Abstract
Epstein–Barr virus (EBV) is an exceptionally successful human viral pathogen maintained as a licensed, plasmid replicon in proliferating cells. We have measured the distributions of EBV-derived plasmids in single live cells throughout the cell cycle in the absence of selection and confirmed the measured rates of duplication and partitioning computationally and experimentally. These analyses have uncovered a striking, non-random partitioning for this minimalist plasmid replicon and revealed additional properties of it and its host cells: (1) 84% of the plasmids duplicate during each S phase; (2) all duplicated plasmids are spatially colocalized as pairs, a positioning that is coupled to their non-random partitioning; (3) each clone of cells requires a certain threshold number of plasmids per cell for its optimal growth under selection; (4) defects in plasmid synthesis and partitioning are balanced to yield wide distributions of plasmids in clonal populations of cells for which the plasmids provide a selective advantage. These properties of its plasmid replicon underlie EBV's success as a human pathogen.
Keywords: Epstein–Barr virus, licensed plasmid replicon, partitioning, replication
Introduction
Multiple human tumor viruses maintain their genomes extrachromosomally using host cell machinery for many of their replicative functions (Chow and Broker, 2006; Fanning and Pipas, 2006; Hammerschmidt and Sugden, 2006; Seeger and Mason, 2006). Epstein–Barr Virus (EBV), which causes both lymphomas and carcinomas, is an extreme example of this cellular parasitism, minimally encoding two cis-acting elements, a Dyad symmetry (DS) as an origin of DNA synthesis, a Family of Repeats (FR) as a maintenance element, and a trans-acting protein, EBV nuclear antigen (EBNA1) that binds these DNA elements to foster their activities (Hammerschmidt and Sugden, 2006). EBV depends on its human host cell for all other functions required for synthesis and partitioning of its plasmids. Studies of EBV plasmids have helped not only to elucidate the mechanisms of their synthesis, but also to understand the cellular activities on which they depend. Examination of EBV's origin of plasmid replication, oriP that consists of FR plus DS (Figure 1A), has shown, for example, that DS binds EBNA1, and with its particular spacing thereby recruits the origin replication complex (ORC) (Bashaw and Yates, 2001; Chaudhuri et al, 2001; Dhar et al, 2001; Schepers et al, 2001; Wang et al, 2006). oriP is also a paradigm for a mammalian autonomously replicating sequence (ARS), which can be efficiently introduced and maintained under selection in cells expressing EBNA1.
Figure 1.
Visualization of EBV-derived plasmids in live cells. (A) A map of an EBV-derived plasmid (pLON-33K). pLON-33K contains oriP, 264 copies of LacO, and a neomycin-resistance gene (neoR). OriP consists of two functional elements, a family of repeats (FR), and a Dyad Symmetry (DS), which contain 20 and 4 EBNA1-binding sites, respectively. (B) Two independent HeLa-EBNA1 clones carrying on average 3–4 copies of pLON-33K plasmids per cell (clones A and B) were isolated. The distributions of the viral plasmids in these clones were measured after treatment with aphidicolin, either by FISH (gray bars) or by live-cell imaging (blue bars). The average copy numbers of EBV plasmids with their standard deviations are shown. The distributions of the plasmids in the cell populations measured by the two assays are overlapping, thus confirming the utility of the live-cell assay.
It is not clear, although, how EBV and plasmids derived from it are maintained and partitioned in proliferating cells. EBNA1 has been proposed to mediate these events by binding FR site specifically and tethering the bound plasmids to chromosomal sites (Marechal et al, 1999; Sears et al, 2003), either by associating with specific cellular proteins (Kapoor et al, 2005) or by binding AT-rich cellular DNA through its AT-hook activity (Sears et al, 2004). This proposal would yield random partitioning; a result not obviously compatible with analyses of the rate at which cells lose oriP vectors following removal of selection and become susceptible to being killed on reapplication of the selective agent (Yates et al, 1984; Kirchmaier and Sugden, 1995). We have resolved this apparent contradiction by monitoring individual EBV plasmids throughout the cell cycle.
DNA sequences can be detected in fixed cells by fluorescence in situ hybridization (FISH) and in live cells by their binding fluorescent proteins. We have used both approaches, modifying the latter to optimize its detection of EBV-derived plasmids, without perturbing their replication. Robinett et al (1996) used a derivative of the Lac repressor fused at its amino terminus to GFP to detect 256 binding sites for the repressor in the dihydrofolate reductase (DHFR) locus with or without its amplification. This derivative, unlike the wild-type repressor, binds DNA in the presence of the inducer, isopropyl-1-thio-β-D-galactopyranoside (IPTG). This property posed a problem in our studies, because long-term occupancy of 264 sites on EBV plasmids selects for their integration into host DNA (J Komano and B Sugden, unpublished findings). We therefore constructed and used wild-type Lac repressor fused at its carboxyl terminus to a tandem dimer of RFP (Campbell et al, 2002) and a nuclear localization signal (NLS), and carried cells in the presence of IPTG to prevent binding of the repressor except when plasmids were visualized. One earlier study visualized EBV-derived plasmids with the GFP-Lac repressor fusion of Robinett et al (1996) before the plasmids became established (Kanda et al, 2001). EBV plasmids are lost at apparent rates of ⩾25% per cell generation for the first 2 weeks following their introduction into cells, after which they are established and lost at apparent rates of 3–5% per cell generation (Leight and Sugden, 2001). We consequently monitored EBV-derived plasmids in cells after their establishment in order to measure their dynamics at a steady state.
Clones of HeLa cells harboring on average two to four copies of EBV-derived plasmids were examined for their individual numbers of plasmids in the absence of selection throughout the cell cycle. These analyses indicated that 84% of all plasmids were duplicated in S phase, and all of these by the end of S phase appeared as colocalized pairs with signal intensities close to two-fold those of the unduplicated plasmids. This colocalization of newly synthesized sister plasmids is without precedent in mammalian cells. A total of 88% of these colocalized pairs partitioned faithfully, separating at anaphase; 12% segregated to only a single daughter cell. The 16% of plasmids that failed to be duplicated segregated randomly, and less than 0.3% of all plasmids detected in G2 failed to being detected in daughter cells following mitosis. These experimentally determined rates were used in computer simulations to predict the evolution of plasmids in cells over time. The distribution of plasmids in a starting population of HeLa cells with an EBV-derived plasmid and in 293 cells with intact EBV were determined, selection removed, and the distributions again measured after 10 and 25 cell generations. These measured distributions were congruent with those predicted by the computer simulations, validating the experimentally determined rates of synthesis and partitioning of the plasmids.
The measurements of EBV plasmids were extended to cells under selection. The distribution of genomic EBV in clones of EBV-immortalized B-cells was measured by FISH. These measurements revealed that selection for EBV plasmids yields cells with widely varying numbers of plasmids, but that there is a heritable, mean optimum for a given number for each clone. The measured distributions of plasmids in cells under selection have also been modeled with computer simulations. These combined analyses support a model that reflects the population dynamics of EBV plasmids in proliferating cells. EBV has evolved its replication with minimal viral contributions to balance the gain and loss of its genome in cells to which it provides a selective advantage.
Results
Measurements of EBV-derived plasmids in HeLa cells in the absence of selection
We followed the fate of individual EBV plasmids in proliferating cells in order to elucidate their synthesis and partitioning. To do so we first developed and characterized EBV-derived plasmids that can be detected visually in live cells. An EBV-derived plasmid (pLON-33K) containing oriP, 264 copies in tandem of Lac operator (LacO), and a neomycin-resistance gene (Figure 1A) was introduced into and established in clones of HeLa cells stably expressing EBNA1 (HeLa-EBNA1). These clones were infected with a retroviral vector expressing wild-type Lac repressor fused to a tandem dimer red fluorescent protein, tdimer2(12) (wtLacI-tdRFP) and an NLS. Different cells expressed different levels of the fusion protein in their nuclei, producing varied levels of a diffuse background. Plasmids were detected as individual dots by their binding of the fluorescent derivative of the Lac repressor, and confirmed as such by their detection in parallel by FISH. Two individual clones (clone A and B) carrying three to four copies of plasmids on average were isolated and used for further experiments. We confirmed that the EBV-derived DNAs in these clones were maintained extrachromosomally dependent upon EBNA1 by Southern blot analysis, by their loss in the absence of selection, and by their replication being inhibited by a dominant-negative derivative of EBNA1 (Supplementary Figures 1A and B). The distributions of the established plasmids in these two cell clones were measured by visualization with wtLacI-tdRFP and by FISH after aphidicolin treatment. Aphidicolin inhibits the replicative DNA polymerases and thereby blocks cells in S phase. Importantly, the distributions of the plasmids in these clones determined by these two different assays ranged from 0 to 30 plasmids per cell and were indistinguishable, validating this visualization of EBV plasmids as a means of enumerating them (Figure 1B). We assayed the number, stability, and distribution of plasmids over time under all the conditions used to visualize them. The presence or absence of IPTG for different times in S, G2/M, or both phases, as well as a double-aphidicolin block had no detectable effect on the maintenance of plasmids or their average numbers per cell (Supplementary Figures 2A, B and C).
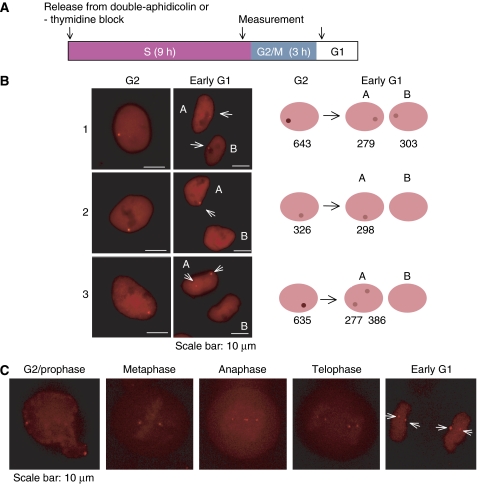
Measurement of the distribution of EBV plasmids from G2 through mitosis
To characterize the partitioning of an EBV plasmid (pLON-33K) during mitosis in detail, distributions of the plasmids were monitored before and after mitosis in live cells. HeLa-EBNA1 clones carrying the plasmids were synchronized by double-aphidicolin or double-thymidine block in the presence of IPTG and G418. These two treatments efficiently blocked cells at the beginning of S phase. After removal of IPTG and the G418, the cells were released from the second synchronization, incubated, and analyzed twice during 12 h, following S phase at 9 h after release from the block and 3 h later following mitosis (Figure 2A). Only healthy cells as judged by their successfully completing mitosis and having intact peripheries without blebs were studied. The distributions of 370 plasmids in 106 mitoses (1–13 plasmids/cell in G2) were characterized by the measurements of the number of dots and the average fluorescent intensities per pixel of the individual dots following S phase and after the subsequent mitosis (Figure 2B). Of the 370 plasmids examined, 310 appeared in G2 as colocalized pairs (Table I). These 310 colocalized plasmids thus appeared as 155 single signals in G2, which following mitosis yielded 310 signals with intensities close to one-half of that measured in the preceding G2 phase. The remaining 60 plasmids appeared in both G2 and following mitosis as single signals whose intensities decreased only slightly, presumably as a result of photobleaching. Seven of these single plasmids in G2 were the only plasmids in the cell, and were passed onto a single daughter cell. Of the 155 pairs colocalized in G2, 130 (88%) partitioned equally to daughter cells following mitosis; 19 partitioned to a single daughter cell yielding two signals in these cells (the daughter cells receiving these pairs are denoted with asterisks in Table I); and the partitioning of six could not be determined. The 53 single plasmids accompanying one or more colocalized pairs in G2 partitioned into daughter cells independently of those pairs. For example, 14 cells in G2 had one colocalized pair and one single plasmid. Following mitosis 13 of the resulting pairs of daughter cells had 1 and 2 plasmids, and one pair of daughters had 3 and 0 plasmids. All of the 370 plasmids detected in G2 were found in cells following mitosis, indicating that the rate of loss of EBV plasmids during mitosis is less than 0.3%.
Figure 2.
Time-lapse analysis of EBV-derived plasmids in synchronized cells. (A) A scheme of the experiment to measure partitioning. HeLa-EBNA-1 clones carrying viral plasmids were synchronized by double-aphidicolin or double-thymidine block in the presence of 10 μM IPTG and 600 μg/ml G418. After removal of IPTG and G418, the cells were released from a second block and incubated for 9 h (S phase) and a subsequent 3 h (G2/M phase). The distributions of 370 plasmids in 106 mitoses (1∼13 plasmids/cell in G2) were characterized by the measurement of the number of dots and the average fluorescent intensities in individual dots in G2 and early in G1. Only cells judged as being healthy by their successful completion of mitosis and having intact peripheries without blebs were studied. (B) Examples of the distributions of pLON-33K in G2 and early in G1. The distributions of the viral plasmids in G2 and early in G1 are categorized into the following three types: (1) plasmids were present as colocalized pairs in G2 and partitioned equally following mitosis; (2) plasmids were localized as single molecules in G2 and partitioned randomly following mitosis; (3) plasmids were present as a colocalized pair in G2 and segregated to only one daughter cell. In these examples, the measured intensities of each signal after correcting for the background are shown under the drawing of each cell to the right. Bars, 10 μm. The varied intensities of the backgrounds in the nuclei likely reflect different levels of expression of wtLacI-tdRFP in them. (C) Spatial–temporal analysis of the segregation of pLON-33K during mitosis. The distribution of the plasmids at each stage of mitosis is shown. Two pairs of colocalized plasmids were segregated at the onset of anaphase, and distributed equally to the daughter cells. Bar, 10 μm.
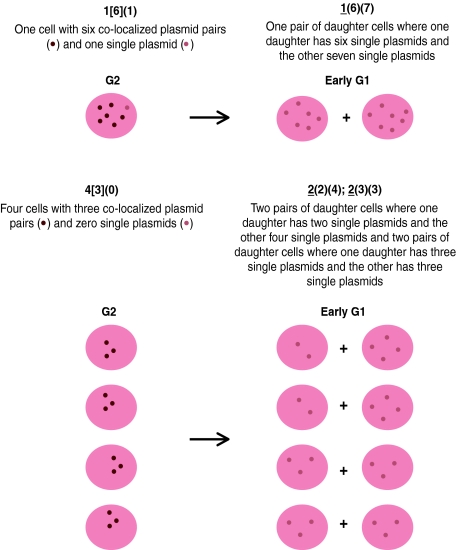
Table 1.
A summary of the distribution of 370 pLON-33K plasmids observed from G2 through M phase, with a cartoon illustrating two examples of the classification system used
| Plasmid content of cells in G2 | Plasmid content of pairs of daughter cells following mitosis | |
|---|---|---|
| N cells with [P] co-localized plasmid pairs and (S) single plasmids | N pairs of daughter cells where one daughter has (i) and the other (j) plasmids N (i) (j) | |
| N [P] (S) | ||
| 1 [6] (1) | → | 1 (6) (7) |
| 1 [5] (0) | → | 1 (5) (5) |
| 2 [4] (0) | → | 2 (4) (4) |
| 4 [3] (1) | → | 4 (3) (4) |
| 4 [3] (0) | → | 2* (2) (4); 2 (3) (3) |
| 2 [2] (2) | → | 1 (2) (4); 1 (3) (3) |
| 8 [2] (1) | → | 3* (1) (4); 5 (2) (3) |
| 18 [2] (0) | → | 2* (1) (3); 16 (2) (2) |
| 2 [1] (3) | → | 2 (2) (3) |
| 5 [1] (2) | → | 3 (1) (3); 2 (2) (2) |
| 14 [1] (1) | → | 1* (0) (3); 13 (1) (2) |
| 35 [1] (0) | → | 11* (0) (2); 24 (1) (1) |
| 3 [0] (2) | → | 3 (1) (1) |
| 7 [0] (1) | → | 7 (0) (1) |
| *Pairs of daughter cells with demonstrably unequally partitioned plasmids derived from plasmids pairs co-localized in G2. | ||
 | ||
Spatial–temporal analysis of 12 colocalized pairs of plasmids during mitosis demonstrated that the plasmids associated with condensed mitotic chromosomes as single signals were segregated at the onset of anaphase, and these pairs partitioned equally to daughter cells. The separated plasmids were frequently localized on the sister chromosomes symmetrically at the end of mitosis (Figure 2C).
Wild-type LacI forms a tetramer and can bind to two LacO sites (Lewis, 2005). It is formally possible that the colocalization of two sister plasmids in G2 reflects their being linked by wtLacI. To test this possibility, we expressed a derivative of wtLacI-tdRFP that can only dimerize and thus bind only one LacO site. Cells were synchronized with aphidicolin, released, and the distributions of 16 plasmids were analyzed before and after mitosis. Of these 16 plasmids, 14 were detected as colocalized pairs (seven signals) in G2, 12 partitioned equally to daughter cells; and two did not (data not shown). The frequency of colocalization of pairs of plasmids bound by a derivative of wtLacI-tdRFP that can only dimerize was the same within experimental error as when the plasmids were bound by the wild-type repressor that can tetramerize (P=0.54, Fisher's one-sided exact test), indicating that forming pairs of EBV plasmids in G2 is independent of the repressor's ability to form tetramers.
Analysis of the distribution of EBV-derived plasmids throughout the cell cycle
To measure the distributions of plasmids throughout the cell cycle, cells were incubated for 24 h without an imposed synchronization and sampled three times. Cells that rounded up for mitosis yielded daughter cells that adhere and flatten at the beginning of G1. The distribution of the plasmids in these cells and the average fluorescent intensities of the pixels in each dot were measured late in G1, late in S, and early in the subsequent G1 (Figure 3A). The intensities were determined for images from the single z-section having the most intense signal measured without gain, and corrected for background by subtracting the average signal of the surrounding pixels. The intensities in each pixel were less than 6% of the maximum of the 14 bit camera we used and thus in its linear range.
Figure 3.
Time-lapse analysis of EBV-derived plasmids in unsynchronized cells. (A) A scheme of the experiment to follow plasmids throughout the cell cycle is shown. After removal of selection HeLa-EBNA1, clones carrying pLON-33K early in G1 were incubated for 12 h (G1) in the presence of 10μg/ml IPTG. After removal of IPTG, the cells were incubated for 9 h (S phase) and 3 h (G2/M phase). The distributions of 18 plasmids in 11 cells (1∼2 plasmids/cell in G2) were characterized by the measurements of the number of dots and the average fluorescent intensities in individual dots late in G1, late in S, and subsequently early in G1. (B) Time-lapse analysis of the average fluorescent intensities of pLON-33K plasmids throughout cell cycle. Representative images of the following two types of the partitioning of the plasmid are shown: (1) the plasmids synthesized in one S phase are colocalized and partitioned faithfully and (2) the plasmids that fail to be replicated in one S phase can not be partitioned faithfully. The top row of images reflects the single z-stack with the most intense signal, from which the intensities were measured and corrected for background. The second row of images reflects deconvolved signals, which are computationally derived and are an indication of the size of the signal. The third row of images is derived by differential interference contrast (DIC) and illustrates the health of the cells. The fourth row are cartoon images giving the average fluorescent intensities of the signals measured late in G1, late in S, and early in G1 after correction for their background is shown under the drawing of each cell. Bars, 10 μm.
A total of 26 of 31 plasmids duplicated in S phase, colocalized as pairs in G2 phase, and partitioned equally during mitosis. Five of the plasmids did not duplicate during S phase, were maintained during G2/M phase, and distributed to one daughter cell. The ratio of the average fluorescent intensities in the plasmids that were duplicated in S phase, colocalized as pairs in G2 phase, and partitioned equally in M phase, increased approximately 1.5-fold during S phase (Figure 3B). On the other hand, the ratio of intensities of the plasmids that failed to duplicate decreased slightly (0.8-fold). These results indicate that 84% of plasmids duplicated successfully, yielded colocalized sister molecules in G2 and early M phase, and usually partitioned faithfully. A total of 16% of the plasmids fail to be synthesized in S phase. This defect in synthesis is the only means by which EBV plasmids are detectably lost from a population of proliferating cells; missegregation, however, can contribute to a daughter cell's failure to acquire a plasmid.
None of these 26 colocalized pairs of sister plasmids followed in this experiment missegregated. This level of fidelity is not statistically different from that found in the experiments (Table I) using imposed synchronies of 12% missegregation (P=0.08, Fisher's exact test). That all 26 colocalized pairs of sister plasmids partitioned without synchrony, faithfully, however, leads us to consider the notion that the imposed synchrony may favor missegregation and thus the 12% is likely an upper limit to this defective partitioning.
Prediction of the distribution of EBV-derived plasmids for multiple generations in the absence of selection by a computer simulation and its experimental confirmation
We predicted the distributions of EBV plasmids for multiple generations in the absence of selection by a computer-based simulation. The distribution of plasmids in a cell clone carried under selection and initially containing 2.0±2.1 copies of plasmids per cell on average was used as starting point for the simulation. Each plasmid in each cell in G1 was given an 84% chance of being duplicated; duplicated plasmids were given an 88% chance of being partitioned equally to two daughter cells and a 12% chance of being partitioned to one daughter cell or the other; unduplicated plasmids were partitioned randomly to either daughter cell. The program was begun with approximately 100 cells having the measured distributions in the analyzed clones and run to simulate 10 and 25 generations. Each run was repeated 40 times with the same initial assumptions, and the 40 results were averaged. We also measured experimentally the distributions of the plasmids in the HeLa clone following removal of selection for 10 and 25 generations. The measured and predicted distributions of EBV plasmids are shown in Figure 4A and closely parallel each other, indicating that the experimentally determined frequencies of duplication and partitioning of the plasmids describe well their fate in proliferating cells in the absence of selection.
Figure 4.
Computer simulations predict the distributions of EBV-derived plasmids in proliferating cells in the absence of selection. (A) A clone of HeLa-EBNA1 cells carrying pLON-33K was cultured after removing the selective agent, G418, for 25 generations. The distributions of the plasmids in the cells before removing G418, day 0, and at day 10, and day 25 after its removal were determined by live-cell imaging (green bars). The distributions of the plasmids in the cells were also predicted (red bars) by computer simulations in which the measured distribution at day 0 was used as the starting point, and the rates of plasmid duplication and partitioning determined experimentally (Figures 2 and 3; and Table I) were used to predict the distribution of the plasmids each cell generation. The computer simulations were repeated 40 times and averaged to provide the smooth curves shown. (B) A clone of 293 cells carrying the intact EBV genome encoding resistance to hygromycin B was cultured after removing the selective agent for 25 generations. The distributions of the plasmids in the cells before removing hygromycin B, day 0, and at day 10, and day 25 after its removal was determined by FISH (green bars) and predicted (red bars) as described above. The average number of plasmids per cell and its standard deviation are shown for each condition.
Similar experiments were performed to test whether the measurements made with pLON-33K in HeLa cells could be extended to intact EBV plasmids in a different host cell, 293. EBV confers a selective advantage on the B-cells it infects, but not on established cells such as 293. Intact EBV plasmids can, however, be maintained in established cells under selection if they encode a drug resistance gene (Delecluse et al, 1998). The distribution of full-length, genomic EBV plasmids encoding resistance to hygromycin B in a clone of 293 cells was measured with FISH, the hygromycin B removed, the cells propagated, and the distributions of plasmids again determined after 10 and 25 cell doublings. In parallel, a computer-based simulation was run, with the distribution of plasmids determined by FISH before removal of the hygromycin B as the starting point. The rates of duplication and partitioning of plasmids measured in HeLa cells were also used in these simulations. The results predicted by these simulations paralleled the measured distributions at 10 and 25 cell generations (Figure 4B), and indicate that the rates of synthesis and partitioning measured for pLON-33K in HeLa-EBNA1 cells are similar to those of intact EBV in 293 cells. When this program was used in simulations with the same starting distribution of EBV as in Figure 4B but with different, assumed rates for its synthesis or partitioning, the predicted distributions differed (Supplementary Figures 3A and B) from those measured (Figure 4B). The sensitivity of the simulations to changes in these rates demonstrates the robustness of this computational approach.
The distribution of EBV plasmids in cells proliferating under selection
We determined the distribution of EBV plasmids in the EBV-transformed lymphoblastoid cell clone 721, which requires EBV for its survival and proliferation (Kennedy et al, 2003). The parental cell population (Kavathas et al, 1980), which has been propagated for more than 100 generations, was synchronized at G1/early S phase or M phase by treatment with aphidicolin or nocodazole, respectively. The distribution of EBV plasmids in the cell population was analyzed by FISH. The numbers of the viral plasmids in the individual cells varied widely in the population. The measurements of the distribution of the viral plasmids are summarized in Figure 5. The distribution was broad, with the mean number of plasmids centered on 9.7±4.1 and 19.5±6.3 in aphidicolin- or nocodazole-treated cells, respectively.
Figure 5.
Computer simulations predict the distribution of EBV genomes in transformed B-cells. A clone of EBV-transformed B-cells, 721, propagated for more than 100 generations, and four of its subclones propagated for approximately 25 generations were analyzed after being blocked in S phase with aphidicolin or in M phase with nocodazole, for their distributions of EBV plasmids per cell by FISH. The average number of plasmids per cell and its standard deviation are shown for each condition. The average numbers of plasmids per cell in all five populations were similar, indicating that there is a genetically determined average number of plasmids per cell that provides the cells an optimal selective advantage. The distribution of plasmids per cell in the parental 721 cells was used as a starting point to simulate the distributions of plasmids in cells after 25 generations. The simulations were modified to include an increasing chance of cells' dying when their number of plasmids decreased from 4 to 0 per cell. These simulated values (red bars) were comparable to the average of the measured values of the four subclones (gray bars).
Four independent subclones were isolated from the parental 721-cell population by limiting dilution, propagated for 25 generations, and their distributions of viral plasmids determined by FISH. The distributions in all four subcloned populations were similarly broad, with the average number of plasmids centered on 6.7–9.6 plasmids in aphidicolin-treated cells, and 11.9–15.1 plasmids in nocodazole-treated cells (Figure 5), indicating that the broad distribution of viral genomes in 721 cells was generated within only 25 generations. The constancy of these mean numbers of plasmids indicates that this mean number of EBV genomes in 721 cells provides this clone and its descendants an optimal selective advantage.
We modeled the behavior of EBV plasmids in 721 cells in order to determine if the frequencies of plasmid duplication and faithful partitioning measured for EBV plasmids in the absence of selection apply to those in 721 cells in which EBV is required both for the survival and proliferation of these cells. In order to model this behavior, it was necessary to understand the relationship between the number of viral genomes in a cell, and the selective advantage those genomes provide it. Two experimental measurements were used to define a cell's dependency on an EBV-derived plasmid for its survival. First, the rate of loss of plasmids from HeLa and 293 cells from which drug selection was removed was measured directly to be 6.7% per generation (Table II). Second, in related, historical experiments in which a selection was removed and after time reapplied, the rate at which cells lost sufficient EBV-derived plasmids to become sensitive to being killed by reapplication of the selective agent has been found to be 3–5% per generation (Yates et al, 1984; Kirchmaier and Sugden, 1995). That these two rates differ indicates that cells dependent on an EBV-derived plasmid lose their selective advantage when their number of plasmids drops below some non-zero, threshold number. Loss of plasmids in cells above this threshold number would not affect the selective advantage conferred on the cell. Because cells dependent on EBV-derived plasmids have a broad distribution of plasmids per cell (Figure 5); more cells can lose some plasmids per generation than succumb to the selective disadvantage. These considerations allowed us to model the behavior of EBV plasmids with our simulation, based on the initial distribution of the number of EBV genomes in individual cells of the 721 parental population. A simulation was made with the rates for duplication and partitioning found for pLON-33K in the HeLa clones. In this simulation the initial conditions were for a single cell with a specified number of plasmids. The simulation was repeated for the number of times a cell with that given number of plasmids was found in the measured parental population of parental 721 cells (Figure 5), and repeated for all given numbers in that parental population. These initial conditions thus mimic the cloning of the parental 721 cells that yielded the four subpopulations. An additional parameter was added to simulate a threshold such that five plasmids per cell yielded 100% survival; cells with four plasmids had an 80% chance of survival; the chance for cells with three plasmids was 60%; with two was 40%; with one was 20%; and with none was 0%. We used this parameter of proportional survival because an abrupt threshold with 0% survival yielded an unphysiological distribution of no survivors beyond that threshold. We selected this particular parameter because test simulations in which the threshold for 100% survival was set either to 4 or 6 plasmids per cell, generated distributions with peak values that were either less than or greater than that measured for the average of the 721 subclones (Figure 5). The results of the simulations were averaged over 40 trials and are shown relative to the average of the distributions measured for the four subclones of 721 cells grown for 25 generations (Figure 5). The congruence of these measured and simulated distributions is consistent with the rates measured for the duplication and partitioning of EBV-derived plasmids in live HeLa cells applying to EBV genomes in B-cells transformed by EBV. This conclusion was being further substantiated by testing the dependence of this simulation on the rate of plasmid duplication of 84% per cell cycle, derived from measurements of pLON-33K in HeLa-EBNA1 cells by arbitrarily substituting values of 74 and 94% for it, and running the simulations again (as for intact EBV in 293 cells in Supplementary Figure 3A). Decreasing or increasing the rate of plasmid duplication used in the simulation shifted the center of the predicted distributions to the lower or higher values, respectively, than that using 84%, which does center on the average of the measured distributions (Figure 5).
Table 2.
Measuring the loss of EBV plasmids in cells without selection
| A. Number of pLON-33K plasmids in HeLa cells over time | ||||
|---|---|---|---|---|
| Clone | Selection | Day 0 | Day 10 | Day 25 |
| A | + | 2.5 | 2.9 | 2.3 |
| − | 1.6 | 0.3 | ||
| B | + | 2.0 | 3.5 | 2.9 |
| − | 1.5 | 0.74 | ||
| B. Number of intact EBV plasmids in 293 cells over time | ||||
| Selection | Day 0 | Day 10 | Day 25 | |
| + | 9.7 | 9.2 | 9.9 | |
| − | 5.5 | 1.7 | ||
| EBV, Epstein–Barr virus. | ||||
| For each of the three clones studied, an average of the three measured numbers of plasmids per cell in the presence of selection was used as the number present in cells at the time of removing selection. The average loss of plasmids per day for the three clones analyzed is 6.7±1.24%. | ||||
Discussion
EBV uses plasmid replication to maintain itself in proliferating cells. This plasmid replication underlies its pathogenicity in cancers such as Burkitt's lymphoma, where the viral plasmids are present in the proliferating cells characteristic of these diseases; but few viral genes are expressed (Rowe et al, 1987; Hammerschmidt and Sugden, 2004). We have analyzed the synthesis and partitioning of EBV plasmids in order to shed light on the details of this viral plasmid replicon.
One unexpected property of this replicon is the frequency with which it fails to duplicate; 16% of EBV plasmids remain unduplicated in each cell cycle. This frequency is initially surprising, given that EBV's origin of plasmid replication oriP was identified by an ARS assay in mammalian cells, is a licensed replicon, and is the most efficient mammalian licensed ARS characterized yet (Sugden et al, 1985). However, we have found that polymerizing a licensed replicator, Rep*, which functions as does DS, but is less efficient, increases Rep*'s frequency of duplication (Kirchmaier and Sugden, 1998; Wang et al, 2006). There is thus no reason now to expect a priori that DS of oriP is maximally efficient and is always duplicated in each cell cycle.
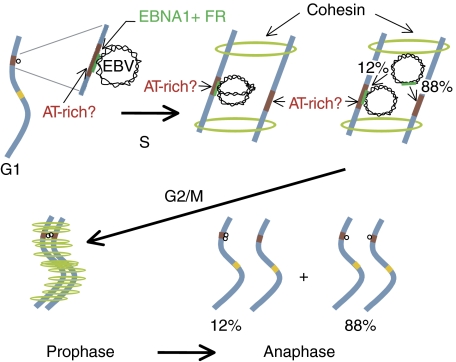
A second fundamental property of EBV's plasmid replicon uncovered here is the colocalization of sister plasmids on being synthesized, and that this positioning in space is coupled to their non-random partitioning. This colocalization of newly synthesized sister plasmids has not been detected before, probably because detection of established EBV plasmids has been carried out with FISH, which requires denaturation and spreading of DNA, conditions that would be expected to dissociate the tethered complexes. For example, approximately twice the number of EBV plasmids is detected by FISH in clones of cells blocked with nocodazole as with aphidicolin (Figure 5). Pairs of EBV plasmids are also frequently found in mitosis to be symmetrically positioned on sister chromatids, when detected by FISH (Supplementary Figure 4) (Delecluse et al, 1993) or by binding tagged EBNA1 (Kanda et al, 2007). In 10 mitotic spreads, for example, 60% of the EBV plasmids were detected as colocalized pairs and 97% of all plasmids were overlapped by chromosomal staining (examples shown in Supplementary Figure 4). These measurements are consistent with EBV plasmids being associated with sister chromatids and the spreading forces used to separate mitotic chromosomes also acting to separate some of the colocalized sister plasmids. This colocalization of sister plasmids in conjunction with EBNA1's ability to tether DNA such as FR to which it binds site specifically to chromosomal DNA leads to a plausible model for the mechanism of EBV's partitioning (Figure 6) as follows: (1) in G1, an EBV plasmid is tethered to a chromosomal site probably directly by EBNA1's AT-hook activity. We favor this mechanism rather than the alternative of indirect tethering by EBNA1's binding the cellular protein, EBP2, because derivatives of EBNA1 that cannot bind EBP2 but have AT-hook activity behave as wild-type EBNA1 in supporting replication. We have found that a fusion of the cellular protein, high-mobility group A (HMGA) 1a, to EBNA1's DNA-binding and dimerization domain, which deletes EBNA1's binding site for EBP2 (Altmann et al, 2006), or a second derivative of EBNA1 that cannot bind EBP2, 2XLR1 (Mackey and Sugden, 1999; Shire et al, 1999), supports wild-type levels of plasmid synthesis and partitioning (Supplementary Figure 5). Both derivatives do have AT-hook activity (Sears et al, 2004); (2) during S phase, the chromosomal site and the tethered EBV undergo synthesis synchronously by their spatial colocalization, placing them in the same replication compartment (Sadoni et al, 2004; Kitamura et al, 2006). This synchrony needs be only within an hour or so, though; synthesis through FR is slow, leading it to being scored as a ‘replication fork barrier' that protracts its own synthesis to be longer than that required to synthesize 160 000 bp of EBV unidirectionally (Gahn and Schildkraut, 1989); (3) newly synthesized EBV sister plasmids bind to the same nearby sites on each sister chromatid, or to the original site if it can accommodate them both. Our measurements indicate that sister plasmids individually associate with sister chromatids 88% of the time. Cohesin holds the sister chromatids together (Nasmyth, 2005) and we hypothesize indirectly holds the tethered EBV plasmids together within the resolution of fluorescence microscopy. EBV plasmids are found in association with chromosomes early in prophase and embedded in mitotic chromosomes detected by staining for cohesin consistent with this hypothesis (Supplementary Figures 4 and 6); (4) when the sister chromatids separate at anaphase, the tethered EBV sister plasmids do so too.
Figure 6.
A model for the licensed duplication and non-random partitioning of EBV plasmids. In G1, EBV plasmids are tethered to chromosomes by EBNA1 binding to FR of oriP and possibly to AT-rich chromosomal sites by EBNA1's AT-hook activity. Duplication of a plasmid is approximately synchronous with that of the chromosomal site to which it is tethered, because the shared location brings both to one replication compartment synchronously. Cohesin holds the newly synthesized chromatids in close proximity so that on the plasmid's eventual release from FR following a ‘stalled fork progression' through FR, in 88% of the cases the duplicated plasmids bind to the duplicated sites on the adjacent sister chromatids. Their close proximity means that their fluorescent detection yields one single, but double-intensity, signal until they separate at anaphase.
The third fundamental property revealed in these studies pertains both to EBV-derived plasmids and their host cells. When these plasmids provide their host cell a selective advantage, the number of plasmids in a cell that affords the cell an optimal advantage is a clonally inherited trait. Four subclones of an EBV-immortalized B-cell generate wide distributions of plasmids per cell, but each clonal population retains a similar mean number of plasmids per cell (Figure 5). This finding is supported by experiments in which the average number of oriP plasmids per cell was determined by nucleic acid hybridization, hygromycin B or G418, to which the plasmids encoded resistance was removed for 20–30 cell generations, the drug was reapplied, resistant subclones isolated, and the average number of plasmids measured in them (Kirchmaier and Sugden, 1995). In these experiments, subclones from five of six different parents had within experimental error the same average number of plasmids as did their parents. This property likely underlies the variation observed in the average number of copies of EBV between different cell lines (Sternas et al, 1990), and provides a selection for a given mean number of plasmids per cell in a clonal population of cells.
The fourth unexpected, fundamental property of EBV plasmids identified here is that the defects in duplication of these plasmids are approximately balanced by a defect in their partitioning. In each generation, 16% of the plasmids fail to be duplicated, leading to a net loss of 8% of the plasmids from a population of cells. Each generation 12 of the 84% of duplicated plasmids partition unequally, leading to approximately half of these daughter cells (5%) having more plasmids than their parents. Clearly some daughter cells also have fewer plasmids than their parents, and if they have fewer plasmids than their threshold number, they will be at such a selective disadvantage as to be lost from the population. This rate of loss is 3–5% of the cells per generation, while the remaining cells that retain sufficient plasmids can be afforded selective advantages such as a fostering of their survival and proliferation as, for example, in the case of B-cells infected by EBV.
The rates of synthesis and partitioning of EBV-derived plasmids are surprisingly efficient when compared with those of derivatives of human chromosomes. These rates for linear replicons engineered from the human X-chromosome are inversely proportional to their length (Spence et al, 2006). Derivatives of the human X-chromosome that are fives times the length of EBV's genome and 25 times that of pLON-33K are lost from human cells in culture at rates of 3–4% per generation, and missegregate at rates up to 16% per generation (ibid.). That EBV-derived plasmids have similar rates and are so much smaller in length indicates that the virus has evolved a particularly efficient mechanism to persist in proliferating cells.
EBV has evolved a simple, flexible scheme to maintain itself in a proliferating population of cells to which it provides a selective advantage. EBV itself drives proliferation of newly infected B-cells through its LMP1 protein (Kaye et al, 1993; Gires et al, 1997; Dirmeier et al, 2005); it sustains frank Burkitt's lymphomas by preventing apoptosis and driving proliferation requiring its EBNA1 protein (Kennedy et al, 2003). These selective advantages when coupled to its mode of synthesis and partitioning, insure that EBV is maintained in infected normal and malignant proliferating B-cells.
Materials and methods
Plasmids, cell culture and transfection, and retrovirus infection are described in the Supplementary data.
Fluorescent in situ hybridization
FISH analysis was performed based on the protocol of Lawrence et al (1989), with some modifications. Briefly, 721 cells, HeLa-EBNA1, and 293 clones carrying EBV plasmids were synchronized by treatment with 15 μM aphidicolin (Sigma) or 100 ng/ml nocodazole (Sigma) for 16 h. Cells were treated with 0.075 M KCl for 20 min at 37°C, fixed in methanol:acetic acid (3:1) for 30 min at room temperature, and spread on the slides. Slides were treated with 4 × SSC (1 × SSC: 0.15 M NaCl, 0.015 M sodium citrate) containing 0.5% (v/v) Nonidet P-40 (Sigma) for 30 min at 37°C, dehydrated in a cold ethanol series (70, 80, 90%) for 2 min each, air dried, and denatured in 70% formamide–2 × SSC for 2 min at 72°C. Slides were dehydrated in a cold ethanol series and air dried. Hybridization probes for detection of EBV plasmids were generated by nick translation using biotin-11-dUTP (Roche). A 20-μg weight of a probe was precipitated by ethanol in the presence of 6 μg salmon sperm DNA (Eppendorf) and 4 μg human Cot-1 DNA (Invitrogen), resuspended in CEP hybridization buffer (Vysis), and incubated for 10 min at 70°C, for 5 min at 4°C, and for 1 h at 37°C. A hybridization mix containing 5 ng probe was placed on each sample and incubated overnight at 37°C in a moist chamber. Slides were washed in 2 × SSC containing 50% formamide for 30 min at 50°C and in 2 × SSC for 30 min at 50°C. Hybridized probe was revealed by incubation with 30 μl detection solution containing streptavidin conjugated to Cy3 (Cytocell) for 20 min at 37°C. Slides were washed twice in 4 × SSC containing 0.05% Triton X-100 (Sigma) for 5 min at room temperature. The chromosomes were counterstained by a mounting medium containing diamidino-2-phenilindole (DAPI) (Vector). The images were acquired by an inverted fluorescence microscope (Axiovert 200 M, Zeiss) equipped with a digital CCD camera (Axiocam HRm, Zeiss) and a z-motor. Cy3 and DAPI were visualized using specific individual filter sets for Texas-red and DAPI. Images were collected by a × 63, 1.4 NA oil objective lens (Plan Prochromaro, Zeiss) with 5–10 slices of z-stacks, with exposures of 0.01 and 1 s to detect Cy3 and DAPI, respectively. AxioVision software (Zeiss) was used for acquisition and computation of the images. For high-resolution images, captured raw images were deconvolved with AxioVision software (Zeiss) using an inverse filter algorithm. All images were digitally processed for presentation with Adobe Photoshop.
Imaging EBV plasmids in live cells
HeLa-EBNA1 clones were grown in a 35-mm diameter glass-bottomed Petri dish (MatTek). Cells were synchronized after removal of G418 by treatment with 15 μM aphidicolin or 2 mM thymidine for 16 h. They were washed twice in the complete medium and grown in the absence of aphidicolin or thymidine for 8 h. They were treated again with 15 μM aphidicolin or 2 mM thymidine for 16 h. After releasing cells from the second block, their culture medium was replaced with phenol red-free DMEM and covered with a layer of mineral oil (Sigma). Approximately 50% of the cells re-entered the cell cycle after removing the second block. The cells were incubated for 12 h (9 h for S phase, 3 h for G2/M phase) in a temperature-controlled chamber, which was maintained at 37°C with a humidified atmosphere of 5% CO2 in air. Images were collected in G2 and early in subsequent G1 to record the partitioning of plasmids from G2 through M phase. Only those cells that progressed through these phases of the cell cycle were analyzed.
Cells in cytokinesis or early in G1 were incubated for 24 h in a 5% CO2-equilibrated chamber maintained at 36.5–37°C on the microscope stage to monitor the plasmids throughout a cell cycle. Images were collected late in G1, late in S, and early in subsequent G1. All images were acquired after first manually scanning the cell for all signals, insuring that no two signals were on top of one another in different focal planes, and then taking 10–15 optical sections in 0.1-μm steps, spanning the signal with exposure times of 0.1 s using a filter set for Texas red. The intensities of the signals were measured without added gain. The signal in each pixel did not exceed 6% of the maximal level of the 14-bit AxioCam HRm camera, and thus was in its linear range. The intensities in each pixel for a signal (9–16 pixels) in the one section with the highest signal intensity were measured and normalized by subtracting from their average the average intensity of the surrounding pixels using AxioVision software (Zeiss). All images were digitally processed for presentation with Adobe Photoshop.
Time-course analysis of the distribution of pLON-33K and genomic EBV
HeLa-EBNA1 clones carrying pLON-33K were cultured in the absence or presence of selection for 25 generations. The distributions of the plasmids in >100 cells were analyzed at generations 0, 10, and 25. IPTG was removed 2 h before the measurements. A clone of 293 cells carrying genomic EBV DNA encoding resistance to hygromycin B was cultured in the absence or presence of selection for 25 generations, and analyzed at generations 0, 10, and 25.
Computer simulations
A program was written in C to permit flexible modeling of plasmid synthesis and partitioning. The likelihood of each plasmid in each cell to duplicate, and randomly to partition or not, can be varied in the program; so can the dependence of each cell on its number of plasmids for survival be varied. An experimentally measured distribution of plasmids in cells is used as the starting point for a simulation usually beginning with 100 cells. The program then determines the fate of each plasmid, whether it duplicates and how the plasmid and its potential sister plasmid partition, in each cell, in each ensuing generation. In the simulations used here, the populations were followed for 10 and 25 simulated generations and the simulations were averaged over 40 trials to calculate mean distributions. In order to limit the computational time, simulations were run consecutively for no more than 14 generations, at which time the fraction of cells with each number of plasmids was used to calculate a representative population of 1000 cells and used to seed the next generation. This adjustment in the size of the population was made such that the final number of consecutive generations was no smaller than five and no greater than 14. This program can be found at http://mcardle.oncology.wisc.2du/sugden/simulation.txt.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information
Supplementary Figure Legends
Acknowledgments
We thank Drs Belmont, Murray, and Tsien for the plasmids essential for our work. We also thank many of our colleagues, particularly Norman Drinkwater, Jun Komano, Kathryn Norby, and Gerhard Oertel for their experimental insights and suggestions. We thank Drs Jim Pawley and John White for their guidance in using the AxioCam HRm camera. This work was supported by a grant from the NIH, CA22443. Bill Sugden is an American Cancer Society Research Professor.
References
- Altmann M, Pich D, Ruiss R, Wang J, Sugden B, Hammerschmidt W (2006) Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc Natl Acad Sci USA 103: 14188–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw JM, Yates JL (2001) Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol 75: 10603–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc Natl Acad Sci USA 98: 10085–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Broker TR (2006) Papillomavirus. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 609–625. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press [Google Scholar]
- Delecluse HJ, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm GW (1993) Episomal and integrated copies of Epstein–Barr virus coexist in Burkitt lymphoma cell lines. J Virol 67: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W (1998) Propagation and recovery of intact, infectious Epstein–Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA 95: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A (2001) Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell 106: 287–296 [DOI] [PubMed] [Google Scholar]
- Dirmeier U, Hoffmann R, Kilger E, Schultheiss U, Briseno C, Gires O, Kieser A, Eick D, Sugden B, Hammerschmidt W (2005) Latent membrane protein 1 of Epstein–Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 24: 1711–1717 [DOI] [PubMed] [Google Scholar]
- Fanning E, Pipas JM (2006) Polyomavirus. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 627–644. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press [Google Scholar]
- Gahn TA, Schildkraut CL (1989) The Epstein–Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58: 527–535 [DOI] [PubMed] [Google Scholar]
- Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W (1997) Latent membrane protein 1 of Epstein–Barr virus mimics a constitutively active receptor molecule. EMBO J 16: 6131–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B (2004) Epstein–Barr virus sustains Burkitt's lymphomas and Hodgkin's disease. Trends Mol Med 10: 331–336 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B (2006) Epstein–Barr virus. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 687–705. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press [Google Scholar]
- Kanda T, Kamiya M, Maruo S, Iwakiri D, Takada K (2007) Symmetrical localization of extrachromasomally replicating viral genomes on sister chromatids. J Cell Sci 120: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Kanda T, Otter M, Wahl GM (2001) Coupling of mitotic chromosome tethering and replication competence in Epstein–Barr virus-based plasmids. Mol Cell Biol 21: 3576–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Lavoie BD, Frappier L (2005) EBP2 plays a key role in Epstein–Barr virus mitotic segregation and is regulated by aurora family kinases. Mol Cell Biol 25: 4934–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavathas P, Bach FH, DeMars R (1980) Gamma ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc Natl Acad Sci USA 77: 4251–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Kieff E (1993) Epstein–Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA 90: 9150–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G, Komano J, Sugden B (2003) Epstein–Barr virus provides a survival factor to Burkitt's lymphomas. Proc Natl Acad Sci USA 100: 14269–14274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B (1995) Plasmid maintenance of derivatives of oriP of Epstein–Barr virus. J Virol 69: 1280–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B (1998) Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein–Barr virus. J Virol 72: 4657–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU (2006) Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM (1989) Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell 57: 493–502 [DOI] [PubMed] [Google Scholar]
- Leight ER, Sugden B (2001) Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol Cell Biol 21: 4149–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M (2005) The lac repressor. C R Biol 328: 521–548 [DOI] [PubMed] [Google Scholar]
- Mackey D, Sugden B (1999) The linking regions of EBNA1 are essential for its support of replication and transcription. Mol Cell Biol 19: 3349–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC (1999) Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol 73: 4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (2005) How might cohesion hold sister chromatids together? Philos Trans R Soc Lond B Biol Sci 360: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M, Rowe DT, Gregory CD, Young LS, Farrell PJ, Rupani H, Rickinson AB (1987) Differences in B cell growth phenotype reflect novel patterns of Epstein–Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J 6: 2743–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D (2004) Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117: 5353–5365 [DOI] [PubMed] [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20: 4588–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Kolman J, Wahl GM, Aiyar A (2003) Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein–Barr nuclear antigen 1. J Virol 77: 11767–11780, Erratum in: (2004) J Virol 78: 5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A (2004) The amino terminus of Epstein–Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78: 11487–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Mason WS (2006) Hepadnavirus. In DNA Replication and Human Disease, DePamphilis ML (ed), pp 729–744. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press [Google Scholar]
- Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L (1999) EBP2, a human protein that interacts with sequences of the Epstein–Barr nuclear antigen 1 important for plasmid maintenance. J Virol 73: 2587–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JM, Mills W, Mann K, Huxley C, Farr CJ (2006) Increased missegregation and chromosome loss with decreasing chromosome size in vertebrate cells. Chromosoma 115: 60–74 [DOI] [PubMed] [Google Scholar]
- Sternas L, Middleton T, Sugden B (1990) The average number of molecules of Epstein–Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein–Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol 64: 2407–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B, Marsh K, Yates J (1985) A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein–Barr virus. Mol Cell Biol 5: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lindner SE, Leight ER, Sugden B (2006) Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol Cell Biol 26: 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B (1984) A cis-acting element from the Epstein–Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA 81: 3806–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Information
Supplementary Figure Legends