Abstract
Studies of primary cells from malignant brain tumors such as glioblastomas are limited by the small size of surgically resected specimens. However, glioblastomas are also frequently debulked via ultrasonic aspiration. In this study, we examined the functional competence and growth of their aspirated cells. Cells from minced tissue and aspirations were comparable in migration, formation of pseudopodia, development of cellular spheres with radial outgrowth, and neuroectodermal features. Cultures were maintained for more than six weeks without fibroblastic overgrowth. Our observations show that ultrasonically aspirated specimens contain cells useful for studies of tumor migration and growth of tumorspheres.
Keywords: Ultrasonic aspiration technique, brain tumor, glioblastoma, cell migration, pseudopodia, tumorspheres
Introduction
Glioblastomas are common, invasive brain tumors that resist all therapies and are almost always lethal. Although they cannot be entirely resected due to their infiltrative growth pattern, they are often debulked. At the time of surgery, once sufficient tumor tissue has been resected for a pathologic diagnosis, an ultrasonic surgical aspirator is frequently utilized to debulk the tumors. This technique removes tissue as single cells from tumors by vibration, saline irrigation, and suction. Due to their disaggregated nature, the aspirated cellular specimens are usually discarded. However, ultrasonic aspirations have been previously noted to yield viable tumor cells [1–4], with immunoreactivity for vimentin, S-100 protein, and glial fibrillary acidic protein (GFAP) [5], that are appropriate for potential diagnostic utility [6, 7]. However, functional competence of ultrasonically aspirated cells, other than growth in culture, has not been determined. We evaluated the ability of cells from ultrasonic aspirations and surgically dissected tumor tissue to form pseudopodia and migrate through gelatin-coated filters. This model recapitulated in vivo glioblastoma cell migration through small spaces formed by webs of cellular extensions in brain parenchyma. Hepatocyte growth factor, HGF, the most powerful chemoattractant for glioblastoma cells [8], and serum were used in our model to stimulate the cells. Also in culture the tumor cells were observed to form small spheres with rapid outgrowth of cellular monolayers. These studies indicate that the cells obtained with ultrasonic aspiration retain functional competence of importance for investigational studies of glioblastomas, including tumor stem cell potential
Materials and Methods
Materials
All materials were obtained from Sigma (St. Louis, MS, USA) unless otherwise stated. Antibodies to nestin and neuronal class III β-tubulin (TUJ1) were obtained from Covance (Berkeley, CA, USA). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for immunoblotting and anti-GFAP for immunoblotting and immunofluoresence were obtained from Abcam, Inc. (Cambridge, MA, USA). Anti-GFAP for immunoperoxidase staining was obtained from DakoCytomation, Inc. (Carpinteria, CA, USA). Anti-Oct 3/4 was obtained from R & D Systems, Inc. (Minneapolis, MN). Alexa-Fluor-labeled anti-rabbit, -mouse, and -goat secondary antibodies were obtained from Molecular Probes, Invitrogen (Carlsbad, CA, USA). The biotinylated mouse and rabbit secondary antibodies used were included in the Universal DakoCytomation Labelled Streptavidin-Biotin2 System, Horseradish Peroxidase (DakoCytomation, Inc.).
Intraoperative smears
Samples of brain tumor tissue from three patients were sent to the Department of Pathology for intraoperative consultations. Fragments of tissue, 0.5 – 1 mm in diameter, were smeared between two glass slides, fixed in ethanol, stained with hematoxylin and eosin, and examined microscopically to confirm that the available tissues were adequate for diagnosis. High-grade gliomas were diagnosed at the time of surgery and prior to donation of specimens to the university’s brain tumor bank according to the guidelines of the Institutional Review Board (IRB). Routine histological sections of each specimen were diagnosed as glioblastoma (also known as glioblastoma multiforme or astrocytoma Grade IV, WHO classification system).
Preparation of media for cell migration and formation of pseudopodia
Additives to Minimal Essential Media (MEM) (Cellgro, Mediatech, Inc., Herndon, VA, USA) were 1% fetal bovine serum (FBS) (Cellgro) and 0.1% bovine serum albumin (BSA) for cell suspensions. Chemoattractants, HGF and serum, were added as indicated.
Preparation of conditioned media for culture of cells
MEM containing 10% FBS, 0.4 mM sodium pyruvate (Cellgro), non-essential amino acid solution for MEM (Cellgro), and antibiotics were conditioned by cultures of human astrocytoma cell lines (U87, LN229, and F98) and rat astrocytoma cells (C6), American Type Culture Collection, ATCC (Manassas, VA, USA) for one to five days and then removed. Conditioned MEM from the cell lines were combined and filtered (0.2 μm pore) prior to feeding the cultures of primary tumor cells.
Preparation of tumor specimens for cell assays and culture
Permission was obtained from the IRB to conduct this study. After tissue sufficient for diagnosis was obtained, solid fragments of tissue (0.5 – 1 cm diameter) were dissected from the midregion of each tumor and tumors were then debulked with an ultrasonic suction aspirator (Integra Neurosciences, Plainsboro, NJ). Cell suspensions in saline and blood were generated. The specimens were carried on ice to the laboratory within two hours of removal from each patient. The surgically resected tissue was minced with No. 11 disposable scalpels (Feather Safety Razor Co., Japan) yielding single cells and a few fragments that were less than 0.5 mm in diameter in 5 ml of MEM. Enzymatic digestion was avoided so that studies of cell migration and formation of pseudopodia could be performed immediately and to eliminate a potential difference with the aspirated specimens. Also, the lack of connective tissue in brain tumor tissue permitted adequate mechanical dispersal of cells without the use of enzymes. Two ultrasonic aspiration specimens were centrifuged for five minutes at 500 g to remove blood and saline and their pellets were each resuspended in 10 ml of MEM. The third aspirated specimen was tested without centrifugation.
Cell migration
Three ml of a resuspended ultrasonic aspiration specimen were compared with 1 ml of the cell suspension generated from minced tissue. Their media was supplemented with1% FBS and 0.1% BSA. The specimens were separately filtered through 40 μm nylon mesh filters (Falcon Cell Strainers, BD Biosciences Discovery Labware, Bedford, MA, USA). The bottom wells of four-well Boyden chambers (Neuro Probe, Gaithersburg, MD, USA) were filled with MEM containing 0.1% BSA and chemoattractants (5% FBS and HGF, 5 ng/ml). The chambers were assembled with 7 μm thick polycarbonate filters (Neuro Probe, Gaithersburg, MD, USA) containing 8 μm diameter pores (1,000 pores/mm2) and coated with 0.01% porcine gelatin. Cell suspensions were then added to the upper wells (three with aspirated cells and one with cells from minced tissue). The chambers were incubated at 37°C for six hours in 5% CO2.Upon completion of the assays, the filter was removed, stained with Diff Quik (Fisher Scientific, Pittsburgh, PA, USA), and placed on a glass slide with migrated materials between the semi-transparent filter and glass. Unmigrated cells were completely wiped away from the upper surface of the filter. Migrated cells were examined with an Olympus BH2 microscope (Olympus Corp. Lake Success, NY, USA) and photographed with an Olympus FV II digital camera using Olympus Microsuite Five Software for Imaging Applications (Soft Imaging System, Lakewood, CO, USA) to capture images. Migrated nuclei were counted in all 100X microscopic fields of the filter exposed to cells from minced tissue (one well), and in one third of the fields on the filter exposed to ultrasonically aspirated cells (three wells). The total area examined for migrated cells in each specimen was 248 mm2. The straight line distances of migrated cell nuclei from their nearest pore were measured in micrometers with a ruler on photographs.
Formation of pseudopodia
A method previously described for harvesting pseudopodia [9] was adapted for ultrasonically aspirated specimens. Cell suspensions were not filtered to remove tissue fragments to maximize the yield of pseudopodia. The few, scattered tissue fragments present were less than 0.5 mm in diameter. Cell solutions from minced tissue and ultrasonically aspirated cells were loaded in separate upper wells of four-well Boyden chambers that were assembled as described above except that the filters were 9 μm thick, pore diameters were 3 μm, and the pore density was 20,000 /mm2. The chambers were incubated as described above. The filters were removed, immersed in methanol for 15 seconds, and then each was placed on a glass slide with migrated pseudopodia trapped between each filter and glass. Non-migrated cell materials were completely wiped from the top of each filter. Fresh Kimwipes were used to press filters firmly against the glass slides to promote adherence of migrated pseudopodia to the glass. The filters were peeled off the slides without removing pseudopodia attached to the glass. Pseudopodia from the first and second specimens stained with Diff Quik were examined microscopically and photographed as described for the migrated cells. Pseudopodia from the first specimen were reexamined for their expression of GFAP and a B cell marker (L26) using immunoperoxidase after removal of the coverslip. Endogenous peroxidase was quenched with 3% H2O2. Blocking with Protein Blocking Agent (Shandon Immunon, Pittsburgh, PA, USA) was followed by application of anti-GFAP (1:350) and anti-L26 (1:100) in Antibody Diluent (Bio Genex, San Ramon, CA, USA) for 60 minutes. Bound immunoglobulin (IgG) was detected with biotinylated goat anti-rabbit IgG or anti-mouse IgG and peroxidase-labelled streptavidin in the Universal DakoCytomation Labelled Streptavidin-Biotin2 System, Horseradish Peroxidase, followed by development with 3-3′ diaminobenzidine Substrate Chromogen (DakoCytomation, Inc.). Sections were counterstained with Mayer’s hematoxylin.
Preparation of pseudopodial immunoblots
Pseudopodia and unmigrated cells from the second specimen were stored at −80° C on glass slides and were later prepared as urea-based lysates in a procedure previously described [9]. Lysates were electrophoresed in a 10% gel under standard reducing conditions. Gel contents were transferred to a polyvinylidene difluoride membrane (Invitrogen) and blocked (Detector Block, Protein Detector Western Blot Kit Lumi-GLO System, Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA). Following an initial reaction of the blot with anti-GAPDH, it was stripped with Restore Plus Western Blot Stripping Buffer according to the manufacturer’s instructions (Pierce, Rockford, IL, USA) and reprobed with anti-GFAP. Applications of rabbit anti-GAPDH (1:000), and mouse anti-GFAP (1:000) were followed by secondary antibodies, horseradish peroxidase-labeled anti-mouse and anti-rabbit, 1:1000 (Kirkegaard & Perry Laboratories). Immunoreactive bands were visualized via horseradish peroxidase and a luminol-based solution to produce chemiluminescence and reacted blots were scanned (Epson Perfection 4990 PHOTO, Epson America, Long Beach, CA, USA) to generate digital images. Relative reactivities of proteins were quantified in digitized bands of chemiluminescence with correction for background using densitometry (UN-SCAN-It gel, Silk Scientific, Orem, UT, USA).
Growth of cultured cells
Cells harvested with ultrasonic aspiration and from minced tissue were cultured separately in filtered, conditioned media from ATCC glioblastoma cell lines. Erythrocytes, unattached cells, and minute fragments of floating tissue were progressively removed with frequent changes in the culture media. Cultures were examined microscopically at least twice a week. At the end of three weeks the cultures were photographed on an inverted Nikon Eclipse TS100 phase contrast microscope with a RT Slider Spot digital camera and software (Diagnostic Instruments, Inc., Sterling Heights, MI, USA).
Immunofluorescent staining
Cells obtained from the ultrasonic aspirations and minced tissues were grown on glass coverslips in chamber wells using tumor conditioned media for six weeks. Cells were fixed in pre-warmed 3.7% formalin. Following three washes with PBS, cells were permeabilized in 0.1% Triton X-100 for 15 min at room temperature. Cells were blocked with 0.3% BSA for all antibodies and with 1% goat serum for anti-nestin, antiβIII-tubulin, and anti-GFAP, and with 1% donkey serum when anti-Oct 3/4 was used. The slides were incubated in primary antibodies diluted in blocking solution, rabbit anti-nestin (1:200), mouse anti-βIII-tubulin (1:200), mouse anti-GFAP (1:100), and goat anti-Oct 3/4 (1:100), overnight at 4°C. Slides were washed in PBS three times. Slides were then incubated in Alexa-Fluor labeled secondary antibodies, anti-goat, anti-rabbit, and anti-mouse IgG (1:200). Nuclei were stained by including Hoechst 33342 trihydrochloride trihydrate (Invitrogen),1:1000, in each solution of secondary antibody. The cells were washed and coverslipped with Vectashield mounting media (Vector Laboratories, Burlingame, CA, USA). Slides were examined using a BHS-RFC reflected light fluorescence attachment on the Olympus BH2 microscope (Olympus) with appropriate ultraviolet light filters. Images of individual emitted fluorescent signals were captured as described earlier and Adobe Photoshop CS2 (Adobe Systems, Inc., San Jose, CA, USA) was used to merge the fluorescent signals.
Statistical Methods
Measurements are stated as means with 95% Confidence Intervals. Comparisons are made with the student t test for two tailed distributions and two samples with unequal variance.
Results
Examination of fresh tumor materials
Intraoperative smears of the three brain tumors demonstrated tumor cellularity, glial processes, marked nuclear pleomorphism, and vascular profiles. A representative smear from one case is shown (Fig. 1A). Ultrasonically aspirated cells obtained from the same tumor were stained with Diff Quik. Their glial processes and pleomorphic nuclei are shown (Fig. 1B). Well-preserved blood vessels were also present. FIGURE 1.
Figure 1.
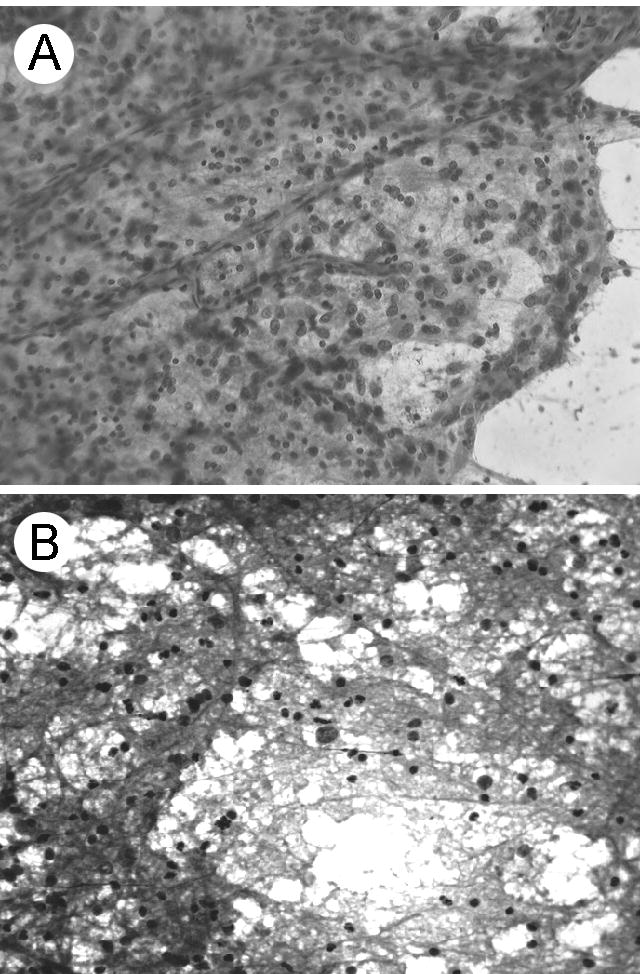
The morphology of the first tumor specimen on the day of surgery. A. Resected tumor material that was smeared and stained for intraoperative consultation. Increased cellularity, glial processes, nuclear pleomorphism, and prominent vascular profiles were representative features of the glioblastoma. The other tumors yielded similar diagnostic material. Hematoxylin and eosin stain. B. Ultrasonically aspirated tumor cells stained three hours after surgery. Tumor cells with glial processes were present. Diff Quik stain. Original magnification X 200.
Cell migration
The migration response of cells from an ultrasonically aspirated specimen (Fig. 1B) was compared to cells from its companion minced tissue specimen. The turbidity in the cell suspension obtained from minced tissue was comparable to 1.5 – 2 million cells per ml of cultured cells in suspensions used routinely in this laboratory for cell migration but cell clumping prevented accurate counts in either type of specimen. In both types of specimens, whole cells migrated through 8 μm diameter pores of 7 μm thick filters. The densities of migrated nuclei from the cell suspensions of aspirated and minced tissue were 12.1 ± 2.14 and 24.9 ± 3.11 per mm2, respectively. In the total volumes of the samples from the first specimen there would have been 29,890 ± 5285 and 30,910 ± 3860 migrated cells from the ultrasonic aspiration and minced tissue, respectively. Nuclei of cells from ultrasonically aspirated and minced tissue were comparable in size (11.4 ± 0.9 and 12.2 ± 1.4 μm diameters, respectively). Migrated cells from the aspirated specimen were detected at 7.7 ± 3.5 μm from the nearest pore, which was a greater (p < 0.05) distance compared to cells from minced tissue (3.6 ± 1.6 μm) in the representative microscopic fields shown in Figs. 2A & B, respectively. A few of the migrated nuclei of cells from the minced tissue overlapped. FIGURE 2
Figure 2.
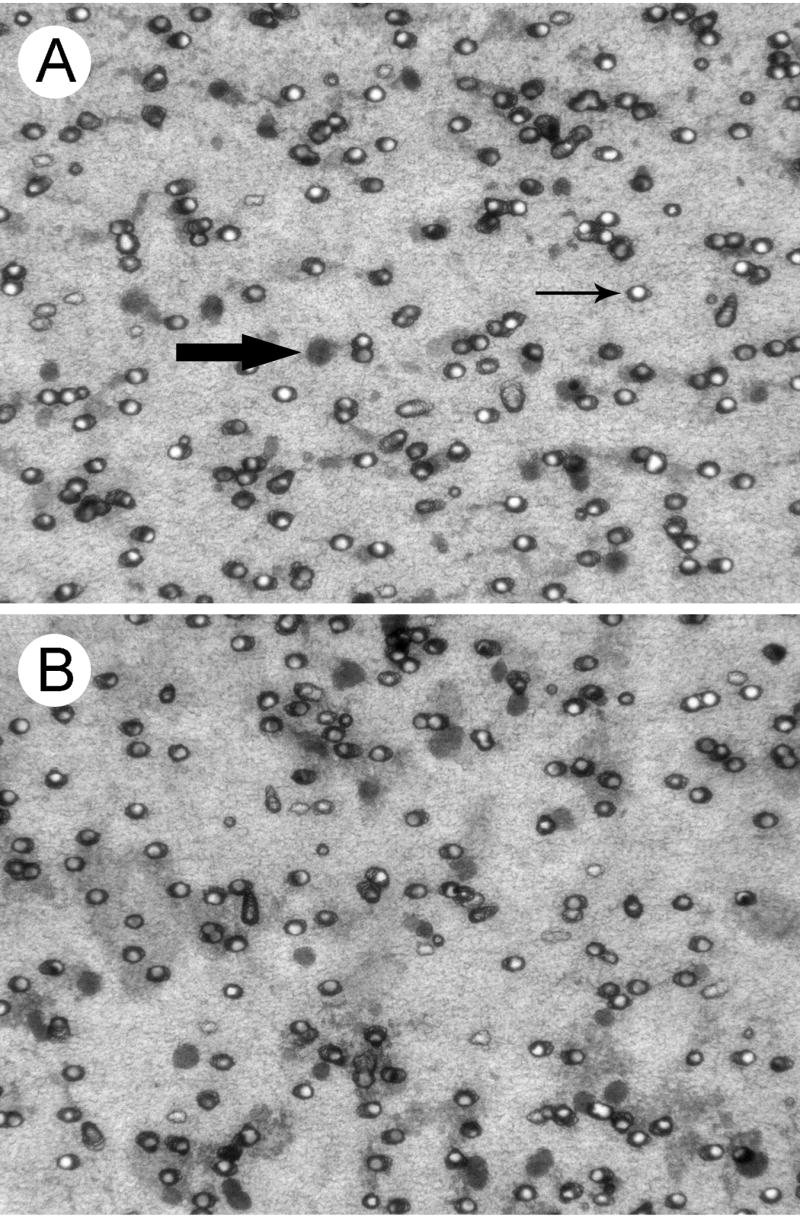
Results of migration assays for primary glioblastoma cells responding to HGF and serum. A. Migrated tumor cells from an ultrasonic aspiration of the first tumor specimen. Tumor cells retained their migratory potential following ultrasonic aspiration. Examples of migrated nuclei (large arrow) and clear lumens of pores (small arrow) are indicated. Semi-transparent filters covered the migrated cells on glass slides. B. Migrated cells that were obtained from minced tumor tissue. The darkly stained, rounded nuclei of tumor cells from aspirations and minced tissue were comparable in morphology and size, with diameters of 11.4 ± 0.9 and 12.2 ± 1.4 μm, respectively. Diff Quik stain. Magnification is indicated by the 8 μm diameter of the pores. These cells were derived from the tumor shown in Fig. 1.
Formation of pseudopodia
In response to HGF and serum, cells from the ultrasonic aspirations (Fig. 1B) and minced tissue formed pseudopodia on filters with 3 μm diameter pores. Only the pseudopodia that extended through the 9 μm thick filters were able to adhere to the glass slides for isolation [9]. The pseudopodia with elongated shapes formed by ultrasonically aspirated cells were shorter than most of those formed by cells from the minced tissue, Figs. 3A. & B., respectively. The predominant shape of pseudopodia was round. The diameters of the largest round pseudopodia from ultrasonically aspirated cells and minced tissue obtained from the first specimen were 7.8 ± 0.8 and 7.2 ± 0.5 μm, respectively (Figs. 3C. & D.). Pseudopodia obtained from the second specimen were also predominantly round (not shown). The cytoplasm of pseudopodia stained homogeneously with Diff Quik except for condensations at sites consistent with locations of pores that were removed when filters were peeled away. Confluent pseudopodia in a few small foci most likely corresponded with overlying tissue fragments that were also removed with the filters. Only a few nuclei migrated through the 3 um pores and one dark round nucleus is shown near the top and middle of Fig. 3B. Proteinaceous material consistent with serum in the third ultrasonically aspirated specimen (not centrifuged) obscured profiles of pseudopodia if they were present (not shown). FIGURE 3
Figure 3.
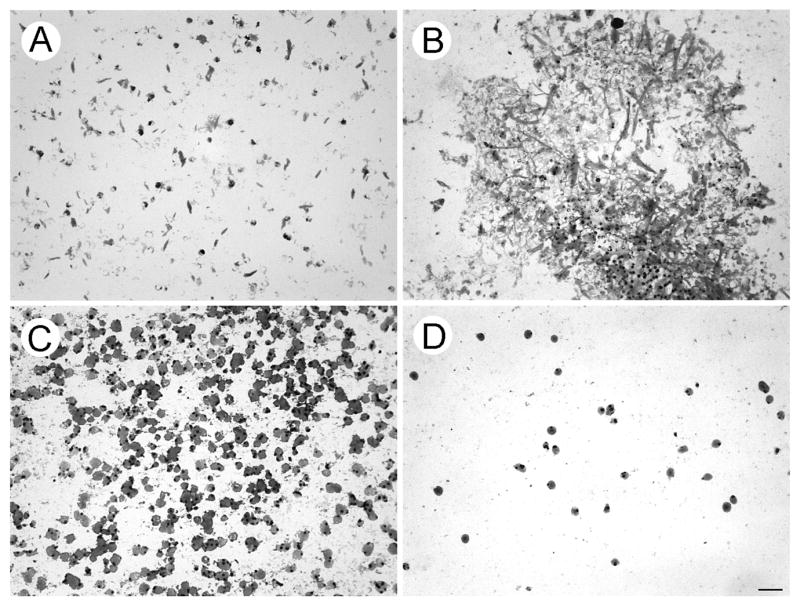
Pseudopodia of tumor cells responding to HGF and serum. Cytoplasmic projections extruded through 3 μm pores were fixed and attached to glass slides prior to filter removal and staining. A. & B. demonstrate elongated pseudopodial profiles formed by cells from ultrasonic aspirations and minced tumor, respectively. C. & D. demonstrate rounded pseudopodial profiles formed by cells from ultrasonic aspirations and minced tumor, respectively. The pseudopodia stained homogeneously with Diff Quik except for condensations consistent with locations of pores prior to removal of filters. The assay in Fig. 2 demonstrated that the leading edge of a migrating cell population was included in the pseudopodia shown. Since the pore density of the filters varied according to pore size (to maintain consistent tensile strength), 20-fold greater numbers of pores were present in filters used for formation of pseudopodia compared to the filter used for cell migration. Only a few nuclei migrated through the 3 μm pores and a darkly stained, migrated nucleus is located in the mid region and near the top of panel B. Diff Quik stain. The magnification bar represents 25 μm.
Expression of GFAP in pseudopodia
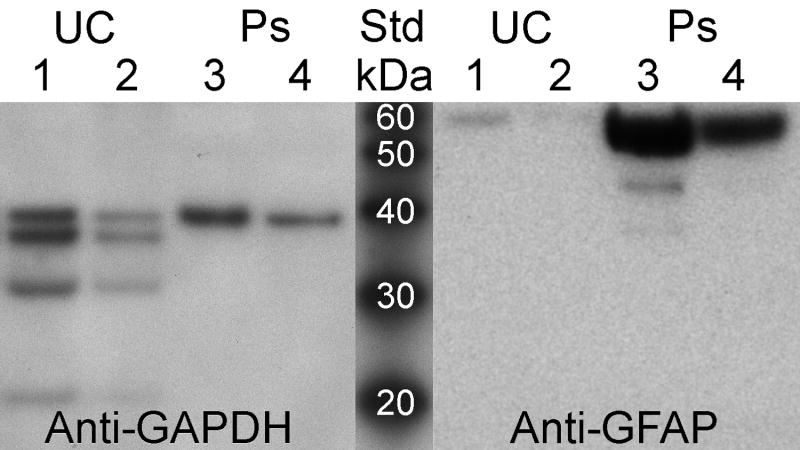
Pseudopodia formed by aspirated cells from the first specimen that were shown in Fig. 3 stained diffusely positive for GFAP (Fig. 4A) and negatively for L26, a B cell marker (Fig. 4B). FIGURE 4 Pseudopodia formed by aspirated cells from the second specimen were harvested as lysates and studied on immunoblots. Pseudopodia formed by cells from minced tissue were insufficient for preparing lysates. A lysate of unmigrated material (tumor cells, fragments of blood vessels, blood cells, etc.) from the second specimen was also generated. The immunoblots demonstrated 9.43-fold enrichment of GFAP (predicted molecular weight of 55 kDa) in the pseudopodia (Lane 4) compared to the unmigrated material (Lane 1). The non-degenerated form of GAPDH (top band with a predicted molecular weight of 36 kDa) shown for comparison to indicate loading (Fig. 5). The mixed population of unmigrated aspirated cells included degenerated cells based on the presence of several low molecular weight forms of GAPDH in the lysate. Expression of only the single band of intact GAPDH as well as GFAP in the pseudopodia suggested their formation was restricted to viable tumor cells. FIGURE 5
Fig. 4.
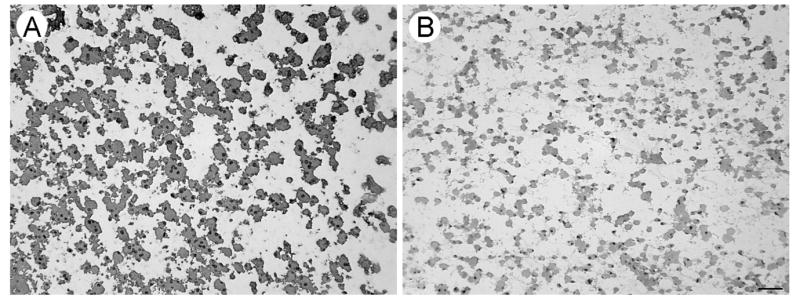
Expression of a glial marker in pseudopodia from the first specimen. A. Diffuse expression of GFAP (brown in a color image) was found in pseudopodia from the ultrasonic aspiration specimen shown in Fig. 3. B. The pseudopodia stained negatively for a B-cell marker (L26). Mayer’s hematoxylin counterstain. The magnification bar represents 25 μm.
Fig. 5.
Enriched expression of a glial marker in lysates of pseudopodia harvested from the second specimen. Aspirated cells formed pseudopodia that were harvested as an urea-based lysate for comparision with unmigrated aspirated cellular material. Expression of GFAP (predicted molecular weight of 55 kDa) was increased in pseudopodia compared to unmigrated cells with GAPDH (predicted molecular weight of 36 kDa) present to indicate loading. Although lower molecular weight bands for GAPDH in lysates of unmigrated cell materials suggested the presence of degeneration, only the intact form of GAPDH was found in the lysates of pseudopodia. Lanes 1 and 4 with approximately equivalent amounts of intact GAPDH demonstrated a 9.43-fold increase of GFAP in pseudopodia compared to unmigrated cells. Hemoglobin in the lysates precluded measurements of their protein content. Almost all of the pseudopodial lysate obtained from the second specimen was loaded in the two lanes (3 & 4) shown.
Morphology of cultured cells
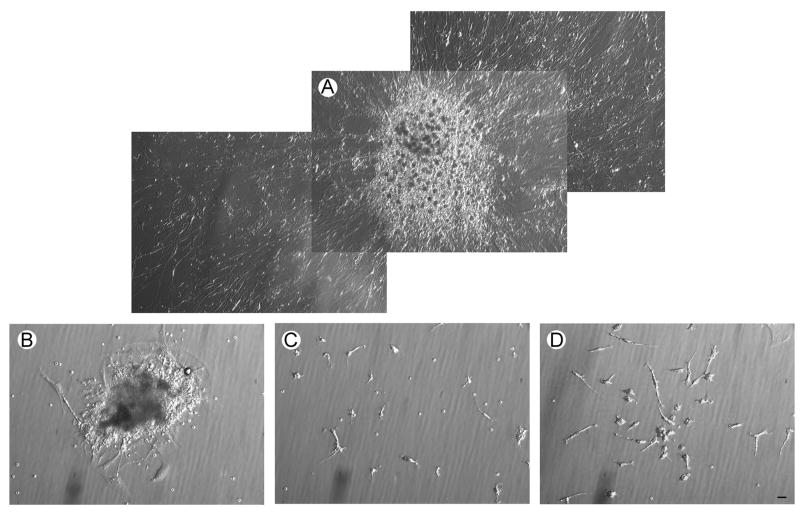
Cells from ultrasonic aspirations and minced tissue were grown separately in culture with filtered media conditioned by glioblastoma cell lines. By two weeks, several loosely attached spheres of very small cells were formed in cultures of both types of specimens. These structures sprouted rapidly expanding radial monolayers of spindled cells over the next few weeks (Fig. 6A.). Tumorspheres with stem cell potential displayed a distinct morphology that differed greatly from irregular aggregates of cells (Fig. 6B.). Single cells with spindled processes also grew from the ultrasonic aspirations and minced tissue (Fig. 6C. & D., respectively). Scattered erythrocytes remained in the cultures through numerous changes of the media. The morphology, including proliferating spheres, of cells from ultrasonic aspirations and minced tissues of the second and third tumors were the same as illustrated for the first tumor. FIGURE 6
Fig. 6.
The morphology of tumor cells in culture for three weeks viewed with phase contrast microscopy. Filtered conditioned MEM medium obtained from cultured ATCC glioblastoma cell lines supported cells obtained from ultrasonic aspirations and minced tissue. A. Merged photographs show the morphology of a representative proliferating tumorsphere formed by ultrasonically aspirated cells. The radial cellular outgrowth that developed rapidly from each tumorsphere indicated that progenitor cells were included in these structures. B. An irregular aggregate of cells from the cultured ultrasonically aspirated specimen is shown. C.& D. Many single cells were present in cultures of cells from ultrasonic aspirations and minced tissue, respectively. The cultures of both types of specimens were similar in morphology. The magnification bar represents 25 μm.
Immunoreactivity of cultured cells
Glass coverslips with adherent cultured cells were stained to investigate their differentiation. Almost all cells obtained with ultrasonic aspiration and from minced tissue were positive for nestin, βIII-tubulin, and GFAP at six weeks, indicating that they were of neuroectodermal lineage. The nuclei were stained with Hoechst. Although each nucleus had three-five bright spots of Oct 3/4 staining, no homogenous staining of entire nuclei was identified (Fig. 7). No significant fibroblastic growth was noted. FIGURE 7
Fig. 7.
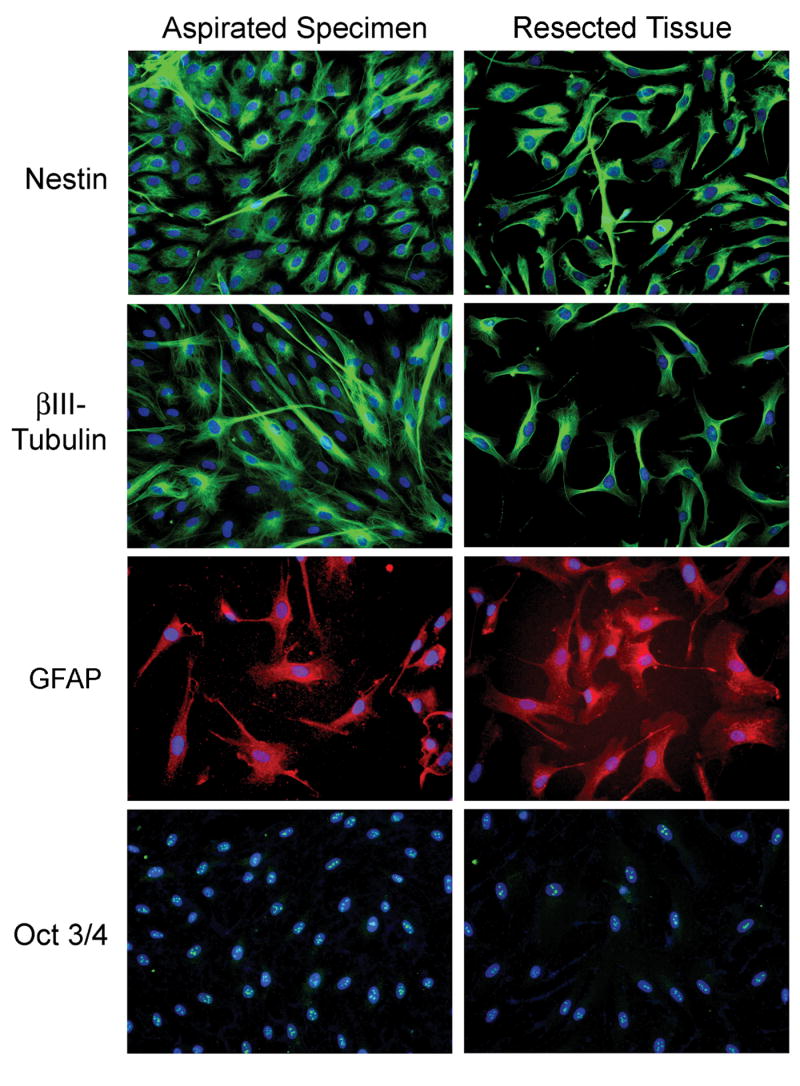
Differentiation of cultured cells determined by immunofluorescent stains. The cultures of ultrasonically aspirated cells (first column) and cells from minced tissue (second column) were similar. Almost all cells stained positive for nestin (green, top row) indicating that they were of primitive neuroectodermal lineage. Cells stained for βIII-tubulin, a neuronal marker (green, second row). The cells also stained for GFAP, a glial marker (red, third row). Nuclei were stained with Hoechst (blue). Each nucleus had three to five bright spots of Oct 3/4 staining (green, bottom row). No homogeneous staining of entire nuclei was identified in either type of specimen. Tumorspheres did not develop on the coverslips used for immunofluorescent stains. Original magnification X 200.
Discussion
Most migration and proliferation studies of tumor cells have been performed on cells grown in tissue culture conditions for at least several generations. Although fresh, intact tissue samples from brain tumors are not usually readily available, primary tumor cells can be obtained from ultrasonically aspirated specimens that would otherwise be discarded. Ultrasonically aspirated cells exposed to chemoattractants (HGF and serum) produced abundant pseudopodia and migrated as whole cells through porous filters. Migrated cells with their nuclei appeared on the undersurfaces of filters only after they had traversed pores that were 7 and 8 μm in length and diameter, respectively. The aspirated specimen tested contained approximately 30,000 migratory cells. The density of migrated cells in the ultrasonic aspiration specimen tested would have yielded at least several hundred migrated nuclei in migration chamber wells that allow cell contact with 50 mm2 of filter. Although the density of migrated cells would probably be too low to quantify with densitometry, the cells could be counted manually. The numbers of migrated cells should be sufficient for several replicates to be used in statistical comparisons of treatments with drugs, inhibitors, etc. in future studies. Thus if these specimens are used for evaluating drug treatments, etc., both test and control replicates in the assays would be obtained from each patient’s tumor to correct for variables, such as exposure to ultrasound, saline, blood, etc. The morphologic appearances of nuclei in ultrasonically aspirated cells and their pseudopodia were comparable to those of cells obtained from the minced tumor tissue. The cells formed elongated and rounded pseudopodia. Although the profiles of pseudopodia in our earlier studies of cell lines were predominantly elongated [9, 10], pseudopodia formed by leukocytes have a similar rounded appearance [11]. Thickness of the filters and the diameter of their pores, 9 and 3 μm, respectively, generated tunnels that prevented isolation of non-specific, random cell projections. Similarity between ultrasonic aspiration and minced tissue samples indicated that cellular competence for cell migration was not destroyed by ultrasonic retrieval.
Each ultrasonic aspiration also provided several proliferating cellular spheres. Components of the aspirated tumor samples , i.e. serum, endothelial cells, and astrocytic cells, have been noted to support and modulate spheres of progenitor cells in other types of cultures [12–14]. The malignant features of tumor progenitors may also help to explain the development of proliferating tumorspheres despite the lack of media designed for stem cells. Although others have obtained tumorspheres with stem cell potential from resected glioblastomas [15, 16], their retrieval following ultrasonic aspiration has not been reported. High-energy shock waves with a wide range of energy and number of shots (0.22 to 0.43 mJ/mm2 and from 200 to 1500 shots) have previously been shown to not interfere with expansion and engraftment of cord blood stem cells and actually improved early progenitor cell expansion [17]. In this study, ultrasonically aspirated cells were exposed to 24 kiloHertz which did not destroy progenitor cells that formed tumorspheres in our cultures. Although proliferating tumorspheres can be generated from glioblastoma cell lines [18], the availability of large numbers of ultrasonically aspirated cells from routine resections of brain tumors provides a novel and widely applicable approach for obtaining primary tumorspheres.
The rapid outgrowth of cell monolayers from the tumorspheres confirmed the presence of progenitor cells. We did not attempt to maintain the progenitor cells in a state of self-renewal or promote their differentiation into multiple lineages. Most of the cells that grew in culture contained neuronal markers, nestin and βIII-tubulin, and a glial marker, GFAP. In adult tissues, the distribution of βIII-tubulin is almost exclusively neuron-specific. However, transient expression of βIII-tubulin is also present in putative neuronal and glial precursor cells. Glioblastoma cells can also express βIII-tubulin [19, 20]. Nestin is a good neural progenitor marker [21]. It is an intermediate neurofilament expressed early in CNS and muscle development that is replaced by other neurofilaments and GFAP but nestin can also be re-expressed in glioblastomas [22–25]. Expression of Oct-3 and Oct-4 transcription factors that are characteristic of pluripotent cells [26–28] can also be found in glioblastomas [29]. The study of differentiation in tumor progenitor cells and their progeny is proposed as a model to understand the malignant features of brain tumors [21, 30] which potentially can be applied to ultrasonic aspirations.
The availability of ultrasonically aspirated specimens expand the options of investigating tumors from individual patients. These specimens yield functional tumor cells to study therapeutic targets, including migrated cells, pseudopodia, and tumorspheres [31–34]. The availability of ultrasonically aspirated primary tumor cells for future studies should broaden and boost our efforts in understanding and suppressing the malignant behavior of glioblastomas.
Acknowledgments
We thank the University of Pittsburgh Brain Tumor Bank for procurement of specimens with the cooperation of R Madhok, MD and DL Lunsford, MD, Department of Neurological Surgery, University of Pittsburgh. We thank Jing He, PhD, Children’s Hospital of Pittsburgh, for advice and technical assistance with phase contrast microscopy. We thank Judith A. Burnham, Department of Pathology, University of Pittsburgh, for technical assistance with immunoperoxidase staining of pseudopodia. We thank The Nick Eric Wichman Foundation, The Beez Foundation, and The Walter L. Copeland Fund for Cranial Research of the Pittsburgh Foundation for financial support and encouragement.
Abbreviations
- ATCC
American Type Culture Collection
- BSA
bovine serum albumin
- FBS
fetal bovine serum
- GAPDH
glyceraldehydes-3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- HGF
hepatocyte growth factor
- IgG
immunoglobulin
- IRB
institutional Review Board
- MEM
Minimal Essential Media
- PBS
phosphate buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oakes WJ, Friedman HS, Bigner SH, Bullock NH, Bigner DD. Successful laboratory growth and analysis of CUSA-obtained medulloblastoma samples. Technical note. J Neurosurg. 1990;72:821–823. doi: 10.3171/jns.1990.72.5.0821. [DOI] [PubMed] [Google Scholar]
- 2.Oosterhuis JW, Lung PF, Verschueren RC, Oldhoff J. Viability of tumor cells in the irrigation fluid of the Cavitron Ultrasonic Surgical Aspirator (CUSA) after tumor fragmentation. Cancer. 1985;56:368–370. doi: 10.1002/1097-0142(19850715)56:2<368::aid-cncr2820560227>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Richmond IL, Hawksley CA. Evaluation of the histopathology of brain tumor tissue obtained by ultrasonic aspiration. Neurosurgery. 1983;13:415–419. doi: 10.1227/00006123-198310000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Thompson MA, Adelson MD, Jozefczyk MA, Coble DA, Kaufman LM. Structural and functional integrity of ovarian tumor tissue obtained by ultrasonic aspiration. Cancer. 1991;67:1326–1331. doi: 10.1002/1097-0142(19910301)67:5<1326::aid-cncr2820670510>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 5.Finley JL, Silverman JF, Dickens MA. Immunocytochemical evaluation of central nervous system tumors obtained by the Cavitron ultrasonic surgical aspirator. Diagn Cytopathol. 1990;6:308–312. doi: 10.1002/dc.2840060504. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra V, Malik R, Gondal R, Beohar PC, Parkash B. Evaluation of histological appearance of tissues removed by cavitron ultrasonic surgical aspirator (CUSA) Acta Neurochir (Wien) 1986;81:132–134. doi: 10.1007/BF01401235. [DOI] [PubMed] [Google Scholar]
- 7.Silverman JF, Jones FD, Unverferth M, Berns L. Cytopathology of neoplasms of the central nervous system in specimens obtained by the Cavitron Ultrasonic Surgical Aspirator. Acta Cytol. 1989;33:576–582. [PubMed] [Google Scholar]
- 8.Brockmann MA, Ulbricht U, Gruner K, Fillbrandt R, Westphal M, Lamszus K. Glioblastoma and cerebral microvascular endothelial cell migration in response to tumor-associated growth factors. Neurosurgery. 2003;52:1391–1399. doi: 10.1227/01.neu.0000064806.87785.ab. [DOI] [PubMed] [Google Scholar]
- 9.Beckner ME, Chen X, An J, Day BW, Pollack IF. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Lab Invest. 2005;85:316–327. doi: 10.1038/labinvest.3700239. [DOI] [PubMed] [Google Scholar]
- 10.Beckner ME, Zhang Z, Agostino NR, Day BW, Pollack IF. Albumin marks pseudopodia of astrocytoma cells responding to hepatocyte growth factor or serum. Lab Invest. 2006;86:1103–1114. doi: 10.1038/labinvest.3700470. [DOI] [PubMed] [Google Scholar]
- 11.Fawcett DW. The Cell, Its Organelles and Inclusions, An Atlas of Fine Structure. W. B. Saunders; Philadelphia, PA: 1966. p. 393. [Google Scholar]
- 12.Barkho BZ, Song H, Aimone JB, Smrt RD, Kuwabara T, Nakashima K, Gage FH, Zhao X. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 2006;15:407–421. doi: 10.1089/scd.2006.15.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 14.Stojkovic P, Lako M, Przyborski S, Stewart R, Armstrong L, Evans J, Zhang X, Stojkovic M. Human-serum matrix supports undifferentiated growth of human embryonic stem cells. Stem Cells. 2005;23:895–902. doi: 10.1634/stemcells.2004-0326. [DOI] [PubMed] [Google Scholar]
- 15.Kang SK, Park JB, Cha SH. Multipotent, dedifferentiated cancer stem-like cells from brain gliomas. Stem Cells Dev. 2006;15:423–435. doi: 10.1089/scd.2006.15.423. [DOI] [PubMed] [Google Scholar]
- 16.Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 17.Berger M, Frairia R, Piacibello W, Sanavio F, Palmero A, Venturi C, Pignochino Y, Berta L, Madon E, Aglietta M, Fagioli F. Feasibility of cord blood stem cell manipulation with high-energy shock waves: an in vitro and in vivo study. Exp Hematol. 2005;33:1371–1387. doi: 10.1016/j.exphem.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci USA. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsetos CD, Del Valle L, Geddes JF, Assimakopoulou M, Legido A, Boyd JC, Balin B, Parikh NA, Maraziotis T, de Chadarevian JP, Varakis JN, Matsas R, et al. Aberrant localization of the neuronal class III beta-tubulin in astrocytomas. Arch Pathol Lab Med. 2001;125:613–624. doi: 10.5858/2001-125-0613-ALOTNC. [DOI] [PubMed] [Google Scholar]
- 20.Katsetos CD, Legido A, Perentes E, Mork SJ. Class III beta-tubulin isotype: a key cytoskeletal protein at the crossroads of developmental neurobiology and tumor neuropathology. J Child Neurol. 2003;18:851–866. doi: 10.1177/088307380301801205. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SC. Neural subtype specification from embryonic stem cells. Brain Pathol. 2006;16:13–12. doi: 10.1111/j.1750-3639.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrmann J, Kolar Z, Mokry J. Nestin as a diagnostic and prognostic marker: immunohistochemical analysis of its expression in different tumours. J Clin Pathol. 2005;58:222–223. doi: 10.1136/jcp.2004.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutka JT, Ivanchuk S, Mondal S, Taylor M, Sakai K, Dirks P, Jun P, Jung S, Becker LE, Ackerley C. Co-expression of nestin and vimentin intermediate filaments in invasive human astrocytoma cells. Int J Dev Neurosci. 1999;17:503–515. doi: 10.1016/s0736-5748(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 24.Schiffer D, Manazza A, Tamagno I. Nestin expression in neuroepithelial tumors. Neurosci Lett. 2006;400:80–85. doi: 10.1016/j.neulet.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Veselska R, Kuglik P, Cejpek P, Svachova H, Neradil J, Loja T, Relichova J. Nestin expression in the cell lines derived from glioblastoma multiforme. BMC Cancer. 2006;6:32. doi: 10.1186/1471-2407-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh YH, Wu Z, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 27.Morrison GM, Brickman JM. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development. 2006;133:2011–2012. doi: 10.1242/dev.02362. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto Y, Hasegawa K, Suemori H, et al. Molecular cloning and function of Oct-3 isoforms in Cynomolgus monkey embryonic stem cells. Stem Cells Dev. 2006;15:566–574. doi: 10.1089/scd.2006.15.566. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber E, Merchant RE, Wiestler OD, Fontana A. Primary brain tumors differ in their expression of octamer deoxyribonucleic acid-binding transcription factors from long-term cultured glioma cell lines. Neurosurgery. 1994;34:129–135. [PubMed] [Google Scholar]
- 30.Steindler DA. Redefining cellular phenotypy based on embryonic, adult, and cancer stem cell biology. Brain Pathol. 2006;16:169–180. doi: 10.1111/j.1750-3639.2006.00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo T. Brain cancer stem-like cells. Eur J Cancer. 2006;42:1237–1242. doi: 10.1016/j.ejca.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Nakano I, Kornblum HI. Brain tumor stem cells. Pediatr Res. 2006;59:54R–58R. doi: 10.1203/01.pdr.0000203568.63482.f9. [DOI] [PubMed] [Google Scholar]
- 33.Tunici P, Irvin D, Liu G, Yuan X, Zhaohui Z, Ng H, Yu JS. Brain tumor stem cells: new targets for clinical treatments? Neurosurg Focus. 2006;20:E27. doi: 10.3171/foc.2006.20.4.17. [DOI] [PubMed] [Google Scholar]
- 34.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]