Abstract
SUMMARY
Brassinosteroids (BRs) are essential hormones for plant growth and development. BRs regulate gene expression by inducing dephosphorylation of two key transcription factors BZR1 and BZR2/BES1 through a signal transduction pathway, which involves cell surface receptors (BRI1 and BAK1) and a GSK3 kinase (BIN2). How BR-regulated phosphorylation controls the activities of BZR1/BZR2 is not fully understood. Here we show that BIN2-catalyzed phosphorylation of BZR1/BZR2 not only inhibits DNA binding but also promotes binding to the 14-3-3 proteins. Mutations of a BIN2-phosphorylation site in BZR1 abolish 14-3-3 binding and lead to increased nuclear localization of BZR1 protein and enhanced BR-responses in transgenic plants. Further, BR-deficiency increases cytoplasmic localization and BR treatment induces rapid nuclear localization of BZR1/BZR2. Thus 14-3-3 binding is required for efficient inhibition of phosphorylated BR transcription factors largely through cytoplasmic retention. This study demonstrates that multiple mechanisms are required for BR regulation of gene expression and plant growth.
INTRODUCTION
Steroids are used as hormones in both animals and plants. Many steroid biosynthetic enzymes are conserved and steroids regulate many of the same physiological and developmental processes in both kingdoms, including gene expression, cell division/expansion, reproductive development, and aging/senescence. However, plant and animal steroid hormones appear to regulate gene expression through distinct signaling mechanisms (Thummel and Chory, 2002). In animals, steroid hormones bind the nuclear receptor family of transcription factors, which are retained in the cytoplasm by interaction with the HSP90 complex; ligand binding disrupts such interaction and leads to nuclear localization of the transcription factors and altered gene expression (Thummel and Chory, 2002). In plants, brassinosteroids (BRs) bind to a cell-surface receptor kinase BRI1, which initiates a phosphorylation cascade that regulates the activities of key transcription factors by phosphorylation/dephosphorylation (Vert et al., 2005).
BRs play essential roles in plant growth and development (Clouse and Sasse, 1998). Deficiency in BR biosynthesis or signal transduction causes severe growth defects, including dwarfism, male sterility, delayed senescence and flowering, and light-grown phenotype in the dark (Li and Chory, 1997; Li et al., 1996). BR binding to the extracellular domain of BRI1 activates its kinase activity, induces dimerization with and activation of another cell-surface receptor kinase (BAK1) (Li et al., 2002; Nam and Li, 2002), and causes disassociation of BKI1, a novel protein that represses BRI1 (Wang and Chory, 2006). BR-activation of the receptor kinases is believed to inhibit BIN2, a glycogen synthase kinase-3 (GSK3)-like kinase (Li and Nam, 2002), or to activate BSU1 (Mora-Garcia et al., 2004), a phosphatase. BIN2 and BSU1 control the phosphorylation status of two homologous transcription factors, BZR1 and BZR2/BES1, which bind to specific promoter sequences to mediate BR-responsive gene expression (He et al., 2005; Yin et al., 2005). Similar to β-catenin in metazoans, which is inhibited by GSK3β-mediated phosphorylation and activated by Wnt-induced dephosphorylation (Stadeli et al., 2006), BZR1 and BZR2/BES1 are inhibited by BIN2-mediated phosphorylation and activated by BR-induced dephosphorylation. Whereas inhibition of GSK3 by Wnt signaling leads to nuclear translocation of dephosphorylated β-catenin, it has been controversial whether a similar mechanism is important for BR regulation of BZR1 and BZR2/BES1.
BZR1 and BZR2/BES1 are key effectors of BR action. The dominant bzr1-1D and bes1-D mutations that effectively stabilize each protein suppress the phenotypes of BR-deficient or insensitive mutants. BZR1 and BZR2/BES1 have been shown to directly bind to promoters of BR-responsive genes, but with different binding site sequence specificities and transcriptional activities. Extensive protein-DNA interaction studies have demonstrated that BZR1 has an optimal binding site of CGTG(T/C)G sequence, which was named the BR response element (BRRE). The BRRE is conserved in the promoters of BR-repressed genes such as CPD, DWF4, ROT3, and BR6OX (He et al., 2005). BZR1 functions as a transcriptional repressor in vivo to mediate feedback inhibition of BR-biosynthetic genes. In contrast, BZR2/BES1 was shown to interact with a bHLH type transcription factor BIM1, and together they bind to the E-box (CANNTG) elements in the promoter of SAUR-AC1 and activate gene expression (Yin et al., 2005). Such differences between BZR1 and BZR2/BES1 are somewhat surprising given their high sequence similarity (88% identity) but are consistent with the opposite cell elongation phenotypes of bzr1-1D and bes1-D mutant plants grown in the light.
How BR-regulated phosphorylation controls the activities of BZR1 and BZR2/BES1 is a key question for understanding BR action. Previous studies have yielded conflicting results about whether phosphorylation by BIN2 alters the nuclear localization of BZR1 and BZR2/BES1 (Wang et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Zhao et al., 2002). BR treatment has been shown to induce rapid dephosphorylation of BZR1-CFP and BZR2/BES1-GFP fusion proteins, followed by an increase of the protein levels in the nuclei of hypocotyl cells (Wang et al., 2002; Yin et al., 2002). It was further shown that the bin2-1 mutant, which encodes a hyperactive kinase, increased phosphorylation and reduced accumulation of BZR1 and BZR2/BES1 proteins (He et al., 2002; Yin et al., 2002). These results suggested that phosphorylation of BZR1 and BZR2/BES1 reduces their protein levels in the nucleus (He et al., 2002; Yin et al., 2002). In contrast, it has been reported recently that BZR1-GFP and BZR2/BES1-GFP were constitutively localized in the nuclei (Vert and Chory, 2006; Zhao et al., 2002), while phosphorylation inhibited the DNA-binding activity of BZR2/BES1 in vitro and reduced its transcriptional activity in yeast (Vert and Chory, 2006). These observations supported a revised model that BIN2-catalyzed phosphorylation inhibits BZR2/BES1 primarily by reducing its DNA-binding and transcriptional activities rather than altering its localization or accumulation (Vert and Chory, 2006). However, all these previous studies only quantified the nuclear signals but not the cytoplasmic signals of the fusion proteins, and thus a change of localization could not be discerned from a change of total protein level. Besides these discrepancies, functional importance of these possible mechanisms has not been studied in vivo. BZR1 and BZR2/BES1 each contain 25 putative BIN2-phosphorylation sites; how each phosphorylation site affects the function of the transcription factors remains unknown.
14-3-3 proteins are phosphopeptide-binding proteins highly conserved in all eukaryotes. They participate in various signal transduction and regulatory processes by interacting with diverse target proteins in a sequence-specific and phosphorylation-dependent manner (Bridges and Moorhead, 2005; Muslin et al., 1996). The first plant 14-3-3 protein was identified as part of a DNA-protein complex (Lu et al., 1992), indicating a role of plant 14-3-3s in regulation of transcription. Further studies showed functions of 14-3-3s in regulating the activities of metabolic enzymes and transcription factors (Sehnke et al., 2002). The Arabidopsis genome contains fifteen 14-3-3 genes, 12 of which are expressed (Sehnke et al., 2002). A large number of 14-3-3 target proteins have been identified in plants (Schoonheim et al., 2007a), suggesting roles of plant 14-3-3s in a wide range of cellular processes. However, a direct role of plant 14-3-3 proteins in mediating signal transduction is yet to be illustrated.
In this study, we demonstrate an essential role for 14-3-3 proteins in BR signal transduction in Arabidopsis. We show that 14-3-3 proteins interact specifically with BZR1 protein that has been phosphorylated by BIN2. Mutations of a BIN2-phosphorylation site in BZR1 that abolish its binding to 14-3-3 proteins result in constitutive BR-response phenotypes similar to bzr1-1D. Such mutant BZR1 proteins are constitutively localized in the nucleus, but are not affected in either protein accumulation or DNA binding. Moreover, we present convincing evidence that BR regulates nuclear localization of BZR1 and BZR2 proteins. Our results demonstrate an important role of 14-3-3 proteins in regulation of the key transcription factors by BR signal transduction.
RESULTS
BR Induces Rapid Nuclear Localization of BZR1
To clarify whether BR regulates nuclear localization of BZR1, we analyzed the relative levels of nuclear and cytoplasmic signals of a BZR1-yellow fluorescent protein (BZR1-YFP) using Spinning-disk confocal microscopy. The results show that the ratio of nuclear to cytoplasmic BZR1-YFP signals is reduced by brassinazole (BRZ), an inhibitor of BR biosynthesis, but increased by brassinolide (BL), the most active brassinosteroid (Figure 1A). BL-induced nuclear localization of BZR1-YFP was observed both when transiently expressed in tobacco leaves (Figure S1) and when stably expressed in transgenic Arabidopsis (Figure 1A). In time course experiments, nuclear accumulation of BZR1-YFP increased rapidly, within 4 minutes of BL treatment, and the nuclear/cytoplasmic ratio continued to increase for 14 min (Figure 1B and movies SM1, SM2). Such kinetics of BR-induced BZR1 nuclear accumulation is consistent with the kinetics of dephosphorylation (He et al., 2002). In contrast, mock treatment did not significantly increase the nuclear/cytoplasmic signal ratio of BZR1-YFP.
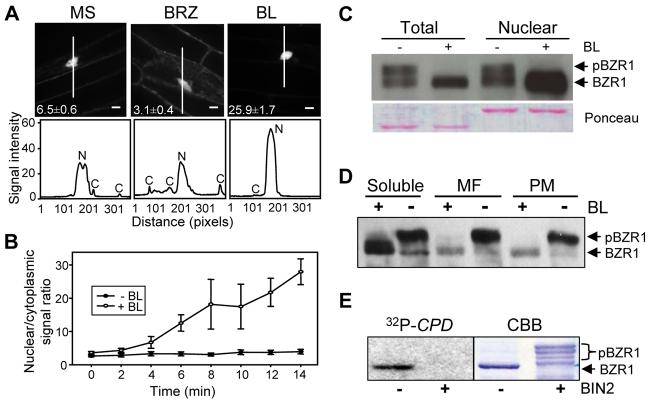
Figure 1. BR Induces Nuclear Localization of BZR1.
(A) Effect of brassinazole (BRZ) and brassinolide (BL) on the subcellular localization of BZR1-YFP. Transgenic Arabidopsis expressing BZR1-YFP were grown on MS or MS + 2 μM BRZ medium in the dark for 4 days. Seedlings grown on BRZ medium were treated with mock or 100 nM BL for 1 hr and images of the YFP signal (top) were obtained using confocal microscopy. Numbers in each image show the average ratios between nuclear and cytoplasmic signal intensities and standard errors calculated from seven cells for each treatment. The white lines inside the images show the areas used for line scan measurements that yielded plot profiles shown in the lower panels. N, nuclear signal, C, cytoplasmic signal. Fluorescent intensity is 1000x and scale bar is 10 μm.
(B) Kinetics of the BL-induced nuclear accumulation of BZR1-YFP. BZR1-YFP seedlings grown on 2 μM BRZ were treated with BL or mock solution (−BL), and images were acquired at each time point to measure the nuclear/cytoplasmic ratios. Error bars are ± standard errors of mean.
(C) Unphosphorylated BZR1 is enriched in the nuclear fraction. Immunoblot of unphosphorylated and phosphorylated BZR1-CFP proteins in total and nuclear fractions from mock- or BL-treated seedlings.
(D) Phosphorylated BZR1 is more enriched in membrane fractions than unphosphorylated BZR1. Transgenic plants expressing the BZR1-CFP protein were treated with (+BL) or without (−BL) 1 μM BL, and the soluble, microsomal (MF), and plasma membrane (PM) fractions were analyzed by immunoblot.
(E) BIN2 phosphorylation inhibits DNA binding activity of BZR1. Gel blot of unphosphorylated and BIN2-phosphorylated MBP-BZR1 proteins was probed with radiolabeled CPD promoter DNA. CBB, Coomassie Brilliant Blue stained gel. Unphosphorylated BZR1 (BZR1) and phosphorylated BZR1 (pBZR1) proteins are marked by arrows (C-E).
We further tested the effect of BR on nuclear localization of BZR1-YFP using subcellular fractionation experiments. These experiments demonstrated that unphosphorylated but not the phosphorylated BZR1-CFP was enriched in the nucleus. Furthermore, BL treatment resulted in a significant increase of the unphosphorylated BZR1 in the nuclear fraction (Figure 1C). Phosphorylated BZR1-CFP protein was also more associated with cell membranes than the unphosphorylated protein (Figure 1D). In subcellular fractionation experiments, both microsomal and plasma membrane fractions contained similar levels of phosphorylated but much lower amounts of unphosphorylated BZR1-CFP compared to the soluble protein fraction (Figure 1D). After BR treatment, the amount of BZR1-CFP associated with membranes was greatly reduced as BZR1 was dephosphorylated. These results collectively indicate that some phosphorylated BZR1 protein is retained in the cytoplasm most likely through interaction with membranes, and BR-induced dephosphorylation of BZR1 leads to its movement from cytoplasm into the nucleus.
Phosphorylation inhibits the DNA binding activity of BZR1
A recent study showed that phosphorylation inhibits DNA binding of BZR2/BES1 (Vert and Chory, 2006). We tested whether phosphorylation by BIN2 inhibits the DNA binding activity of BZR1. A maltose binding protein (MBP)-BZR1 fusion protein was phosphorylated by BIN2 and separated on a SDS-PAGE gel. When the gel blot was probed with radiolabeled DNA containing a BZR1 binding site (BRRE; He et al., 2005), only the unphosphorylated, but not the phosphorylated MBP-BZR1 showed DNA binding, indicating that phosphorylation by BIN2 inhibits BZR1's DNA binding activity (Figure 1E).
Phosphorylation of BZR1 by BIN2 Promotes Binding to the 14-3-3 Proteins
To further understand the molecular mechanisms of BZR1 regulation, we performed yeast two-hybrid screen for BZR1-interacting proteins. Out of 98 positive clones identified, 80 clones encode members of the 14-3-3 protein family. They represent five of the 12 isoforms in Arabidopsis (Sehnke et al., 2002), namely 14-3-3λ, 14-3-3κ, 14-3-3ε, 14-3-3ϕ and 14-3-3ω. Further yeast two-hybrid assays indicated that 14-3-3λ interacts with BZR1, bzr1-1D, and the C-termini (amino acids 90-336) of BZR1 and bzr1-1D, but not with BRI1, BIN2 or bin2-1 (Figure 2A).
Figure 2. BIN2-Phosphorylated BZR1 Interacts With 14-3-3 Proteins.
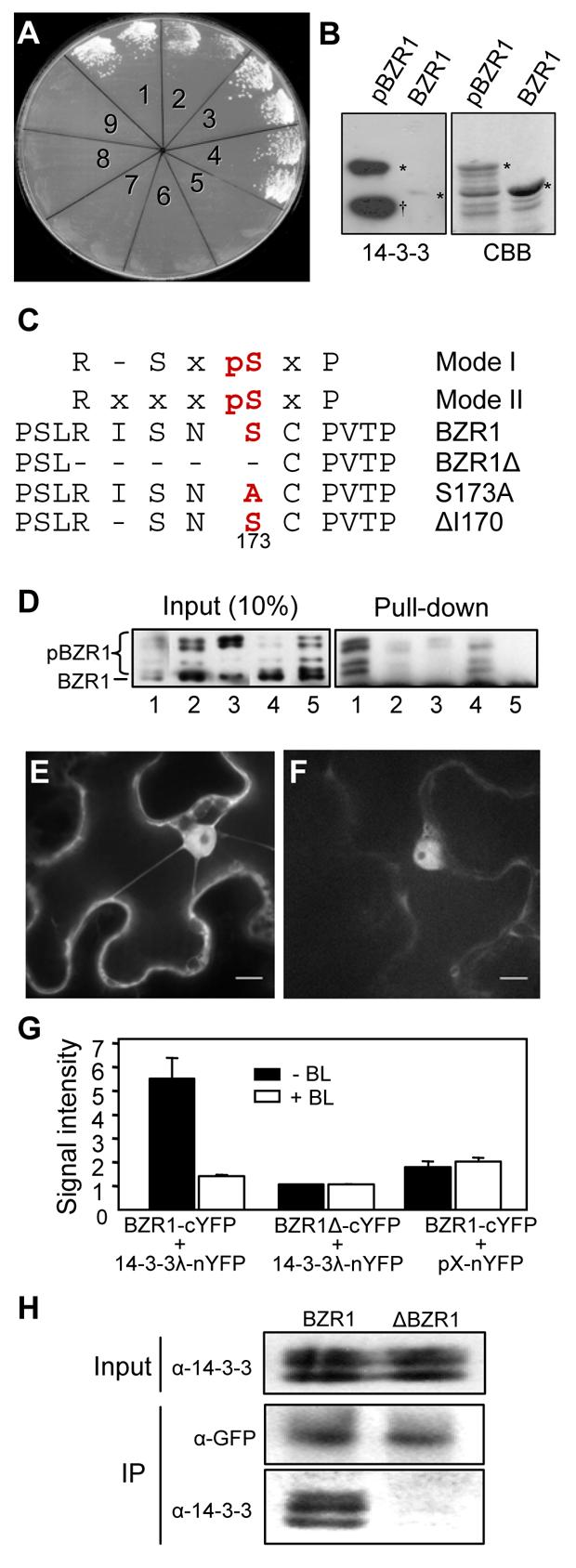
(A) BZR1 interacts with 14-3-3λ in yeast two-hybrid assays. Each yeast clone contains the pGAD-14-3-3λ prey construct and one of the following genes in the pGBKT7 vector: 1, BZR1, 2, bzr1-1D, 3, BZR1C, 4, bzr1-1DC, 5, BIN2, 6, bin2-1, 7, BRI1-KD, 8, p53, 9, no gene insert. Growth of yeast cells indicates interaction between the test protein and 14-3-3λ.
(B) 14-3-3λ specifically interacts with phosphorylated BZR1. Recombinant MBP-BZR1 protein was phosphorylated by GST-BIN2, gel blotted, and probed with GST-14-3-3λ and anti-GST antibody. Right panel shows the Coomassie Brilliant Blue (CBB) stained gel. Asterisks mark the unphosphorylated (BZR1) and phosphorylated (pBZR1) MBPBZR1 bands. Dagger sign (δ) marks the GST-BIN2 band.
(C) BZR1 contains a putative 14-3-3 binding site. Two known 14-3-3 binding site sequences (Mode I and Mode II) are aligned against BZR1 sequence. ‘x’ represents any given amino acid. Amino acids are presented as single letters namely: R, Arginine; I, Isoleucine; pS, phosphoSerine; N, Asparagine; C, Cysteine; P, Proline. Conserved Serine residue at -3 position that's crucial for 14-3-3 binding was numbered and marked in bold. Various mutations created in the 14-3-3 binding sites are also shown. Hyphen (-) represents deletion of an amino acid residue.
(D) Effects of mutations of the 14-3-3-binding site of BZR1 on the binding of 14-3-3λ. Wild type and mutant MBP-BZR1 proteins were phosphorylated by GST-BIN2 and then mixed with unphosphorylated proteins before pull-down assay (right panel) with glutathione agarose beads containing GST-14-3-3λ (lanes 1 to 4) or GST (lane 5). Lanes 1 to 5: MBP-BZR1, MBP-BZR1Δ, MBP-BZR1S173A, MBP-BZR1ΔI170, and MBPBZR1.
(E) In vivo interaction between BZR1 and 14-3-3λ detected by BiFC. The N-terminal and C-terminal halves of YFP were fused to 14-3-3λ and BZR1, respectively, and the constructs were co-transformed into tobacco leaf cells and fluorescence images were obtained using confocal microscopy. Scale bar is 10 μm.
(F) The BZR1-cYFP and the 14-3-3λ-nYFP constructs were co-transformed into tobacco cells with 1 μM BL. YFP fluorescence images were obtained using a confocal microscope. Scale bar is 10 μm.
(G) Quantification of BiFC signals. Each pair of indicated constructs was co-transformed into tobacco cells with (+BL) or without (−BL) 1 μM BL. Fluorescence signal intensity (x1000) was average of measurements from at least 7 transformed cells after background subtraction. Error bars are ± standard error of mean.
(H) Mutations in 14-3-3 binding site abolish in vivo interaction of BZR1 with 14-3-3s. Co-Immunoprecipitation (Co-IP) was carried using anti-GFP antibody coupled to Protein A Sepharose beads with tissue material from either BZR1-YFP or ΔBZR1-YFP transgenic plants. Western blot was probed with α-GFP and α-14-3-3 antibodies.
14-3-3s are known to bind specific sequences of target proteins in a phosphorylation-dependent manner (Muslin et al., 1996). To determine if 14-3-3 binding requires phosphorylation of BZR1 by BIN2, purified MBP-BZR1 fusion protein was phosphorylated by a GST-BIN2 protein in vitro, separated on a SDS-PAGE gel, and blotted to nitrocellulose membrane. Incubation of the blot with GST-14-3-3λ and anti-GST antibodies detected strong binding to the phosphorylated MBP-BZR1 and very weak binding to the unphosphorylated form (Figure 2B), indicating that phosphorylation by BIN2 facilitates BZR1 binding to 14-3-3s. The interaction in yeast is likely due to phosphorylation of BZR1 by the yeast GSK3 kinases.
In an attempt to understand the function of 14-3-3 proteins, we obtained single and double knockout mutants of 14-3-3λ and 14-3-3κ, two closely homologous genes that were identified most frequently in our yeast two-hybrid screen. Neither single nor double knockout mutants displayed phenotypes or altered BR responses (data not shown), most likely due to redundancy of the gene family. 14-3-3 RNAi plants also did not show any noticeable differences in phenotypes (data not shown). Apparently, an experimental approach that specifically disrupts BZR1:14-3-3 interaction is required to understand how 14-3-3 binding affects BZR1 function.
14-3-3 proteins are known to bind mostly two types of specific sequences: mode I RXXpSXP or mode II RXXXpSXP (X= any amino acid, R= Arginine, pS= phosphoSerine, and P= Proline), in which the S residue needs to be phosphorylated (Aitken, 2006; Muslin et al., 1996). A potential mode II type 14-3-3 binding site sequence was identified in BZR1 at amino acid 169-175 (RISNSCP) (Figure 2C). Mass spectrometric analysis of BIN2-phosphorylated BZR1 protein showed that serine-173 in the putative 14-3-3 binding site of BZR1 is phosphorylated by BIN2 (Figure S2). When the binding site (RISNS, at position 169-173) was deleted (BZR1Δ) or the serine 173 mutated to alanine (S173A) (Figure 2C), 14-3-3 binding was greatly reduced (Figure 2D). In contrast, deletion of isoleucine-170 (ΔI170) of BZR1 that changes the sequence from a mode II to a mode I 14-3-3 binding site (Figure 2C) did not abolish 14-3-3 binding (Figure 2D). These results indicate that 14-3-3 proteins bind to BZR1 through the conserved binding site that contains phosphorylated serine 173.
The in vivo interactions of wild type and mutant BZR1s with 14-3-3λ were studied using a Bi-Molecular Fluorescence Complementation system (BiFC). BiFC is based on the principle that a functional fluorescence complex is observed when two proteins of interest fused to the N- and C- terminal halves of YFP interact in vivo. Co-transformed BZR1-cYFP and 14-3-3λ-nYFP produced strong YFP fluorescence (Figures 2E and 2G). The fluorescence signal was reduced by BR treatment (Figures 2F and 2G), which induces BZR1 dephosphorylation. By contrast, cotransformation of BZR1Δ-cYFP with 14-3-3λ-nYFP or BZR1-cYFP with a non-fusion nYFP yielded very low fluorescence signal (Figure 2G). Consistent with the BiFC data, co-immunoprecipitation experiments (Co-IP) using transgenic Arabidopsis plants demonstrated that wild type BZR1 and BZR1ΔI170, but not ΔBZR1 or BZR1S173A, interacted with 14-3-3 proteins in vivo (Figure 2H and data not shown). These results indicate that phosphorylated BZR1 interacts with 14-3-3 in vivo and that this interaction is inhibited by BR-induced BZR1 dephosphorylation.
14-3-3 Binding Inhibits BZR1's Activity
To understand the function of BZR1:14-3-3 interaction in BR signaling, we generated transgenic plants expressing BZR1 containing 14-3-3 binding site mutations. Interestingly, transgenic plants expressing BZR1Δ (16/92; 17%) or BZR1S173A (14/104; 13%) showed phenotypes similar to bzr1-1D (Figure 3), including shorter petioles, rounder and curled leaves (Figures 3A and 3B), delayed flowering (Figure 3C), bending of the stem at the branch junction (Figure 3D), long hypocotyls when grown on BR biosynthesis inhibitor BRZ in the dark (Figure 3E), and suppression of bri1-5 (Figure 3F). In contrast, none of the transgenic plants transformed with wild type BZR1 (0/157) or mutant BZR1ΔI170 (0/118) showed any bzr1-1D-like phenotype (Figure 3). Consistent with the morphological phenotypes, the expression of BZR1-target genes CPD and DWF4 (He et al., 2005) is greatly reduced in BZR1Δ as well as bzr1-1D plants (Figure 3G). Expression of BZR1S173A using the native BZR1 promoter also caused bzr1-1D-like phenotype (Figure S3). These results indicate that 14-3-3 proteins interact with BZR1 to inhibit its function and abolition of such interaction leads to increased BZR1 activity.
Figure 3. 14-3-3 Binding Inhibits BZR1 Activity.
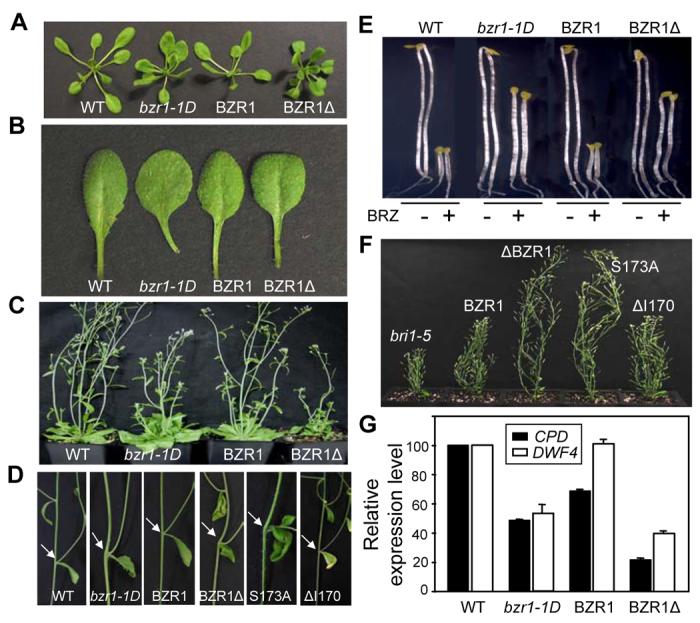
(A-D) Phenotypes of BZR1Δ and BZR1S173A transgenic plants compared with wild type Columbia (WT) and bzr1-1D. Three-week old plants (A) and leaves (B), five-week old plants (C) and bending of the stems at branch junction (pointed by arrows) (D).
(E) BZR1Δ and BZR1S173A mutations suppress BR-deficiency phenotypes. Seedlings were grown in the dark on MS medium with or without 2 μM BRZ for 4 days. Two representative seedlings for each treatment are shown.
(F) Mutations in BZR1 that reduce 14-3-3 binding suppress bri1-5 phenotype. Plants shown from left to right are bri1-5, bri1-5/BZR1-YFP, bri1-5/BZR1Δ-YFP, bri1-5/BZR1S173A-YFP and bri1-5/BZR1ΔI170-YFP.
(G) BR biosynthesis genes are down regulated in BZR1Δ plants. Expression levels of CPD and DWF4 genes were measured by Quantitative Real Time PCR. The data was normalized first to UBC and then to Col. Error bars are ± standard errors of mean.
Mutations in 14-3-3 Binding Site Do Not Affect Accumulation, Dephosphorylation, or DNA Binding Activity of BZR1
The dominant gain-of-function mutation in bzr1-1D mutant causes accumulation of BZR1 protein leading to constitutive BR-response phenotypes (Wang et al., 2002). To determine whether the phenotypes of BZR1Δ and BZR1S173A plants were due to altered BZR1 protein accumulation or phosphorylation, we performed immunoblot analysis. Despite causing strong phenotypes, mutations of the 14-3-3 binding site did not increase the accumulation or dephosphorylation of BZR1 protein (Figure 4A). South-Western Blot analysis showed that these mutations did not affect BZR1's ability to bind DNA, because phosphorylation by BIN2 abolished the DNA binding activity of the wild type as well as the mutant BZR1 proteins (Figure 4B). Therefore, the bzr1-1D-like phenotypes displayed by BZR1Δ and BZR1S173A plants are unlikely due to a difference in the accumulation or DNA binding activity of the BZR1 protein.
Figure 4. Mutations in 14-3-3 Binding Site Do Not Affect Stability or DNA Binding Activity of BZR1.
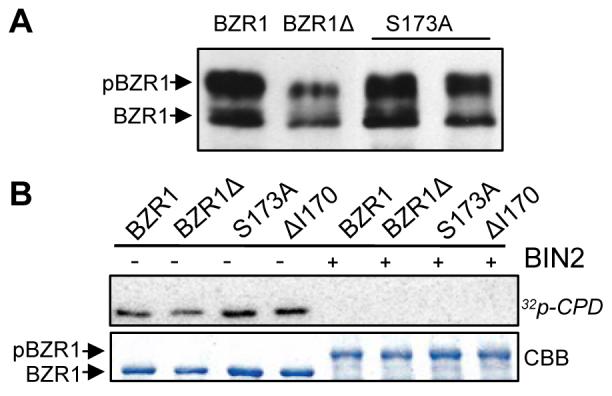
(A) Immunoblot of BZR1-YFP, BZR1Δ-YFP, and BZR1S173A-YFP (two independent lines) proteins in the transgenic plants. BZR1-YFP was detected using a α-GFP antibody.
(B) Mutations in 14-3-3 binding site do not alter the DNA binding activity of BZR1. Gel blot of unphosphorylated and BIN2-phosphorylated MBP-BZR1 proteins was probed with radio-labeled CPD promoter DNA. CBB, Coomassie Brilliant Blue stained gel.
14-3-3 Binding Increases Cytoplasmic Localization of BZR1
14-3-3 proteins have been shown to negatively regulate nuclear localization of target proteins in both metazoans and plants (Igarashi et al., 2001; Muslin and Xing, 2000). Considering that BR induces nuclear localization of BZR1, we examined whether BZR1Δ- and BZR1S173A-YFP have altered subcellular localization (Figure 5). Indeed, BZR1Δ-YFP and BZR1S173A-YFP proteins showed significantly increased nuclear localization in Arabidopsis and tobacco cells (Figures 5A, 5B and S4), whereas BZR1ΔI170-YFP was localized in both cytoplasm and nucleus similar to wild type BZR1-YFP (Figures 5A and S4). Quantification of the nuclear/cytoplasmic signals confirmed these microscopic observations (Figure 5B). BRZ increased the cytoplasmic signal of wild type BZR1-YFP, but had little effect on BZR1Δ-YFP (Figures 1A and S5). When transiently co-expressed in tobacco leaf cells, both BZR1-CFP and bzr1-1D-CFP proteins showed identical localization patterns with that of cotransformed BZR1-YFP, indicating that the bzr1-1D mutation, which increases the protein accumulation, does not affect the subcellular localization of the protein (Figure 5C). In contrast, BZR1Δ-CFP showed much reduced levels of cytoplasmic signal as compared to BZR1-YFP (Figure 5C). This difference in localization disappeared when cells were treated with BR, presumably due to BR-induced nuclear localization of BZR1-YFP (Figures 5C and S1). Consistent with microscopic observations, subcellular fractionation showed that BZR1S173A-YFP protein levels are reduced in the cytosolic fraction compared to the BZR1ΔI170-YFP (Figure S6). These results demonstrate that mutation of the 14-3-3 binding site increases the nuclear localization of BZR1.
Figure 5. 14-3-3 Binding Reduces Nuclear Localization of BZR1.
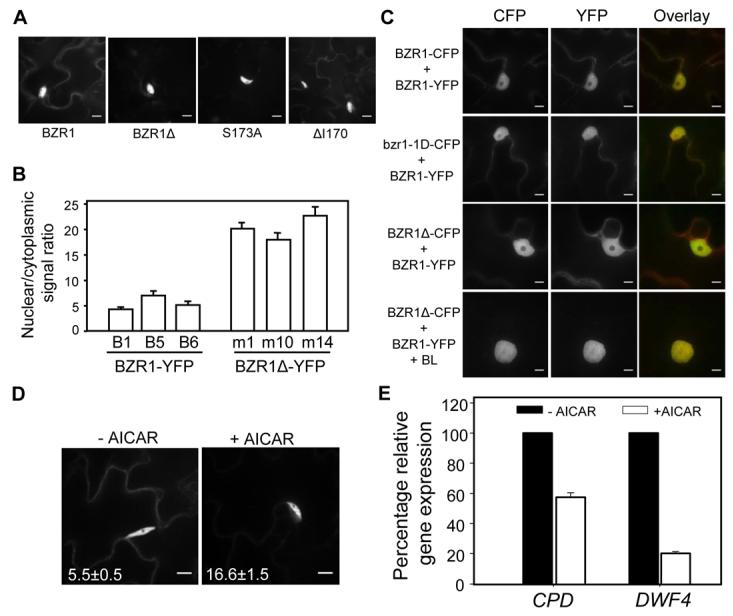
(A-B) Subcellular localization of BZR1-, BZR1Δ-, BZR1S173A-, and BZR1ΔI170-YFP proteins in the leaves of transgenic Arabidopsis plants. Representative images of YFP fluorescence (A) and quantification of nuclear/cytoplasmic signal ratios (B) of three independent lines from BZR1-YFP and BZR1Δ-YFP are shown. Error bars are ± standard errors of mean.
(C) BZR1Δ-CFP protein predominantly localizes in the nucleus. Pairs of indicated YFP and CFP fusion constructs were co-transformed into tobacco leaves with or without 1 μM BL. Images in CFP and YFP channels were false-colored as green and red, respectively, in overlay. Scale bar is 10 μm.
(D) AICAR treatment decreases cytoplasmic localization of BZR1. Transgenic plants expressing BZR1-YFP were treated with either mock solution or 20 mM AICAR for 1 hr, and images were obtained using confocal microscope. Scale bar is 10 μM. Numbers in each image show the average ratios between nuclear and cytoplasmic signal intensities and standard errors calculated from seven cells for each treatment.
(E) AICAR treatment down regulates BZR1-target gene expression. Transgenic plants expressing BZR1-YFP were treated with either mock solution or 20 mM AICAR for 3 hr and expression of CPD and DWF4 was measured using qRT-PCR.
Previous studies have shown that 14-3-3 interactions with the target proteins can be disrupted by 5-Aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) (Paul et al., 2005). We observed that AICAR treatment increased the nuclear/cytoplasmic ratio of BZR1, and reduced the expression of BZR1-target genes CPD and DWF4 (Figures 5D and 5E), further supporting that disrupting 14-3-3 binding increases BZR1's nuclear localization and activates its function. Together these results strongly support a model that phosphorylated BZR1 is retained in the cytoplasm by binding to 14-3-3 proteins and BR-induced dephosphorylation abolishes 14-3-3 binding to allow nuclear localization and transcriptional regulation of BZR1.
14-3-3 Binding Negatively Regulates BZR2/BES1 Function
Yeast two-hybrid screen also showed interaction between BZR2/BES1 and 14-3-3 proteins (Figure 6A). To determine if subcellular localization of BZR2/BES1 is also regulated by BR and 14-3-3 proteins, we measured the nuclear/cytoplasmic signal ratio of BZR2/BES1-GFP in transgenic Arabidopsis plants using confocal microscopy. We observed that, similar to BZR1-YFP (Figures 1A and 1B), BZR2/BES1-GFP localization in the nucleus was decreased by BRZ treatment and increased by BL treatment (Figure 6B). BZR2/BES1 contains a putative 14-3-3 binding site (Figure S7), and mutation of S171A in BZR2/BES1 (equivalent to S173A in BZR1) increased the nuclear localization of the protein (Figures 6C and S7). These results collectively indicate that 14-3-3 proteins function in BR signal transduction by regulating the subcellular localization and activity of both BZR1 and BZR2/BES1.
Figure 6. BR Treatment and Mutation of 14-3-3 Binding Site Increase Nuclear Localization of BZR2/BES1.
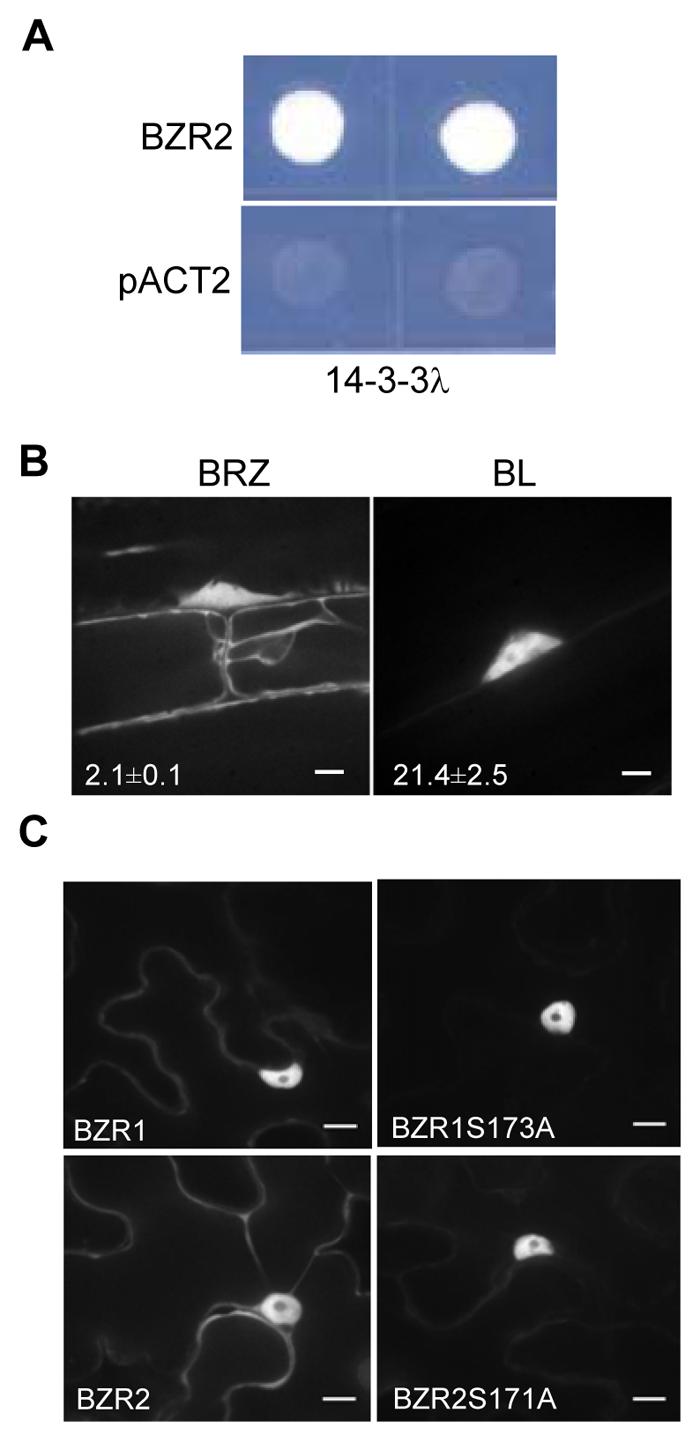
(A) 14-3-3λ interacts with BZR2 in yeast two-hybrid assays. pACT2 is the empty prey vector as a negative control.
(B) BL treatment increases the nuclear localization of BZR2. BZR2-GFP transgenic seedlings were grown on media containing 2 μM BRZ or 10 nM BL in the dark for 4 days, and BZR2-GFP subcellular localization was visualized by confocal microscopy. Numbers in each image show the average ratios between nuclear and cytoplasmic signal intensities and standard errors calculated from seven cells for each treatment. Error bars are ± standard errors of mean.
(C) YFP fusion constructs of BZR1, BZR1S173A, BZR2 or BZR2S171A were transiently expressed in tobacco leaves and subcellular localization of YFP was observed. Scale bar is 10 μm.
DISCUSSION
In the present study, we illustrate a critical role of 14-3-3 proteins in BR signal transduction. Our data demonstrate that BIN2-catalized phosphorylation inhibits the function of the BZR1 and BZR2/BES1 proteins not only by inhibiting DNA binding but also by promoting binding to the 14-3-3 proteins, which increases cytoplasmic retention of BZR1 and BZR2/BES1. Together with previous studies, we have shown that phosphorylation can inhibit BZR1 and BZR2/BES1 through at least four mechanisms: increasing proteolysis, inhibiting DNA binding, promoting 14-3-3 binding, and cytoplasmic retention (Figure 7). These multiple mechanisms of regulation are consistent with the presence of large numbers of putative BIN2-phosphorylation sites in BZR1 and BZR2/BES1, and likely act together to confer efficient and precise regulation of key transcription factors required for normal BR response and plant growth.
Figure 7. A Model for the BR Signaling Pathway in Arabidopsis.
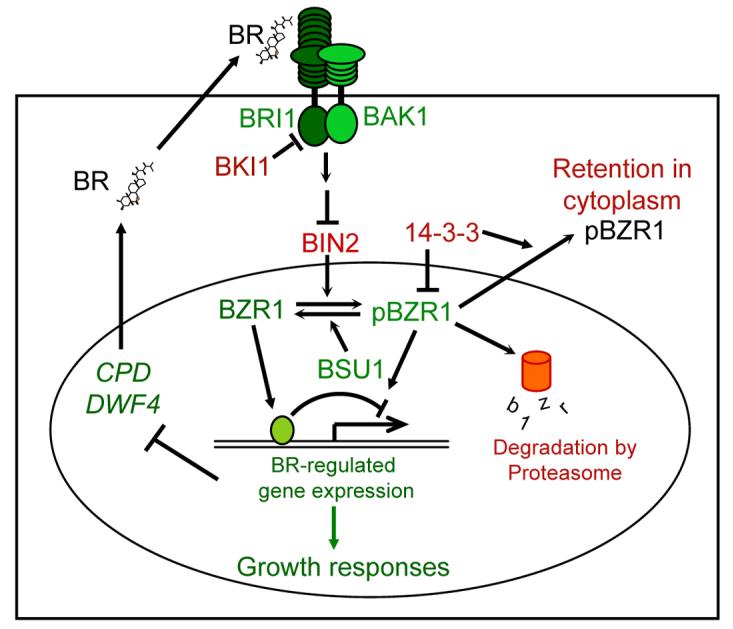
BR signaling involves a cell-surface receptor complex (BRI1/BAK1), a GSK3 kinase (BIN2), a phosphatase (BSU1) and two homologous transcription factors (BZR1 and BZR2/BES1). Green and red colors represent positive and negative functional roles in BR signaling, respectively. Arrows and bars represent actions of promotion and inhibition, respectively. In the absence of BR, BKI1 suppresses BRI1, and BIN2 phosphorylates and inhibits BZR1 and BZR2/BES1 (not shown). BR binds to the extracellular domain of BRI1 to activate its kinase, which leads to disassociation of BKI1 from BRI1 and dimerization with and activation of BAK1. Then, through an unknown mechanism, the activated receptor kinases inhibit BIN2 or activate BSU1, yielding dephosphorylated BZR1 and BZR2/BES1, which directly regulate BR-responsive gene expression. BIN2 phosphorylation negatively regulates BZR1 by increasing proteasomal degradation and BZR2/BES1 by inhibiting DNA binding. The current study shows that BR promotes nuclear localization of BZR1 and BZR2/BES1. BIN2 phosphorylation of BZR1 not only inhibits its DNA binding but also promotes its binding with 14-3-3 proteins, which is required for cytoplasmic retention and efficient inhibition of phosphorylated BZR1 in vivo. 14-3-3 proteins also regulate subcellular localization of BZR2/BES1. We propose that the large numbers of BIN2-phosphorylation sites in BZR1 and BZR2/BES1 allow BIN2 to inhibit BZR1/BZR2 through multiple mechanisms, conferring an efficient control of gene expression and plant growth by BR signaling.
Multiple mechanisms of regulation of key transcription factors have been observed in signaling pathways in animals. For example, the Ci/Gli transcription factor of the Hedgehog (Hh) pathway is controlled by multi-layered regulatory mechanisms. It is believed that tissue-, and developmental stage-specific expression of some of the additional regulators in vertebrates provides a mechanism for ensuring precision in spatial and temporal control of Hh signaling (Jiang, 2006). Similarly, multiple mechanisms of BZR1 regulation may provide higher levels of efficiency and precision than a single mechanism in BR signaling. One might expect that inhibiting DNA binding should be sufficient to turn off the transcription factors. However, BZR1Δ-YFP and BZR1S173A-YFP, but not the wild type BZR1-YFP, cause bzr1-1D-like phenotypes in the transgenic plants, despite the fact that phosphorylation inhibits their DNA binding activity in vitro. Apparently phosphorylation per se is not sufficient and binding to the 14-3-3 proteins is required for efficient inhibition of BZR1 activity in vivo. This is not surprising since genome-wide surveys of in vivo DNA binding by transcription factors have revealed discrepancies between in vitro and in vivo DNA binding specificities of transcription factors (Biggin, 2001). In vivo DNA binding by transcription factors is not only influenced by their intrinsic sequence-specific recognition properties, but also affected by other factors, such as chromatin structure or cooperative interactions with other DNA binding proteins. It is conceivable that when not bound by 14-3-3s, phosphorylated BZR1 and BZR2/BES1 are localized in the nucleus and can regulate target genes by interacting with partner DNA binding proteins, such as BIM1(Yin et al., 2005).
Multiple mechanisms would also allow fine-tuning of BR sensitivity by other environmental or developmental signals. For example, sugars globally affect 14-3-3 binding to target proteins (Cotelle et al., 2000), and stresses and abscisic acid (ABA) increase expression of plant 14-3-3 proteins (Chen et al., 2006; Schoonheim et al., 2007b), which could potentially contribute to the antagonistic interaction between BR and ABA (Friedrichsen et al., 2002). Such regulation of 14-3-3s by environmental and developmental cues might explain the previous conflicting observations of constitutive nuclear accumulation of BZR1 and BZR2/BES1 (Vert and Chory, 2006; Zhao et al., 2002). In fact, we observed strong nuclear-localization response in young growing tissues, but weak response in hypocotyls of older seedlings (Figure S8) and no obvious response in fully developed mature leaves that have finished cell elongation (Figure S8). Therefore, 14-3-3s provide a potential point of cross-talk for other pathways to modulate BR response, which is important for optimizing growth according to physiological and environmental conditions. Nevertheless, the strong bzr1-1D-like phenotypes shown by the BZR1Δ and BZR1S173A transgenic plants grown in both light and dark support a ubiquitous and critical role for 14-3-3 proteins in regulating BZR1.
14-3-3 proteins increase the cytoplasmic retention of phosphorylated BZR1. Disrupting 14-3-3 binding, either by mutating the binding site in BZR1 or treatment with AICAR, leads to increased nuclear localization of BZR1. BR induced nuclear localization correlates well with BZR1 dephosphorylation and inhibition of 14-3-3 binding. These results strongly support an important role of 14-3-3s in BR-regulation of subcellular localization of BZR1. BIN2 phosphorylation promotes cytoplasmic retention of BZR1 by promoting 14-3-3 binding, and BR-induced dephosphorylation increases nuclear localization of BZR1 by abolishing 14-3-3 binding.
The importance of cytoplasmic retention for inhibition of phosphorylated BZR1 is supported by the strong bzr1-1D-like phenotypes and altered expression of BZR1 target genes that are associated with increased BZR1 nuclear localization. It is possible, however, that 14-3-3 proteins may also inhibit other aspects of BZR1 function, such as BZR1's transcriptional activity or its interaction with other partner proteins. However, such additional effects on transcriptional activity would be difficult to discern in the presence of altered nuclear localization. It should be noted that the nuclear BiFC signal of BZR1:14-3-3λ interaction suggests that 14-3-3s might inhibit the phosphorylated BZR1 in the nucleus, although it is possible that BZR1 cytoplasmic retention is mediated by other members of the 14-3-3 family rather than 14-3-3λ in vivo. Four additional 14-3-3 isoforms interacted with BZR1 in our yeast two-hybrid screens and at least three isoforms co-immunoprecipitated with BZR1-YFP. In fact, 14-3-3λ is predominantly localized in the nucleus while 14-3-3ω, which interacts with BZR1 in yeast, 14-3-3ϕ and 14-3-3υ all show wide distribution in the cytoplasm, at least in certain cell types (Paul et al., 2005; Sehnke et al., 2002). Therefore, it is unclear whether a particular 14-3-3 isoform(s) mediates cytoplasmic retention of BZR1 and BZR2/BES1 in vivo. Although cytoplasmic retention may not be the only mechanism of 14-3-3 inhibition of BZR1, it certainly contributes to the inhibition.
Similar functions of 14-3-3 proteins in regulating protein nuclear localization have been observed in several other systems. For example, in tobacco, 14-3-3s mediate gibberellin (GA) regulation of nuclear localization of transcriptional activator REPRESSION OF SHOOT GROWTH (RSG) (Ishida et al., 2004), in a manner similar to the regulation of BZR1. However, it remains unclear how GA signaling, which is mediated by a soluble receptor that controls degradation of a nuclear component, regulates the RSG-14-3-3 interaction (Ishida et al., 2004). In yeast, phosphorylation of Cdc25 by Chk1 kinase and MSN2 by TOR kinase creates binding sites in the transcription factors for 14-3-3s, which promote nuclear export of Cdc25 and MSN2, leading to DNA-damage checkpoint and inhibition of carbon-source-regulated genes, respectively (Beck and Hall, 1999; Lopez-Girona et al., 1999). Interestingly, 14-3-3 family proteins have also been shown to differentially regulate the glucocorticoid receptor (GR). 14-3-3η is known to bind to GR and function as a positive regulator of GR by blocking its degradation (Kim et al., 2005), whereas 14-3-3σ binds to GR and function as a negative regulator by retaining GR in the cytoplasm (Kino et al., 2003). A more striking parallel to the BR pathway is the regulation of heat shock transcription 1 (HSF1) by GSK3 phosphorylation and 14-3-3 binding in humans (Wang et al., 2003; 2004). It remains unclear whether BR signaling pathway is evolutionarily related to any animal signaling pathways, such as the heat shock pathway, the Wnt signaling pathway, or cell-surface receptor-mediated steroid signaling pathways. The use of evolutionarily conserved signaling proteins such as GSK3 and 14-3-3s supports that BR signal transduction pathway is an ancient steroid signaling pathway.
In summary, this study illustrates a molecular mechanism by which 14-3-3 proteins mediate phosphorylation-dependent control of the BZR1 and BZR2/BES1 transcription factors in the BR signaling pathway of Arabidopsis. A similar mechanism of 14-3-3 regulation of rice BZR1 homolog has also been observed (Bai and Wang, unpublished), suggesting that the regulation of localization and activity of BZR1 by 14-3-3s is a conserved mechanism for steroid regulation of gene expression in higher plants. 14-3-3 proteins may also regulate other components of the BR signaling pathway. Recent studies have shown that 14-3-3s co-purify with BRI1 (Karlova et al., 2006), and interact directly with BAK1's Arabidopsis homolog SERK1 (Rienties et al., 2004) and barley homolog HvBAK1 (Schoonheim et al., 2007a). How 14-3-3s regulate the receptor kinases remains to be elucidated, and the physiological implications of 14-3-3 regulation of BR signaling is yet to be fully appreciated by future studies.
EXPERIMENTAL PROCEDURES
Plant Material
Arabidopsis thaliana ecotype Columbia-0, bzr1-1D, bri1-5, transgenic Arabidopsis plants harboring either full length YFP fusions or Bi-Molecular Fluorescence Complementation (BiFC) constructs and Nicotiana benthamiana (tobacco) plants were all grown in green houses under 16 hr light long day conditions.
Dark-Grown Phenotypic Analysis
Arabidopsis seeds were sterilized with bleach and germinated on half strength Murashige and Skoog (MS) medium supplemented with or without 2 μM BRZ. Seeds were stratified for 3 days at 4°C, kept under white light for 6 hr before moving into the dark. Hypocotyl lengths from 4-day old vertically grown seedlings were measured using ImageJ software available at the NIH website (http://rsb.info.nih.gov/ij/).
Isolation of Nuclear Fraction
Transgenic seedlings expressing BZR1-CFP were grown in liquid medium containing half strength MS and 1% sucrose for 9 days under continuous light. They were treated with 100 nM BL or mock solution for 1 hr and samples were harvested. Nuclei protein fractions (Figure 1C) were extracted according to a published protocol (Bae et al., 2003). For the data shown in Figure S6, nuclei fractions were isolated using the following protocol. Plant material was ground in NEB buffer (20 mM HEPES, pH7.5; 40 mM KCl; 10 mM MgCl2; 1% Triton X-100; 1 mM EDTA; 10% Glycerol) and filtered through Miracloth (Calbiochem, San Diego, CA) to obtain the total extract. It was then centrifuged at 5000×g for 2 min to separate cytosolic fraction (supernatant) from nuclei fraction (pellet). SDS gel loading buffer was added to each fraction, heated at 65°C for 5 min, centrifuged at 20,000×g for 5 min, and analyzed on a 4-12% gradient SDS- PAGE gel (Invitrogen).
Plasma Membrane Fractionation
One-week old liquid-grown seedlings treated with either mock or 100 nM BL for 2 hr, were processed for plasma membrane (PM) preparation (Figure 1D). Seedlings were ground in 2 vol. of Buffer H (100 mM HEPES-KOH, pH 7.5; 330 mM Sucrose; 10% (v/v) Glycerol; 5 mM EDTA; 5 mM Ascorbic acid; 0.6% (w/v) PVP 40; 5 mM DTT; 25 mM NaF; 2 mM Imidazole; 1 mM Sodium molybdate and Protease inhibitors) with a Powergen 700D homogenizer at 5,000 rpm for 3 min. Homogenate was filtered through a single layer of miracloth, and centrifuged at 10,000×g for 10 min to remove cell debris and organelles. Microsomal fraction was collected by centrifuging at 60,000×g for 30 min. An aliquot of supernatant containing the soluble protein was snap frozen in liquid nitrogen. The microsomal pellet was resuspended in Buffer R (5 mM Potassium phosphate, pH 7.8; 330 mM Sucrose; 3 mM KCl; 0.1 mM EDTA; 1 mM DTT and Protease inhibitors). Similarly, an aliquot of microsomal fraction was loaded onto a two-phase partitioning system containing 6% polymers and 8 mM KCl, and partitioning was done according to Larsson et al. (1994). U3 upper phase was diluted with 10 vol. of Buffer R and centrifuged at 200,000×g for 1 hr to pellet PM vesicles. After the final centrifugation, PM vesicles were resuspended in a buffer containing 5 mM Potassium phosphate, pH 7.8; 250 mM Sucrose; 3 mM KCl; 0.1 mM EDTA. All protein samples were quantified using Bradford assay (Bio-Rad, Hercules, CA) and equal amount of proteins were loaded on the SDS-PAGE gels for immunoblots, which were probed using a polyclonal anti-GFP antibody (Wang et al., 2002).
Southwestern Blot
Phosphorylation of MBP-BZR1 by GST-BIN2 was carried out in the presence of 100 μM ATP as described previously (He et al., 2002). Unphosphorylated and phosphorylated MBP-BZR1 proteins were separated on SDS-PAGE gels and blotted on nitrocellulose membrane. Blot was incubated with a CPD probe that is chemically synthesized (5'-aaaaggtcctactttatgcagaaaccccccgtgtgcccactctccccttc-3') and labeled with 32P-dTTP.
Overlay Western Blot
Gel blot containing unphosphorylated and phosphorylated MBP-BZR1 proteins was incubated with 20 μg of recombinant GST-14-3-3λ protein, washed, and then probed with anti-GST antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Mass Spectrometry
MBP-BZR1 protein phosphorylated by GST-BIN2 in vitro was subjected to standard in-solution alkylation/tryptic digestion. Briefly, 1000 molar excess DTT over total proteins in 25 mM ammonium bicarbonate was added and the reduction reaction was carried out for 1 hr at 37°C. Then, 5.5x molar excess iodoacetamide over DTT was added in 25 mM ammonium bicarbonate and the alkylation reaction was carried out at 25°C in the dark for 1 hr. Low molecular weight compounds in the reaction mixture were reduced by using Amicon Ultra-4 10K (Millipore, Billerica, MA) and proteins were in a buffer containing 25 mM ammonium bicarbonate and 20% acetonitrile. Tryptic digestion was carried out overnight by 1% (w/w) modified trypsin (Promega, Madison, WI) and was stopped by adding formic acid to a final concentration of 1% in the solution.
The digestion mixture was diluted in water to a concentration of 100 fmole/μL and 1 μL of the aliquot was injected into an UltiMate Capillary LC system via a FAMOS Autosampler (LC Packings, Sunnyvale, CA). Peptides were separated on a 75μm × 15cm C18 reverse phase capillary column at a flow rate of 300 nL/min while running a 3-32% acetonitrile gradient with 0.1% formic acid. The HPLC eluent was connected directly to an LTQ/FT instrument (Thermo Finnigan, Bremen, Germany). The LC-MS/MS method consisted of one survey scan in FT mode with a resolution of 25,000 followed by three CID scans in LTQ. The raw LC-MS/MS data was converted to ASCII peak list by Mascot Distiller software (Matrix Science, Boston, MA) with carbamidomethylation as a fixed modification and phosphorylation and methionine oxidation as variable modifications.
In Vitro Pull-Down Assay
Unphosphorylated and phosphorylated MBP-BZR1 proteins were mixed together, and were incubated with beads containing either GST-14-3-3λ or GST alone in Binding buffer (50 mM Tris-Cl, pH 7.5; 5 mM MgCl2). After incubation for 1 hr, beads were washed with Binding buffer, and 14-3-3-binding proteins were eluted with elution buffer (50 mM Tris-Cl, pH 7.5, 5 mM Glutathione) and detected using a polyclonal anti-MBP antibody in Western Blot.
Transient Transformation
Agrobacterium cells containing either full-length or BiFC expression vectors were washed and resuspended in the induction medium (10 mM MES buffer, pH 5.6; 10 mM MgCl2 and 150 μM Acetosyringone) and were infiltrated into young leaves of four-week old tobacco plants. Thirty-six hours after infiltration, expression of various fluorescent proteins was analyzed by confocal microscopy. The data generated using this system was also confirmed in stable transgenic Arabidopsis plants.
Co-Immunoprecipitation (IP)
Plant material was ground in NEB buffer (20 mM HEPES, pH 7.5; 40 mM KCl; 1mM EDTA; 1% Triton X-100), filtered and centrifuged at 20,000×g for 10 min. One milliliter of supernatant was incubated with anti-GFP antibody coupled to Protein A Sepharose beads for 20 min. Beads were washed four times with wash buffer (20 mM HEPES, pH7.5; 40 mM KCl; 0.1% Triton X-100), and bound proteins were eluted with buffer containing 2% SDS.
Plasmids
The Bi-Molecular Fluorescence Complementation system (BiFC) was engineered based on the 172 N-terminal amino acids (aa1-172) of enhanced Yellow Fluorescent Protein (eYFP), and the 85 C-terminal amino acids (aa154-239) of enhanced Cyan Fluorescent Protein (eCFP) which were subcloned into pPZP312 driven by the CaMV 35S promoter and a CaMV terminator derived from pRT100. These two halves were named nYFP and cYFP, respectively (the cYFP differs from cCFP by two amino acids namely V164A and Y204T). All vectors were converted into gateway-compatible destination vectors by PCR-cloning the Gateway cassette from Gateway Vector Conversion System (Invitrogen, Carlsbad, CA) using the BglII and HindIII restriction sites. Entry clones containing various interacting proteins were recombined with the Gateway-destination vectors using the LR reaction kit (Invitrogen). Full-length coding sequence of 14-3-3λ was cloned directly into binary vectors using XbaI sites engineered into pGREEN0229 and pPZP312 vector backbones. Full-length CFP and YFP fusions of wild type and mutant BZR1 (containing mutations in the 14-3-3 binding site) were made in pEarleyGate (pEG) vectors (Earley et al., 2006) driven by the CaMV 35S promoter. The BZR1:BZR1S173A-CFP construct was created using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the wild type construct as template (Wang et al., 2002). The primers used for site directed mutagenesis PCR are listed in the Supplementary methods. All binary vectors were introduced into Agrobacterium strain GV3101 by electroporation and then transformed into Arabidopsis using the floral dip method. Basta was used for selecting the transgenic plants.
RNA and Protein Analysis
RNA samples were isolated from two-week old transgenic plants using TRIzol reagent (Invitrogen). cDNA was synthesized from total RNA using M-MuLV Reverse Transcriptase (MBI Fermentas, Hanover, MD) and oligo dT. Quantitative Real Time PCR was performed using iQ SYBR Green reagent (Bio-Rad, Hercules, CA) to amplify BZR1-YFP, UBC, CPD and DWF4 gene-specific regions. Primer sequences used for amplification are listed in Supplementary methods. Transgenic lines were compared for transcript abundance and subsequently analyzed by Western Blots. BZR1-YFP protein was extracted in 2x SDS-PAGE buffer and was detected using a polyclonal antibody against GFP (Wang et al., 2002).
Spinning-Disk Confocal Microscopy
Fluorescence of CFP, YFP, and BiFC (YFP) were visualized using a Spinning-Disk Confocal microscope (Leica Microsystems, Heerbrugg, Germany) (Paredez et al., 2006). In brief, CFP was excited by a HeCd laser at 442 nm and YFP was excited by an Argon laser at either 488 or 514 nm, and emission filtering was achieved using band pass filters 480/40, 525/50, 570/65 (Chroma Technology Corp., Rockingham, VT). Both Glycerin (n.a.=1.3) and oil-immersion (n.a.=1.4) 63x objectives were used for confocal microscopy and all images were acquired with a 512b Roper Cascade EMCCD camera using a gain of 3800. Two-frame averaging of image acquisition improved the signal to noise ratio. All images were acquired using MetaMorph software (Molecular Devices, Sunnyvale, CA). Images of BZR1-YFP and mutant BZR1-YFPs were obtained with identical image acquisition settings; YFP was excited at 488 nm and images were obtained with a 500 ms exposure time. For quantifying the effect of BRZ and BL on BZR1-YFP localization, images were obtained with a 200 ms exposure time (Figure 1A). For the time course experiment (Figure 1B), seedlings were grown on 2 μM BRZ in the dark for 4 days, then mounted on a slide in 100 nM BL, and were imaged immediately to obtain the image at time zero. The same field of view was imaged subsequently for the duration of the time course described in Figure 1B. Similar change in nuclear localization was observed when seedlings already under the microscope in liquid MS medium were profused with BL solution. Time course experiments were repeated three times and representative data are shown in Figure 1B. Images of YFP fluorescence in BiFC assays were obtained using 488 nm excitation with a 500 ms exposure time (Figure 2).
Quantification of Fluorescent Protein Signal
All images were analyzed in ImageJ (http://rsb.info.nih.gov/ij/). To measure the ratio between nuclear and cytoplasmic signals of BZR1-YFP for each cell, a small area of fixed size (21 pixels) was drawn and measurements of integrated densities were taken from representative areas within the nucleus, cytoplasm and background (central vacuole) of each cell. Three repeat measurements were performed for each cell and the average of background values was then subtracted from the average values for the nuclear and cytoplasmic signals. Nuclear to cytoplasmic signal ratio (N/C ratio) was then calculated for each cell. Average N/C ratio and standard error for independent T1 transgenic lines were calculated from measurements of at least 7 cells from each plant (Figure 4B). Line scan measurements spanning both the nucleus and cytoplasm were also carried out and representative plot profiles of sample measurements are presented in Figure 1A. For quantifying the BiFC signal, an outline was drawn around each transformed cell and the signal intensity (integrated density) of the whole cell was calculated after subtracting the mean background density measured from an adjacent untransformed cell. Signal intensities from at least seven transformed cells were then averaged (Figure 2G).
Supplementary Material
Supplementary Movies SM1 and SM2. BR induces rapid nuclear localization of BZR1. BZR1-YFP seedlings grown on 2 μM BRZ were treated with BL (SM1) or mock solution (SM2), and images were acquired at every two min intervals. Images were then converted into a stack using ImageJ software.
ACKNOWLEDGEMENTS
We thank W. Briggs and L. West for comments on the manuscript; W. Frommer and D. Moss for providing the BiFC vectors; T. Asami for providing BRZ; J. Chory for providing BES1-GFP vector, Yu Sun for Real Time PCR analysis; and R. Kumar and N. Gendron for technical assistance. This work was supported by grants from NIH (R01 GM66258-01), the U.S. Department of Energy (DE-FG02-04ER15525), NSFC (No. 30328004), NIH (NCRR 01614, A.L.B.), the Post-doctoral Fellowship Program of Korea Science & Engineering Foundation (C00251, T.-W. Kim), and a training grant from NIH (5T32GM007276, J.M.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include supplemental experimental procedures, eight supplemental figures and two supplemental movies, and are available with online version of the paper at http://www.developmentalcell.com.
REFERENCES
- Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bae MS, Cho EJ, Choi EY, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003;36:652–663. doi: 10.1046/j.1365-313x.2003.01907.x. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Biggin MD. To bind or not to bind. Nat Genet. 2001;28:303–304. doi: 10.1038/91045. [DOI] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- Chen F, Li Q, Sun L, He Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 2006;13:53–63. doi: 10.1093/dnares/dsl001. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. Embo J. 2000;19:2869–2876. doi: 10.1093/emboj/19.12.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang Z-Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell. 2004;16:2641–2651. doi: 10.1105/tpc.104.024604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli Signaling by Dual Ubiquitin Pathways. Cell Cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 Protein Complex Includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Jang SW, Sung HJ, Lee HJ, Kim IS, Na DS, Ko J. Role of 14-3-3 eta as a positive regulator of the glucocorticoid receptor transcriptional activation. Endocrinology. 2005;146:3133–3140. doi: 10.1210/en.2004-1455. [DOI] [PubMed] [Google Scholar]
- Kino T, Souvatzoglou E, De Martino MU, Tsopanomihalu M, Wan Y, Chrousos GP. Protein 14-3-3sigma interacts with and favors cytoplasmic subcellular localization of the glucocorticoid receptor, acting as a negative regulator of the glucocorticoid signaling pathway. J Biol Chem. 2003;278:25651–25656. doi: 10.1074/jbc.M302818200. [DOI] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol. 1994;228:451–469. [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA. 1992;89:11490–11494. doi: 10.1073/pnas.89.23.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- Paul AL, Sehnke PC, Ferl RJ. Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol Biol Cell. 2005;16:1735–1743. doi: 10.1091/mbc.E04-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienties IM, Vink J, Borst JW, Russinova E, Vries SC. The Arabidopsis SERK1 protein interacts with the AAA-ATPase AtCDC48, the 14-3-3 protein GF14lambda and the PP2C phosphatase KAPP. Planta. 2004;221:394–405. doi: 10.1007/s00425-004-1447-7. [DOI] [PubMed] [Google Scholar]
- Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, Quatrano RS, de Boer AH. 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 2007b;49:289–301. doi: 10.1111/j.1365-313X.2006.02955.x. [DOI] [PubMed] [Google Scholar]
- Schoonheim PJ, Veiga H, da Costa Pereira D, Friso G, van Wijk KJ, de Boer AH. A Comprehensive Analysis of the 14-3-3 Interactome in Barley Leaves Using a Complementary Proteomics and Two-hybrid Approach. Plant Physiol. 2007a;243:670–683. doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell. 2002;14(Suppl):S339–354. doi: 10.1105/tpc.010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr Biol. 2006;16:R378–385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Chory J. Steroid signaling in plants and insects--common themes, different pathways. Genes Dev. 2002;16:3113–3129. doi: 10.1101/gad.1042102. [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- Wang X, Grammatikakis N, Siganou A, Calderwood SK. Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3 epsilon binding, and cytoplasmic sequestration of heat shock factor 1. Mol Cell Biol. 2003;23:6013–6026. doi: 10.1128/MCB.23.17.6013-6026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grammatikakis N, Siganou A, Stevenson MA, Calderwood SK. Interactions between extracellular signal-regulated protein kinase 1, 14-3-3epsilon, and heat shock factor 1 during stress. J Biol Chem. 2004;279:49460–49469. doi: 10.1074/jbc.M406059200. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movies SM1 and SM2. BR induces rapid nuclear localization of BZR1. BZR1-YFP seedlings grown on 2 μM BRZ were treated with BL (SM1) or mock solution (SM2), and images were acquired at every two min intervals. Images were then converted into a stack using ImageJ software.