-10 mRNA accumulation, adenosine fails to alter the magnitude of these responses, attesting to a translational rather than a transcriptional effect of adenosine on the stimulation of IL-10 production. We confirm this mechanism by showing that the 3′-UTR of the IL-10 mRNA has a substantial inhibitory effect on the translation, but not transcription, of a reporter construct, and that this repressive effect is partially relieved by adenosine. Furthermore, we report that regions of the IL-10 3′-UTR form specific complexes with proteins present in macrophage extracts, and that adenosine enhances the formation of these complexes. Finally, our results reveal that A2B receptors have a critical role in mediating the potentiating effect of adenosine on IL-10 secretion.
Materials and Methods
Cell culture
The mouse macrophage cell line RAW 264.7 was obtained from the American Type Culture Collection. Peritoneal macrophages from male CD-1 mice (Charles River Laboratories) were isolated using thioglycolate elicitation as described previously (24). All mice were maintained in accordance with the recommendations of the “Guide for the Care and Use of Laboratory Animals”, and the experiments were approved by the New Jersey Medical School Animal Care Committee. Cells were grown in DMEM supplemented with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1.5 mg/ml sodium bicarbonate in a humidified atmosphere of 95% air and 5% CO2.
Drugs and reagents
Adenosine, the selective A1 receptor agonist 2-chloro-N6-cyclopentyladenosine (CCPA), A2A receptor agonist 2-p-(2-carboxyethyl)phenethylamino-5′-N-ethyl-carboxamidoadenosine (CGS-21680), A2B receptor agonist 5′-N-ethylcarboxamidoadenosine (NECA), and A3 receptor agonist N6-(3-iodobenzyl)-adenosine-5′-N-methyluronamide (IB-MECA) were purchased from Sigma-Aldrich. SB203580 was purchased from Calbiochem. LPS and inosine were obtained from Sigma-Aldrich (Escherichia coli 055:B5). Stock solutions of the various agonists and antagonists, as well as SB203580, were prepared using dimethylsulphoxide.
DNA constructs
The IL-10 promoter vector constructed by inserting the mouse IL-10 promoter (−1538/+64) into the pGL2B luciferase (Promega) polylinker used in our studies was a gift from Dr. S. T. Smale (University of California, Los Angeles, CA) (6). Luciferase reporter plasmids containing regions of the IL-10 3′-UTR have been described previously (13). In brief, four fragments of cDNA corresponding to AU1 (+727 to +818), AU2 (+807 to +936), AU3 (+1157 to +1239), and AU4 (+727 to +1239) regions of the 3′-UTR of IL-10 mRNA were cloned into an XbaI site (+1934) located between the luciferase gene and the poly(A) signal in the pGL3-control vector carrying a SV40 promoter/luciferase expression unit. A construct containing the 3′-UTR of the TNF-α cDNA inserted into the XbaI site of the pGL3-control vector was provided by Dr. C. Gueydan (Free University, Brussels, Belgium) (27).
The DNA constructs used for in vitro transcription to produce riboprobes for specific regions of the IL-10 3′-UTR were constructed as follows. Fragments corresponding to the AU1, AU2, and AU3 regions were generated by RT-PCR using the TaqPCR Core kit (Qiagen) with RNA isolated from LPS-treated RAW cells. The following primers were used: AU1: 5′-GGACCGGTCCTCTCTCCGAAATATTTATTACC-3′ and 5′-GGGACGTCGGGCTTCTTTCTAAATAGTTCACA-3′; AU2: 5′-GGACCGGTGAAAGAAGCCCAATATTAT-3′ and 5′-GGGACGTCGAAGAACCCCTCCCATCATATA-3′; AU3: 5′-GGACCGGTCTGAAAACTGTGGCCAGTTTG-3′ and 5′-GGGACGTCTACTTCTAGTTAAAAATGAG-3′. Amplified cDNA fragments were inserted into the 3′-U overhangs of the cloning vector pDrive (Qiagen PCR cloning kit). A mutant AU2 DNA fragment lacking the GTATTTATT nonamer was prepared by synthesizing both strands, followed by annealing the two strands, and ligating the resultant DNA into pDrive.
Transient transfection and luciferase activity
For transient transfections, cells were seeded either in a 6- or a 24-well tissue culture dish for IL-10 promoter or IL-10 3′-UTR studies, respectively, 1 day before transient transfection. For the promoter studies, confluent monolayers of cells were transfected with 10 μl/ml FuGENE 6 Transfection Reagent (Roche) in 700 μl of medium per well. The medium contained 3.5 μg/ml IL-10 luciferase promoter construct, and 2.3 μg/ml control Renilla luciferase (pRL-TK; Promega) plasmid. After an overnight transfection, the medium was replaced, and the cells were treated with adenosine (100 μM) or its vehicle (medium), in the presence or absence of LPS (10 μg/ml), for 8 h. Luciferase activity was measured using the Dual Luciferase Reporter 1000 Assay System (Promega), as we have described previously (28).
For studies with IL-10 3′-UTR constructs, cells were transfected with 10 μl/ml FuGENE 6 Transfection Reagent (Roche) in 200 μl of medium per well. The medium contained 3.1 μg/ml IL-10 3′-UTR promoter constructs (AU1, AU2, AU3, and AU4) or the TNF-α-3′-UTR construct, or 1.75 μg/ml pGL3-control. After a 4-h transfection, the medium was replaced with fresh medium, and the cells were treated with adenosine, NECA, or vehicle in the presence or absence of LPS (10 μg/ml) for 5 h. Renilla (control) constructs were not used in these experiments, because in pilot studies we found that inclusion of Renilla vectors caused a trans effect between promoters on these cotransfected plasmids (data not shown). Luciferase activity was measured using the Luciferase Reporter Assay System (Promega), and normalized relative to the micrograms of protein, as we have described previously (29).
Analysis of gene expression by real-time RT-PCR
Total RNA was prepared from cells and reverse-transcribed, as previously described (30). For detection of IL-10 mRNA, real-time PCR commercial kits (Applied Biosystems) specific for IL-10 or rRNA were used, and all data were normalized to constitutive rRNA values. Luciferase transcript levels were measured using the following probes: 5′-ATGGAAGACGCCAAAAACATAAAG-3′ and 5′-ATAGCTTCTGCCAACCGAAC-3′. The Applied Biosystems 7700 sequence detector was used for amplification of target mRNA, and quantitation of differences between treatment groups was calculated according to the manufacturer's instructions.
Preparation of cytosolic extracts for RNA EMSA and UV cross-linking studies
Confluent RAW 264.7 cells were incubated with adenosine or its vehicle (medium) for varying time periods, after which, the cells were rinsed once with ice-cold PBS (Sigma-Aldrich), and then scraped into buffer A, consisting of 10 mM HEPES buffer (pH 7.9), 1.5 mM MgCl2,10 mM KCl, 1 mM DTT, and 1 mM PMSF (all from Sigma-Aldrich). After a brief centrifugation at 600 × g, the pellets were resuspended in two original packed cell volumes of buffer A and transferred into a precooled 1-ml Dounce homogenizer. After lysis of cells by 10 strokes of a B-type pestle, the nuclei were pelleted at 4000 × g for 2 min at 4°C. The cytoplasmic fraction was transferred to new tubes and 0.11 vol of buffer B (0.3 M HEPES (pH 7.9), 1.4 M KCl, 30 mM MgCl2) was added. The mixture was then cleared by centrifugation at 16,000 × g for 15 min, the supernatant transferred to fresh tubes, and 80% glycerol added to a final concentration of 10% (v/v). The cytosolic extracts were immediately frozen at −80°C.
In vitro transcription of RNA substrates
RNA transcripts were synthesized by use of T7 or SP6 RNA polymerase according to the manufacturer's instructions (Promega), in the presence of 50 μCi of [32P]UTP (Amersham Biosciences), as described previously (31). RNAs were gel-purified from 5% polyacrylamide, 8 M urea gels by overnight crush elution in high salt buffer (0.4 M NaCl, 50 mM Tris (pH 8.0), 0.1% SDS) before use in reactions. Eluted RNAs were ethanol-precipitated and resuspended in water.
RNA EMSA
For RNA-cytosolic extract binding reactions, 10 μg of cytosolic extract were incubated with 5 × 104 dpm [32P]RNA sequence at 25°C for 30 min in buffer D consisting of 20 mM HEPES (pH 7.9), 20% (v/v) glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM PMSF, 1 mM DTT. Samples were then subjected to electrophoresis at a constant 200 volts on a 4% native poly acrylamide gel (acrylamide/bisacrylamide ratio of 29:1; Sigma-Aldrich) at 4°C. The radiolabeled RNA-protein complexes were detected by autoradiography on Kodak BioMax MR film, and their relative band intensities determined by densitometric analysis. In RNA competition assays, unlabeled, homologous RNA sequences were added in 100-fold excess to the binding reactions before the addition of radiolabeled RNA probes.
UV cross-linking and SDS-PAGE
RNA-protein binding reactions were conducted using 20 μg of cytosolic extract and 105 dpm of [32P]RNA sequences. Following the binding reaction, samples were irradiated with UV light (2500 μJ) (CL-1000, UV Crosslinker; UVP) on ice for 15 min (32). Samples were briefly boiled in SDS sample buffer and RNA-protein complexes resolved on 8–16% Trisglycine gels (Invitrogen Life Technologies). Gels were then dried for 15 min at 80°C, and exposed to Kodak BioMax MR film and analyzed by autoradiography. Precision Plus Protein Dual Color Protein Standards (Bio-Rad) were used to estimate the m.w. of complexes.
IL-10 ELISA
Cells in 24-well plates were treated with adenosine or various adenosine receptor agonists followed immediately by addition of 10 μg/ml LPS. Supernatants for IL-10 determination were obtained 5 h after stimulation with LPS. IL-10 levels in cell supernatants were determined by ELISA, using the IL-10 Duoset ELISA kit (DuoSet; R&D Systems) according to the manufacturer's instructions.
Preparation of cell membrane extracts and analysis of A2B receptor expression by Western blotting
Cell monolayers were washed in PBS and removed from culture dishes by scraping. Cells were then homogenized in hypotonic buffer (10 mM KCl, 1.5 mM MgCl2, 10 mM Tris (pH 7.4)) using a Dounce homogenizer in the presence of protease inhibitors (1 μg/ml aprotinin, 1 μg/ml leupeptin 1 μg/ml pepstatin, and 1 mM PMSF). Unbroken cells and nuclei were removed by low-speed centrifugation at 2000 × g for 10 min. To pellet the crude membrane fraction, the supernatants were centrifuged at 100,000 × g for 30 min. The membrane pellet was resuspended in TNE buffer (10 mM Tris, 10 mM NaCl, 1 mM EDTA), 30% glycerol, and protease inhibitors. The protein concentration was determined as described above. Equal amounts (100 μg) of protein were separated by electrophoresis on a 10% SDS polyacrylamide gel, and transferred to a nitrocellulose membrane. The membrane was then probed with polyclonal goat anti-A2B adenosine receptor (no. sc-7507; Santa Cruz Biotechnology), followed by incubation with a secondary HRP-conjugated anti-goat Ab (no. A5420; Sigma-Aldrich). Specific immune complexes were detected using the ECL Western Blotting Detection Reagent (Amersham Biosciences).
Results
Adenosine receptor agonists up-regulate IL-10 production by LPS-activated RAW 264.7 macrophages
To study the regulation of IL-10 production by adenosine, we used RAW 264.7 macrophages; these cells have been shown to express adenosine receptors (20, 27, 28, 33), and many aspects of the regulation of IL-10 gene expression have been uncovered using this cell line (6, 7, 13). RAW 264.7 macrophages produce constitutively low levels of IL-10. LPS stimulation for 5 h resulted in an ∼10-fold increase in IL-10 production (Fig. 1A). In LPS-activated cells, adenosine (100 μM) augmented IL-10 production by ∼2.4-fold. Adenosine had no effect on IL-10 production by unstimulated macrophages (Fig. 1A). In addition, treatment of mouse peritoneal macrophages with adenosine immediately before LPS administration potentiated LPS-induced IL-10 production, as measured 5 h after LPS stimulation. The IL-10 concentration was 0.28 ± 0.01 ng/ml in LPS-stimulated cells, and 0.58 ± 0.06 ng/ml in cells cotreated with adenosine and LPS (p < 0.01). Because adenosine exerts most of its cellular effects via occupancy of adenosine receptors, we next sought to determine which adenosine receptor(s) were responsible for the potentiating effect of adenosine on IL-10 production by LPS in RAW 264.7 macrophages. To address this question, we first tested the effect of selective adenosine receptor agonists on IL-10 production. The selective A1 receptor agonist CCPA or the selective A2A receptor agonist CGS-21680 failed to mimic the stimulatory effect of adenosine on LPS-induced IL-10 production. However, both the selective A3 receptor agonist IB-MECA and the A2B receptor agonist NECA increased IL-10 production by LPS-stimulated RAW cells, with NECA being the most potent (Fig. 1B). Although NECA can also bind to all other adenosine receptors, the order of potency of agonists shown in Fig. 1B implicates A2B receptors (34–36) as being responsible for the augmentation of IL-10 production upon adenosine receptor occupancy. To further confirm the role of A2B receptors, the ability of alloxazine, a selective A2B receptor antagonist (36–39), was examined in preventing the stimulatory effect of adenosine on LPS-induced IL-10 production. Fig. 1C demonstrates that alloxazine reversed the stimulatory effect of adenosine on LPS-induced IL-10 production. Previously, we have documented that the mRNA for the A2B receptor is expressed in RAW 264.7 cells and is up-regulated following stimulation with LPS (33). To confirm that the A2B receptor is expressed at the protein level, we subjected cell membrane fractions obtained from unstimulated and LPS-stimulated RAW 264.7 cells to Western blotting using an anti-A2B Ab. Fig. 1D illustrates that the A2B receptor is present in membrane fractions of RAW 264.7 cells and that LPS increases the expression level of A2B receptors.
FIGURE 1.
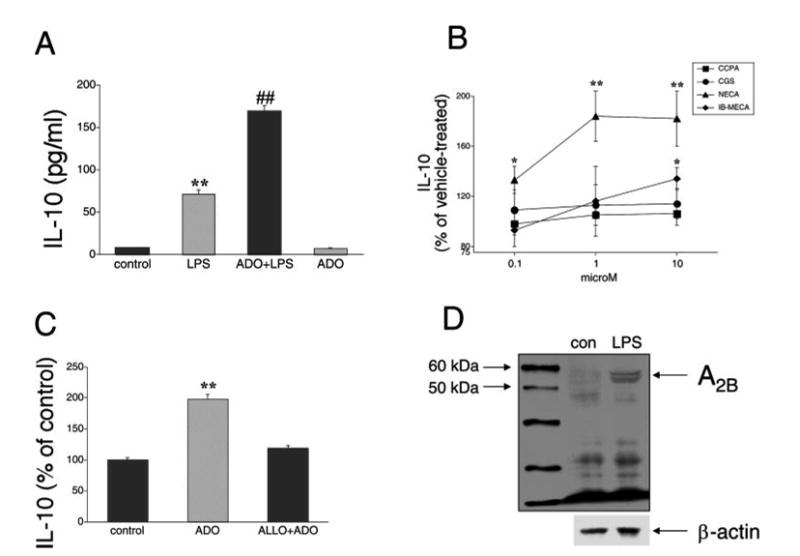
A, Adenosine (ADO) augments LPS-stimulated IL-10 production by LPS-activated RAW 264.7 macrophages. Adenosine (100 μM) was added to cells immediately before immunostimulation with 10 μg/ml LPS. IL-10 concentrations were measured from supernatants taken 5 h after stimulation with LPS. B, Effect of the A1 receptor agonist CCPA, A2A receptor agonist CGS-21680 (CGS), nonselective/A2B receptor agonist NECA, and A3 receptor agonist IB-MECA on LPS-induced IL-10 production by RAW cells. Selective agonists were added to cells immediately before immunostimulation with 10 μg/ml LPS. IL-10 concentrations were measured from supernatants taken 5 h after stimulation with LPS. C, The A2B receptor antagonist alloxazine (ALLO) prevents the effect of adenosine on IL-10 production. Alloxazine (10 μM) was administered 30 min before adenosine (100 μM) and LPS (10 μg/ml) followed by a 5-h-long incubation, after which supernatants were taken for IL-10 ELISA. D, Western blot analysis confirms expression of A2B receptors in membrane fractions from RAW 264.7 macrophages. LPS (10 μg/ml) treatment of the cells for 3 h up-regulates A2B receptor expression. β-actin was used as control for protein loading. Results (mean ± SEM) shown are representative of at least three experiments with n = 4 in each experiment, with the exception of D, where n = 2 for each group. **, p < 0.01.
The stimulatory effect of adenosine on IL-10 production is not associated with increased IL-10 mRNA accumulation or promoter activity
To begin to investigate the intracellular mechanisms responsible for the increased production of IL-10 by LPS-activated macrophages following adenosine administration, we determined IL-10 mRNA levels using real-time PCR, 2.5 and 5 h after addition of adenosine and/or LPS. We found that LPS increased IL-10 mRNA levels by ∼6-fold at 2.5 h and 2000-fold at 5 h. However, adenosine failed to further augment the LPS-induced accumulation of IL-10 mRNA at either time point (Fig. 2, A and B). We also examined the effect of adenosine on LPS-induced IL-10 promoter activity by transfecting cells with a construct in which luciferase expression is driven by the IL-10 promoter (6). Although we wished to determine IL-10 promoter activity 5 h after LPS stimulation, as was the case for IL-10 protein and mRNA measurements, luciferase levels were measured 8 h after LPS treatment, because shorter incubation times produced insufficient signal levels (data not shown). Similar to results reported in previous studies (6, 11), LPS was found to elicit an ∼10-fold increase in IL-10 promoter activity (Fig. 2C). Cotreatment of cells with adenosine failed to further enhance LPS-stimulated IL-10 promoter activity (Fig. 2C). Collectively, the fact that adenosine did not alter either IL-10 mRNA levels or promoter activity in LPS-stimulated macrophages indicates that adenosine exerts its stimulatory effect on IL-10 production by a posttranscriptional mechanism.
FIGURE 2.
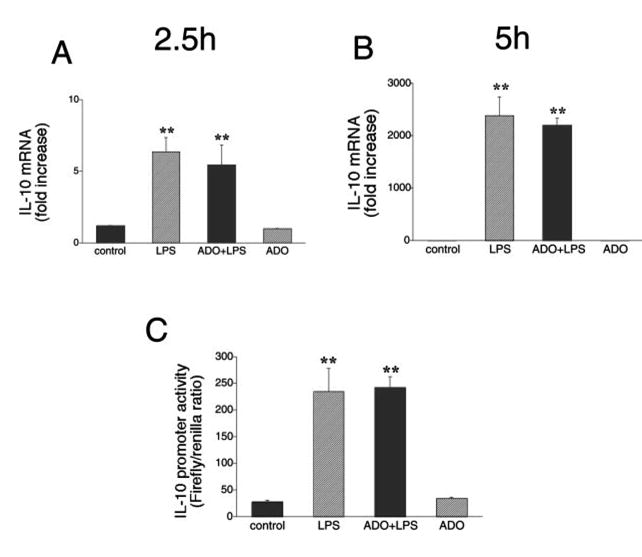
Lack of effect of adenosine (ADO, 100 μM) on LPS (LPS, 10 μg/ml)-induced IL-10 mRNA accumulation in RAW 264.7 cells. Adenosine was added to cells immediately before immunostimulation with LPS. IL-10 mRNA concentrations were measured by real-time PCR using RNA isolated 2.5 (A) or 5 h (B) after stimulation with LPS. C, Adenosine treatment (100 μM) has no effect on LPS (10 μg/ml)-induced IL-10 promoter activity in RAW 264.7 cells. To measure IL-10 promoter activity, cells were transiently transfected with an IL-10 promoter construct (harboring an SV40 promoter) and control Renilla-luciferase vector. Cells were treated with adenosine in the presence or absence of LPS for 8 h. Firefly luciferase reporter activities were normalized against Renilla luciferase activities, and IL-10 promoter activity was expressed as the Firefly:Renilla ratio. Data are mean ± SEM of n = 3 wells. Three experiments with similar results were performed. **, p < 0.01 indicates a significant increase in IL-10 mRNA levels or promoter activity following adenosine administration.
Posttranscriptional regulation by the IL-10 3′-UTR
Before investigating in detail the nature of the posttranscriptional regulation of IL-10 production by adenosine, it was important to gain a deeper insight into how IL-10 is regulated posttranscriptionally in the absence of adenosine. AU-rich elements (AREs) in the 3′-UTRs of cytokine mRNAs are critical for the posttranscriptional regulation of cytokine gene expression. AREs influence cytokine gene expression in a cell type- and stimulus-specific manner (40–46). IL-10 mRNA has recently been reported to contain such regulatory ARE sequences (13). To ascertain the role of these AREs in the posttranscriptional regulation of IL-10, we transfected macrophages with previously described (13) posttranscriptional luciferase reporter constructs containing cDNA sequences corresponding to the entire 3′-UTR of the IL-10 mRNA (AU4, +727 to +1239), or regions of the IL-10 3′-UTR (AU1, +727 to +818; AU2, +807 to +936; and AU3; +1157 to +1239) cloned into the XbaI site of the pGL3-control vector (Fig. 3A). Transcripts from the resulting plasmids therefore contain the luciferase-coding region, plus a part of the IL-10 mRNA 3′-UTR sequence. First, we compared luciferase activities in cells transfected with the pGL3-control vector with those transfected with plasmids AU1, AU2, AU3, or AU4. We observed a dramatic decrease in luciferase activity (100- to 300-fold) with plasmids AU1, AU2, AU3, and AU4 when compared with pGL3-control (Fig. 3B). It is noteworthy that the degree of inhibition conferred by the various IL-10 3′-UTR segments seen in our experiments was substantially greater than reported previously (13). These differences can be explained by the fact that in the previous study, reporter activity was measured 54 h after transfection, whereas in the current study, reporter activity was assessed 9 h after the transfection.
FIGURE 3.
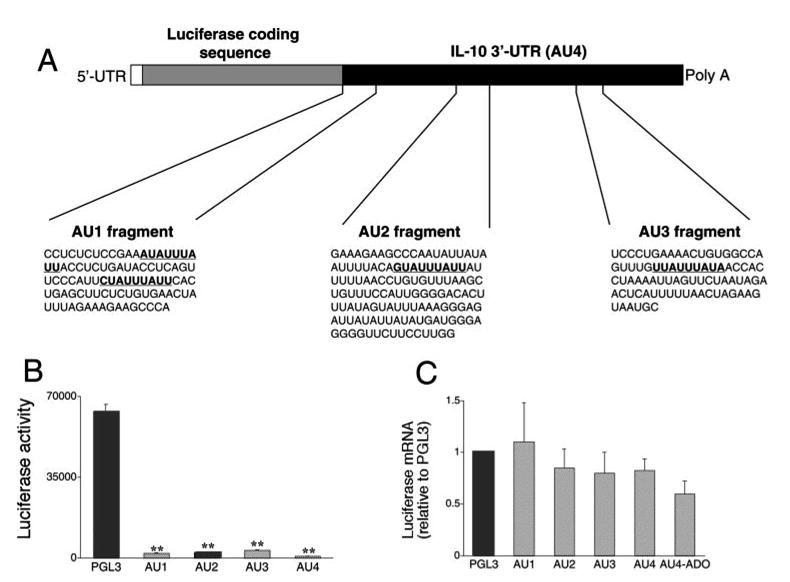
A, Nucleotide sequences of three potential regulatory AU-rich regions (AU1, AU2, and AU3) of the IL-10 3′-UTR (AU4). Highlighted are potential regulatory nonamer motifs. AU1–4 regions were inserted after the luciferase coding sequence of the pGL3-control reporter vector. B, Luciferase activities generated using plasmids containing AU1-AU4 are sharply decreased when compared with that generated by the pGL3-control reporter vector. Luciferase activity was measured from cells lysed 9 h after transfection with the various plasmids and normalized to protein content. C, Luciferase mRNA levels in cells transfected with plasmids containing AU1-AU4 are no different from that found in cells transfected with the pGL3-control reporter vector. Luciferase mRNA concentrations were measured using real-time PCR from cells lysed 9 h after transfection with the various plasmids. Representative data (mean ± SEM of n = 4 wells) are shown. Experiments were repeated at least three times. **, p < 0.01 vs pGL3-control.
Next, it was important to answer the question of whether the lower luciferase activity (protein expression) observed in AU1-, AU2-, AU3-, and AU4-transfected cells as compared with pGL3-control-transfected cells, was due to destabilization of the luciferase mRNA, or to translational repression. To address this issue, we measured luciferase mRNA levels from cells transfected with the various constructs using real-time PCR. If the inserted fragments from the IL-10 3′-UTR contain RNA-destabilizing activity, the luciferase mRNA levels in cells transfected with AU1, AU2, AU3, or AU4 should be lower than luciferase mRNA levels in pGL3-control-transfected cells. Alternatively, the presence of comparable mRNA levels would suggest a translational regulatory mechanism. Fig. 3C illustrates that there were no differences between luciferase mRNA levels in cells transfected with the IL-10 3′-UTR-containing constructs and cells transfected with the control vector, arguing for a translational inhibitory effect of the IL-10 3′-UTR.
Adenosine receptor activation regulates IL-10 3′-UTR function
In the next set of studies, we were interested in determining whether adenosine receptor occupancy would increase the activity of the various IL-10 3′-UTR-luciferase reporters in both LPS-treated and LPS-untreated cells.
We observed a 2-fold increase in luciferase activity using the AU4-containing plasmid following treatment with 100 μM adenosine (Fig. 4A), but adenosine had no effect on cells transfected with the pGL3-control vector (Fig. 4B). This observation, together with the finding that adenosine failed to increase luciferase mRNA levels in AU4-transfected cells (Fig. 3B), indicated that adenosine increases translation of the luciferase mRNA via a mechanism involving the AU4 region. It is noteworthy that the magnitude of the effect of adenosine on this posttranscriptional reporter system is similar to that observed for the effect of adenosine on the biosynthesis of IL-10.
FIGURE 4.
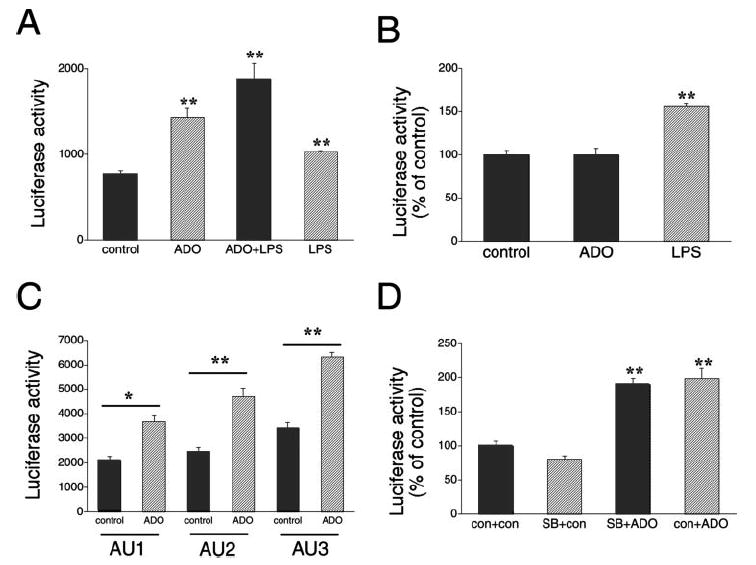
A, Effect of adenosine (ADO, 100 μM) and/or LPS (LPS, 10 μg/ml) on luciferase activity in RAW 264.7 cells transfected with a IL-10 luciferase reporter plasmid containing the entire 3′-UTR of IL-10 (AU4) downstream of the luciferase gene. Luciferase activity was measured from cells lysed 9 h after transfection with AU4 and normalized to protein content. ADO or LPS were administered 4 h after the transfection. B, Effect of ADO or LPS on luciferase activity in RAW 264.7 cells transfected with the pGL3-control luciferase vector. Luciferase activity was measured from cells lysed 9 h after transfection and normalized to protein content. ADO or LPS were administered 4 h after the transfection. C, ADO augments luciferase activity in RAW 264.7 cells transfected with IL-10 luciferase reporter plasmids containing various regions of the 3′-UTR of IL-10 (AU1–3) downstream of the luciferase gene. Luciferase activity was measured from cells lysed 9 h after transfection and normalized to protein content. ADO was administered 4 h after the transfection. D, The p38 MAPK inhibitor SB203580 (SB, 1 μM) cotreatment fails to decrease the ADO-stimulated increase in AU4 luciferase activity. Results (mean ± SEM) shown are representative of at least three experiments with n = 4 in each experiment. **, p < 0.01; con, control.
Fig. 4A shows that LPS can also increase luciferase activity in cells transfected with AU4-containing plasmid. However, because LPS enhanced reporter activity to a similar degree in cells transfected with the pGL3-control vector (Fig. 4B), it can be concluded that the effect of LPS is independent of the AU4 region. Because the SV40 promoter in the PGL3-control vector contains NF-κB sites, it is plausible that the LPS-enhanced reporter activity was due to an effect on these NF-κB sites in both the AU4-containing and empty control vectors.
Having established that cells transfected with the AU4-containing construct, which encompasses the entire IL-10 3′-UTR, were responsive to adenosine, we next explored the effect of adenosine on IL-10 posttranscriptional regulation using luciferase constructs carrying the shorter AU1, AU2, and AU3 regions of the IL-10 3′-UTR. As shown in Fig. 4C, adenosine (100 μM) treatment of cells expressing the luciferase constructs carrying AU1, AU2, and AU3 recapitulated the stimulatory effect of adenosine observed with the AU4 construct. The stimulatory effect of adenosine was greatest when using the AU2 and AU3 luciferase constructs. Overall, the magnitude of the effect of adenosine with the AU1, AU2, and AU3 constructs was similar to that found using the AU4 construct. This observation suggests that all three regions contain elements responsive to adenosine.
The p38 MAPK has been implicated in the posttranscriptional regulation of a number of cytokine genes, including TNF-α, IL-6, and vascular endothelial growth factor (42, 43, 45). We recently documented that adenosine activates p38 MAPK in RAW 264.7 macrophages (28). We explored the role of the p38 MAPK pathway in regulating IL-10 posttranscriptional regulation by testing the effect of the selective p38 MAPK pathway blocker SB203580 (1 μM) on the adenosine-induced increase in AU4 reporter activity. Fig. 4D demonstrates that SB203580 failed to prevent the adenosine-stimulated increase in AU4 activity. Thus, activation of the p38 MAPK pathway does not appear to be necessary for the adenosine-mediated enhancement of IL-10 3′-UTR activity.
Because adenosine is rapidly degraded to inosine, and inosine can alter macrophage function (47, 48), we sought to determine whether the posttranscriptional effects of adenosine are due to its degradation product. Inosine (1000 μM) was unable to reproduce the stimulatory effect of adenosine on luciferase activity in cells transfected with the AU4-containing luciferase construct (data not shown). To further establish the role of adenosine receptor activation in modulating IL-10 3′-UTR-mediated regulation, we examined the responsiveness of RAW 264.7 cells transfected with the AU4-containing plasmid or the pGL3-control vector to NECA. Similar to adenosine, NECA elevated luciferase activity by ∼2-fold in cells with the AU4-containing constructs, but not in cells transfected with the pGL3-control vector (Fig. 5).
FIGURE 5.
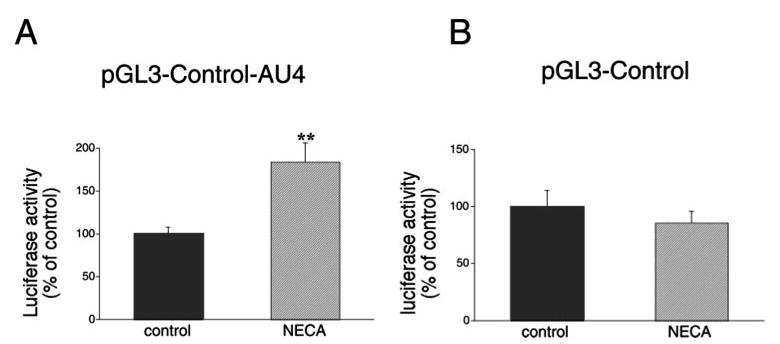
A, The selective A2B receptor agonist NECA (10 μM) increases luciferase activity in RAW 264.7 cells transfected with an IL-10 luciferase reporter plasmid containing the entire 3′-UTR of IL-10 (AU4) downstream of the luciferase gene. Luciferase activity was measured from cells lysed 9 h after transfection with AU4 and normalized to protein content. NECA was administered 4 h after the transfection. B, NECA does not influence luciferase activity in RAW 264.7 cells transfected with the pGL3-control luciferase vector. Luciferase activity was measured from cells lysed 9 h after transfection and normalized to protein content. NECA was administered 4 h after the transfection. Results (mean ± SEM) shown are representative of at least three experiments with n = 4 in each experiment. ** p < 0.01.
Adenosine receptor activation does not affect TNF 3′-UTR function
To investigate whether the stimulatory effect of adenosine receptor activation on the IL-10 3′-UTR reporter constructs was specific for this cytokine, we tested the effect of adenosine receptor stimulation on a TNF-α 3′-UTR reporter. The reporter construct that we used carried cDNA corresponding to the entire 3′-UTR of the TNF-α mRNA, which was inserted into the XbaI site of the pGL3-control vector (27). In accord with results of previous studies using this (27, 49) and other similar (50) constructs, we found that LPS induced a large increase in reporter activity (Fig. 6A), which was far greater (∼20-fold) than that achieved by using the empty pGL3-control vector (∼30% increase; Fig. 4B). These previous studies showed that the increase in reporter activity following LPS administration was, at least in part, representative of the derepressive effect of LPS on the translational blockade conferred by the 3′-UTR of the TNF-α mRNA. Importantly, adenosine did not alter reporter activity using this construct, indicating that the stimulatory effect of adenosine on the IL-10 3′-UTR is specific. Similar to adenosine, NECA was also unable to alter reporter activity in cells transfected with the TNF-α 3′-UTR construct (Fig. 6B).
FIGURE 6.
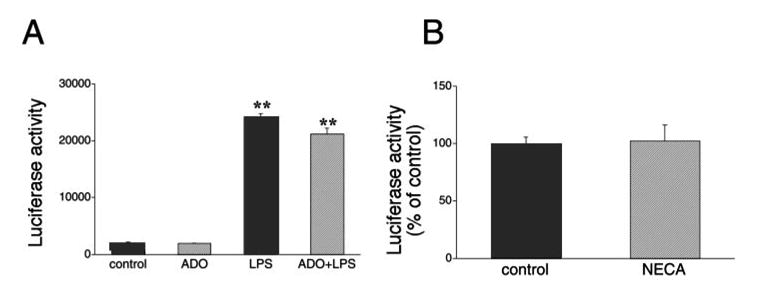
A, Effect of adenosine (ADO, 10 μM) and/or LPS (LPS, 10 μg/ml) on luciferase activity in RAW 264.7 cells transfected with a posttranscriptional TNF-α luciferase reporter plasmid containing an insert corresponding to the entire 3′-UTR of the TNF-α mRNA. Luciferase activity was measured from cells lysed 9 h after transfection and normalized to protein content. ADO or LPS were administered 4 h after the transfection. B, NECA does not influence luciferase activity in RAW 264.7 cells transfected with the pGL3-control luciferase vector. Luciferase activity was measured from cells lysed 9 h after transfection and normalized to protein content. NECA was administered 4 h after the transfection. Results (mean ± SEM) shown are representative of at least three experiments with n = 4 in each experiment. **, p < 0.01.
Adenosine regulates protein binding to the 3′-UTR of the IL-10 mRNA
The posttranscriptional regulation of mRNA is determined in many cases by interactions between specific RNA-binding proteins and cis-acting sequences located in the 3′-UTR of the mRNA. To understand more fully how adenosine regulates function of the 3′-UTR of the IL-10 mRNA, we set out to investigate whether the mRNA of the IL-10 3′-UTR interacts with cellular proteins, and if so, whether adenosine alters these interactions. Because the effect of adenosine in increasing reporter activity was greatest when using the AU2 and AU3 regions (Fig. 4C), we examined protein interactions with these two regions.
Equal amounts of cytoplasmic proteins from RAW cells obtained at various time points following adenosine treatment were incubated with in vitro-transcribed [32P]AU2 RNA. RNA-protein complexes were fractionated on a native polyacrylamide gel. These EMSA experiments demonstrated that two ribonucleoprotein complexes were formed with protein extracts from untreated RAW cells (Fig. 7A). Binding activity of both complexes was reproducibly increased by adenosine at the 30- and 60-min time points (Fig. 7, A and B), but not later (data not shown). Both complexes were specific, because a 100-fold molar excess of cold oligonucleotide completely abrogated both complexes (Fig. 7C). To begin to delineate which fragment of the AU2 sequence was necessary for the formation of these ribonucleoprotein complexes, we mutated the AU2 probe by deleting a 9-nt long region containing the GUAUUUAUU sequence (Fig. 3A). This sequence was chosen because this or similar sequences found in the 3′-UTR of cytokine mRNAs are thought to be responsible for the binding of trans-acting protein factors to the RNA (42, 43). When we used this mutated AU2 probe, we did not observe the two distinct complexes that were present when using the nonmutated probe (Fig. 7C). With the mutated probe, we detected only one complex, which was much more diffuse and less intense than those seen with the nonmutated probe. Adenosine was no longer able to alter the appearance of this complex. These results indicate that the GUAUUUAUU sequence plays an important role in protein binding to the AU2 region of the 3′-UTR of the IL-10 mRNA.
FIGURE 7.
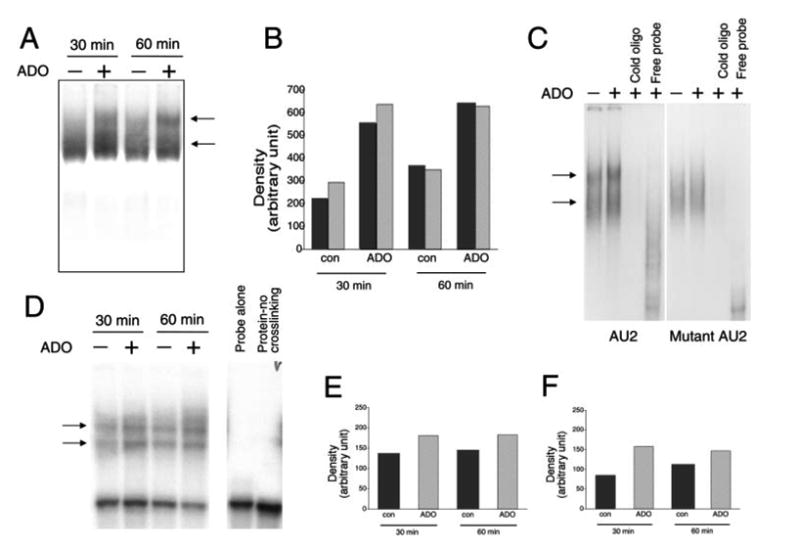
A, Adenosine (ADO) treatment of RAW 264.7 macrophages for 30 or 60 min increases RNA-protein complex formation between the AU2 region of the 3′-UTR of IL-10 mRNA and cellular components of RAW cells. Radiolabeled AU2 RNA probes were incubated with cytosolic fractions of RAW cells obtained at the end of the 30- or 60-min incubation period, complexes were separated by EMSA, and visualized using autoradiography. Two distinct complexes were observed in both untreated and treated cells. B, Densitometric analysis of intensities of the upper (■) and lower (
 ) complexes at 30 and 60 min following adenosine or control (con) treatment. C, Demonstration of the specificity of interactions between the AU2 region of the 3′-UTR of IL-10 mRNA and cellular fractions of RAW cells isolated 60 min after treatment with 100 μM adenosine. A 100× molar excess of cold AU2 oligonucleotide prevents formation of specific complexes in EMSA experiments (left panel). A mutated AU2 RNA probe lacking the GUAUUUAUU sequence forms a diffuse complex with cellular protein fractions of RAW cells as detected by EMSA (right panel). A 100× molar excess of cold mutated AU2 oligonucleotide prevents formation of this complex (right panel). D, UV cross-linking of cytosolic extracts from RAW 264.7 macrophages to an AU2 RNA probe results in formation of two specific complexes. Densitometric analysis indicates that adenosine treatment promotes formation of both the upper (E) and lower (F) complexes when measured using extracts taken 30 and 60 min after adenosine treatment of cells. Shown are representative results of a single experiment of three experiments with similar results.
) complexes at 30 and 60 min following adenosine or control (con) treatment. C, Demonstration of the specificity of interactions between the AU2 region of the 3′-UTR of IL-10 mRNA and cellular fractions of RAW cells isolated 60 min after treatment with 100 μM adenosine. A 100× molar excess of cold AU2 oligonucleotide prevents formation of specific complexes in EMSA experiments (left panel). A mutated AU2 RNA probe lacking the GUAUUUAUU sequence forms a diffuse complex with cellular protein fractions of RAW cells as detected by EMSA (right panel). A 100× molar excess of cold mutated AU2 oligonucleotide prevents formation of this complex (right panel). D, UV cross-linking of cytosolic extracts from RAW 264.7 macrophages to an AU2 RNA probe results in formation of two specific complexes. Densitometric analysis indicates that adenosine treatment promotes formation of both the upper (E) and lower (F) complexes when measured using extracts taken 30 and 60 min after adenosine treatment of cells. Shown are representative results of a single experiment of three experiments with similar results.
To provide further proof of protein binding to the IL-10 AU2 region, [32P]AU2 was incubated with cytosolic extracts from adenosine-treated or untreated macrophages taken at various time points after treatment. The samples were then exposed to short wavelength UV irradiation. Proteins were separated by SDS-PAGE, and the dried gels were autoradiographed to visualize proteins that were cross-linked to the radiolabeled AU2 probe. Fig. 7D demonstrates that, similar to results obtained using EMSA, two bands were detected in samples from both adenosine-treated and untreated cells. The intensity of both bands was higher with extracts from adenosine-treated cells than from untreated cells (Fig. 7, E and F) taken at both 30 and 60 min, but not at a later time point (data not shown). No bands were detected in the absence of UV irradiation (Fig. 7D). These results thus confirm data obtained in the EMSA studies.
Finally, we examined, using EMSA, whether the AU3 region formed any complexes with cellular protein extracts of RAW cells taken from both adenosine-treated and vehicle-treated cells. Similar to our results with the AU2 RNA probe, protein extracts of vehicle-treated RAW cells formed two complexes with the AU3 probe and the intensity of both of these complexes was increased by adenosine (Fig. 8). Both of these complexes were specific, because a 100-fold molar excess of cold oligonucleotide completely abolished both complexes (data not shown).
FIGURE 8.
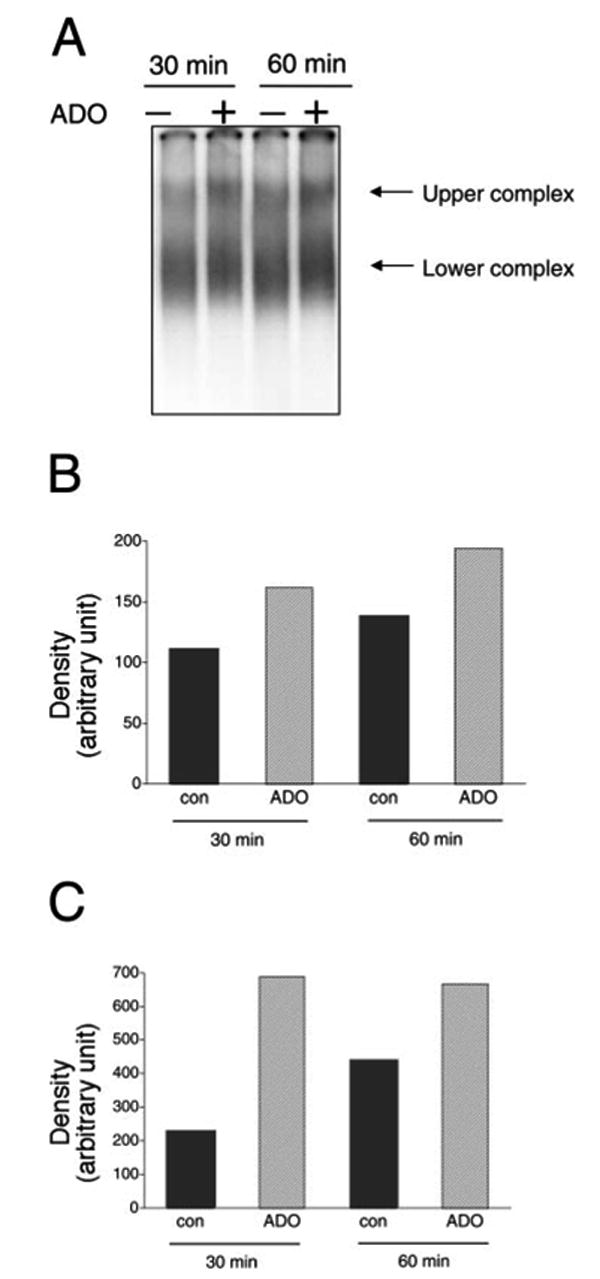
Adenosine treatment of RAW 264.7 macrophages for 30 or 60 min increases RNA-protein complex formation between the AU3 region of the 3′-UTR of IL-10 mRNA and cellular components of RAW cells. Radiolabeled AU2 RNA probes were incubated with cytosolic fractions of RAW cells obtained at the end of the 30- or 60-min incubation period, complexes were separated by EMSA, and visualized using autoradiography. Two distinct complexes were observed in both untreated and treated cells (A). Densitometric analysis indicates that adenosine treatment increases the intensity of both the upper (B) and lower (C) complexes when measured using extracts taken 30 and 60 min after adenosine treatment of cells. The results shown are from a single experiment (representative of three independent experiments with similar results).
Discusssion
In this study, we show that adenosine augments IL-10 production in murine macrophages without altering IL-10 mRNA accumulation or IL-10 promoter activity. In addition, adenosine stimulates reporter activity from IL-10 reporter constructs containing regions of the 3′-UTR of the IL-10 gene. Therefore, we propose that adenosine enhances IL-10 production by a posttranscriptional mechanism.
Despite its biological importance, relatively little is known about the regulation of IL-10 gene expression. Most previous studies of IL-10 gene expression focused on the mechanisms of LPS-induced transcriptional activation of the IL-10 gene through the IL-10 promoter (6–11). Using quantitative real-time PCR, we found that LPS stimulation increased IL-10 mRNA levels by ∼2000-fold, while levels of secreted IL-10 protein were increased by only ∼10-fold. These observations suggest that additional regulatory levels of IL-10 biosynthesis are operational in murine macrophages. A growing body of evidence suggests that regulation of gene expression may be accomplished through posttranscriptional mechanisms, which include regulation of mRNA stability, and translational regulation (42, 43). The discrepancy between IL-10 mRNA and protein levels was observed when measuring both IL-10 mRNA and protein accumulation at the same relatively late, 5-h time point, indicating that a destabilizing effect is unlikely to have a role in reducing IL-10 protein levels relative to mRNA concentrations. The 3′-UTR of the IL-10 mRNA has been shown previously to destabilize IL-10 mRNA in the EL4 T lymphocyte cell line (13). In contrast, we found that the 3′-UTR of the IL-10 mRNA does not exert the same destabilizing effect in RAW 264.7 macrophages, because inserting the 3′-UTR of the IL-10 mRNA downstream of the luciferase coding region in the pGL3-control vector failed to decrease luciferase mRNA levels. On the other hand, there was a dramatic, ∼100-fold drop in luciferase protein levels secondary to insertion of the 3′-UTR of the IL-10 mRNA after the luciferase coding region, confirming the translational regulation of IL-10. Considering that IL-10 mRNA was induced ∼2000-fold at 5 h after LPS stimulation, while IL-10 protein expression was elevated by ∼10-fold at this point, the 100-fold decrease in translation can explain why IL-10 protein is induced ∼200-fold less than IL-10 mRNA. It is, however, important to note that other mechanisms, including transcriptional changes and changes in IL-10 mRNA stability level, might also have influenced the steady-state mRNA levels measured at this time point. The exact contribution of these processes will require further studies to measure transcriptional activity using nuclear run-on assays and to assess the course of mRNA degradation. The 3′-UTR of the TNF-α gene confers a similar translational repressive effect on TNF-α mRNA (27, 50). Interestingly, although the translational repression caused by the TNF-α 3′-UTR is relieved by LPS (this study and Refs. 27 and 50), we find that LPS does not relieve the translational repressive effect of the 3′-UTR of the IL-10 mRNA.
In contrast, adenosine receptor activation by either the endogenous ligand adenosine, or by the stable adenosine analog NECA is able to relieve, at least in part, the translational repressive effect of the 3′-UTR of the IL-10 mRNA. The magnitude of this increase in translational activity (∼2-fold) after adenosine receptor activation is similar to the degree of adenosine- or NECA-induced augmentation of IL-10 protein production. This observation supports the view that the stimulatory effect of adenosine receptor activation on LPS-induced IL-10 production is due to a translational effect involving the 3′-UTR of the IL-10 mRNA. Our data also demonstrate that the translational stimulatory effect of adenosine receptor occupancy is specific for the 3′-UTR of IL-10, because adenosine failed to alter reporter activity from the 3′-UTR of the TNF-α mRNA. In a recent study using DNA microarray analysis (33), we reported that adenosine did not modulate expression of LPS-induced mRNAs in RAW 264.7 macrophages, nor did it alter mRNA expression in LPS-untreated, naive cells. Although the mRNA for TNF-α was among the many cytokine mRNAs whose expression was not changed by adenosine treatment in LPS-activated macrophages, levels of both secreted and intracellular TNF-α were substantially reduced following adenosine treatment (24, 33). Similar to this observation, an earlier study reported that adenosine had no effect on mRNA levels of TNF-α in LPS/IFN-γ-stimulated RAW 264.7 macrophages, despite its inhibitory effect on secreted TNF-α concentrations (51). Together, these data indicate that adenosine appears to regulate cytokine production by posttranscriptional mechanisms.
We also found that treatment of cells with adenosine exerted similar posttranscriptional stimulatory effects on the AU1, AU2, and AU3 subregions of the IL-10 3′-UTR to that observed on the full IL-10 3′-UTR (AU4). AU1, AU2, and AU3 contain related potential regulatory nonamer sequences (40, 43) that might be involved in the posttranscriptional regulatory action of adenosine. AUAUUUAUU and CUAUUUAUU are present in AU1, GUAUUUAUU is found in AU2, and UUAUUUAUA is a part of AU3. These nonamers are thought to control posttranscriptional regulatory events via interactions with specific binding proteins (40, 43). Our results provide insight into the possible role of proteins that bind to the IL-10 3′-UTR in mediating the stimulatory effect of adenosine on the translational regulatory processes involving the IL-10 3′-UTR. We demonstrate that the AU2 and AU3 regions are able to form two different complexes with protein extracts taken from RAW cells, and that adenosine intensifies this complex formation. In addition, deletion of the GUAUUUAUU sequence from the AU2 region abrogated protein binding to AU2, identifying this nonamer motif as a key element in the translational regulation of the AU2 region. Further studies will be required to ascertain the identity of the proteins that bind to this region, and the mechanisms by which adenosine augments complex formation.
Our results indicate that A2B receptors mediate the stimulatory effect of adenosine on IL-10 production in RAW 264.7 macrophages. Several lines of evidence support this conclusion. First, the order of potency of agonists (NECA > IB-MECA > CGS-21680) is typical of A2B receptors (34, 35). Second, the selective A2B receptor antagonist, alloxazine, prevented the adenosine-induced augmentation of IL-10 production. Finally, we detected A2B receptors in membrane protein fractions from RAW 264.7 cells. Our data are partially contradictory to those of a previous study which demonstrated that the A2A receptor agonist CGS-21680 potentiated IL-10 production by human monocytes and that this effect was reversed by a selective A2A antagonist (22). In contrast, because in this previous study NECA was both a more potent (EC50 ∼100 nM for NECA vs EC50 ∼500 nM for CGS-21680) and more efficacious (∼2.5-fold maximal increase with NECA vs ∼1.5-fold maximal increase with CGS-21680) agonist than CGS-21680, a possible role for A2B receptors in the enhancement of IL-10 production in human monocytes can also not be ruled out.
Previous studies have proposed the concept that the differential transcriptional regulation of IL-10 vs TNF-α and other proinflammatory cytokines might ensure the belated induction of IL-10 compared with proinflammatory cytokines (6, 7). In addition, this differential transcriptional regulation might serve to enable macrophages to uncouple IL-10 production from that of proinflammatory cytokines permitting differential responses to extracellular stimuli. Our results showing the differential posttranscriptional regulation of IL-10 and TNF-α in response to adenosine uncover a novel mechanism for uncoupling IL-10 production from that of TNF-α.
Abbreviations used in this paper
- UTR
untranslated region
- CCPA
2-chloro-N6-cyclopentyladenosine
- CGS-21680
2-p-(2-carboxyethyl) phenethylamino-5′-N-ethyl-carboxamidoadenosine
- NECA
5′-N-ethylcarboxamidoadenosine
- IB-MECA
N6-(3-iodobenzyl)-adenosine-5′-N-methyluronamide
- ARE
AU-rich element
Footnotes
This work was supported by National Institutes of Health (NIH) Grants R01 GM66189 (to G.H.), R01-GM57982 (to S.J.L.), R03AR052038-01 (to C.S.L.), as well as by National Science Foundation Grant MCB-0426195 (to C.S.L.), Hungarian National R&D Programme 1A/036/2004 (to E.S.V.), and the Intramural Research Program of NIH, National Institute on Alcohol Abuse and Alcoholism.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Mosmann TR, Schumacher JH, Fiorentino DF, Leverah J, Moore KW, Bond MW. Isolation of monoclonal antibodies specific for IL-4, IL-5, IL-6, and a new Th2-specific cytokine (IL-10), cytokine synthesis inhibitory factor, by using a solid phase radioimmunoadsorbent assay. J Immunol. 1990;145:2938–2945. [PubMed] [Google Scholar]
- 2.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1920. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barsig J, Kusters S, Vogt K, Volk HD, Tiegs G, Wendel A. Lipopolysaccharide-induced interleukin-10 in mice: role of endogenous tumor necrosis factor-α. Eur J Immunol. 1995;25:2888–2893. doi: 10.1002/eji.1830251027. [DOI] [PubMed] [Google Scholar]
- 4.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 5.Gilroy DW, Lawrence T, Perretti M, Rossi AG. Inflammatory resolution: new opportunities for drug discovery. Nat Rev Drug Discov. 2004;3:401–416. doi: 10.1038/nrd1383. [DOI] [PubMed] [Google Scholar]
- 6.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 7.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 8.Ma W, Lim W, Gee K, Aucoin S, Nandan D, Kozlowski M, Diaz-Mitoma F, Kumar A. The p38 mitogen-activated kinase pathway regulates the human interleukin-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J Biol Chem. 2001;276:13664–13674. doi: 10.1074/jbc.M011157200. [DOI] [PubMed] [Google Scholar]
- 9.Liu YW, Tseng HP, Chen LC, Chen BK, Chang WC. Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein β and δ in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J Immunol. 2003;171:821–828. doi: 10.4049/jimmunol.171.2.821. [DOI] [PubMed] [Google Scholar]
- 10.Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HW. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 11.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–3492. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner S, Prosch S, Schenke-Layland K, Riese U, Gausmann U, Platzer C. cAMP-induced interleukin-10 promoter activation depends on CCAAT/enhancer-binding protein expression and monocytic differentiation. J Biol Chem. 2003;278:5597–5604. doi: 10.1074/jbc.M207448200. [DOI] [PubMed] [Google Scholar]
- 13.Powell MJ, Thompson SA, Tone Y, Waldmann H, Tone M. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J Immunol. 2000;165:292–296. doi: 10.4049/jimmunol.165.1.292. [DOI] [PubMed] [Google Scholar]
- 14.Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- 15.Fredholm BB, Ijzerman AP, Jacobson KA, Klotz KN, Linden J. International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol Rev. 2001;53:527–552. [PMC free article] [PubMed] [Google Scholar]
- 16.Hasko G, Cronstein BN. Adenosine: an endogenous regulator of innate immunity. Trends Immunol. 2004;25:33–39. doi: 10.1016/j.it.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Hasko G, Deitch EA, Szabo C, Nemeth ZH, Vizi ES. Adenosine: a potential mediator of immunosuppression in multiple organ failure. Curr Opin Pharmacol. 2002;2:440–444. doi: 10.1016/s1471-4892(02)00172-8. [DOI] [PubMed] [Google Scholar]
- 18.Bouma MG, Stad RK, van den Wildenberg FA, Buurman WA. Differential regulatory effects of adenosine on cytokine release by activated human monocytes. J Immunol. 1994;153:4159–4168. [PubMed] [Google Scholar]
- 19.Haskó G, Szabó C, Németh ZH, Kvetan V, Pastores SM, Vizi ES. Adenosine receptor agonists differentially regulate IL-10, TNF-α, and nitric oxide production in RAW 264.7 macrophages and in endotoxemic mice. J Immunol. 1996;157:4634–4640. [PubMed] [Google Scholar]
- 20.McWhinney CD, Dudley MW, Bowlin TL, Peet NP, Schook L, Bradshaw M, De M, Borcherding DR, Edwards CK., 3rd Activation of adenosine A3 receptors on macrophages inhibits tumor necrosis factor-α. Eur J Pharmacol. 1996;310:209–216. doi: 10.1016/0014-2999(96)00272-5. [DOI] [PubMed] [Google Scholar]
- 21.Sajjadi FG, Takabayashi K, Foster AC, Domingo RC, Firestein GS. Inhibition of TNF-α expression by adenosine: role of A3 adenosine receptors. J Immunol. 1996;156:3435–3442. [PubMed] [Google Scholar]
- 22.Link AA, Kino T, Worth JA, McGuire JL, Crane ML, Chrousos GP, Wilder RL, Elenkov IJ. Ligand-activation of the adenosine A2a receptors inhibits IL-12 production by human monocytes. J Immunol. 2000;164:436–442. doi: 10.4049/jimmunol.164.1.436. [DOI] [PubMed] [Google Scholar]
- 23.Mayne M, Fotheringham J, Yan HJ, Power C, Del Bigio MR, Peeling J, Geiger JD. Adenosine A2A receptor activation reduces proinflammatory events and decreases cell death following intracerebral hemorrhage. Ann Neurol. 2001;49:727–735. doi: 10.1002/ana.1010. [DOI] [PubMed] [Google Scholar]
- 24.Pinhal-Enfield G, Ramanathan M, Hasko G, Vogel SN, Salzman AL, Boons GJ, Leibovich SJ. An angiogenic switch in macrophages involving synergy between Toll-like receptors 2, 4, 7, and 9 and adenosine A2A receptors. Am J Pathol. 2003;163:711–721. doi: 10.1016/S0002-9440(10)63698-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haskó G, Kuhel DG, Chenn JF, Schwarzschild MA, Deitch EA, Mabley JG, Marton A, Szabó C. Adenosine inhibits IL-12 and TNF-α production via adenosine A2a receptor-dependent and independent mechanisms. FASEB J. 2000;14:2065–2074. doi: 10.1096/fj.99-0508com. [DOI] [PubMed] [Google Scholar]
- 26.Khoa ND, Montesinos MC, Reiss AB, Delano D, Awadallah N, Cronstein BN. Inflammatory cytokines regulate function and expression of adenosine A2A receptors in human monocytic THP-1 cells. J Immunol. 2001;167:4026–4032. doi: 10.4049/jimmunol.167.7.4026. [DOI] [PubMed] [Google Scholar]
- 27.Gueydan C, Houzet L, Marchant A, Sels A, Huez G, Kruys V. Engagement of tumor necrosis factor mRNA by an endotoxin-inducible cytoplasmic protein. Mol Med. 1996;2:479–488. [PMC free article] [PubMed] [Google Scholar]
- 28.Nemeth ZH, Leibovich SJ, Deitch EA, Sperlagh B, Virag L, Vizi ES, Szabo C, Hasko G. Adenosine stimulates CREB activation in macrophages via a p38 MAPK-mediated mechanism. Biochem Biophys Res Commun. 2003;312:883–888. doi: 10.1016/j.bbrc.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Nemeth ZH, Deitch EA, Szabo C, Fekete Z, Hauser CJ, Hasko G. Lithium induces NF-κB activation and interleukin-8 production in human intestinal epithelial cells. J Biol Chem. 2002;277:7713–7719. doi: 10.1074/jbc.M109711200. [DOI] [PubMed] [Google Scholar]
- 30.Morimoto M, Morimoto M, Whitmire J, Xiao S, Anthony RM, Mirakami H, Star RA, Urban JF, Jr, Gause WC. Peripheral CD4 T cells rapidly accumulate at the host: parasite interface during an inflammatory Th2 memory response. J Immunol. 2004;172:2424–2430. doi: 10.4049/jimmunol.172.4.2424. [DOI] [PubMed] [Google Scholar]
- 31.Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 32.Lutz CS, Alwine JC. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 33.Nemeth ZH, Leibovich SJ, Deitch EA, Vizi ES, Szabo C, Hasko G. cDNA microarray analysis reveals a nuclear factor-κB-independent regulation of macrophage function by adenosine. J Pharmacol Exp Ther. 2003;306:1042–1049. doi: 10.1124/jpet.103.052944. [DOI] [PubMed] [Google Scholar]
- 34.Feoktistov I, Biaggioni I. Adenosine A2B receptors. Pharmacol Rev. 1997;49:381–402. [PubMed] [Google Scholar]
- 35.Xaus J, Mirabet M, Lloberas J, Soler C, Lluis C, Franco R, Celada A. IFN-γ up-regulates the A2B adenosine receptor expression in macrophages: a mechanism of macrophage deactivation. J Immunol. 1999;162:3607–3614. [PubMed] [Google Scholar]
- 36.Narravula S, Lennon PF, Mueller BU, Colgan SP. Regulation of endothelial CD73 by adenosine: paracrine pathway for enhanced endothelial barrier function. J Immunol. 2000;165:5262–5268. doi: 10.4049/jimmunol.165.9.5262. [DOI] [PubMed] [Google Scholar]
- 37.Peyot ML, Gadeau AP, Dandre F, Belloc I, Dupuch F, Desgranges C. Extracellular adenosine induces apoptosis of human arterial smooth muscle cells via A(2b)-purinoceptor. Circ Res. 2000;86:76–85. doi: 10.1161/01.res.86.1.76. [DOI] [PubMed] [Google Scholar]
- 38.Morrison RR, Talukder MA, Ledent C, Mustafa SJ. Cardiac effects of adenosine in A2A receptor knockout hearts: uncovering A2B receptors. Am J Physiol. 2002;282:H437–H444. doi: 10.1152/ajpheart.00723.2001. [DOI] [PubMed] [Google Scholar]
- 39.Walia B, Castaneda FE, Wang L, Kolachala VL, Bajaj R, Roman J, Merlin D, Gewirtz AT, Sitaraman SV. Polarized fibronectin secretion induced by adenosine regulates bacterial-epithelial interaction in human intestinal epithelial cells. Biochem J. 2004;382:589–596. doi: 10.1042/BJ20040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 41.Hel Z, Skamene E, Radzioch D. Two distinct regions in the 3′ untranslated region of tumor necrosis factor α mRNA form complexes with macrophage proteins. Mol Cell Biol. 1996;16:5579–5590. doi: 10.1128/mcb.16.10.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 43.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukocyte Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- 45.Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton TA, Ohmori Y, Tebo JM, Kishore R. Regulation of macrophage gene expression by pro- and anti-inflammatory cytokines. Pathobiology. 1999;67:241–244. doi: 10.1159/000028101. [DOI] [PubMed] [Google Scholar]
- 47.Hasko G, Kuhel DG, Nemeth ZH, Mabley JG, Stachlewitz RF, Virag L, Lohinai Z, Southan GJ, Salzman AL, Szabo C. Inosine inhibits inflammatory cytokine production by a posttranscriptional mechanism and protects against endotoxin-induced shock. J Immunol. 2000;164:1013–1019. doi: 10.4049/jimmunol.164.2.1013. [DOI] [PubMed] [Google Scholar]
- 48.Hasko G, Sitkovsky MV, Szabo C. Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol Sci. 2004;25:152–157. doi: 10.1016/j.tips.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 49.McMullen MR, Cocuzzi E, Hatzoglou M, Nagy LE. Chronic ethanol exposure increases the binding of HuR to the TNFα 3′-untranslated region in macrophages. J Biol Chem. 2003;278:38333–38341. doi: 10.1074/jbc.M304566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–475. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parmely MJ, Zhou WW, Edwards CK, 3rd, Borcherding DR, Silverstein R, Morrison DC. Adenosine and a related carbocyclic nucleoside analogue selectively inhibit tumor necrosis factor-α production and protect mice against endotoxin challenge. J Immunol. 1993;151:389–396. [PubMed] [Google Scholar]


