Abstract
Type IV pili (Tfp) are widespread filamentous bacterial organelles that mediate multiple virulence-related phenotypes. They are composed mainly of pilin subunits, which are processed before filament assembly by dedicated prepilin peptidases. Other proteins processed by these peptidases, whose molecular nature and mode of action remain enigmatic, play critical roles in Tfp biology. We have performed a detailed structure/function analysis of one such protein, PilX from Neisseria meningitidis, which is crucial for formation of bacterial aggregates and adhesion to human cells. The x-ray crystal structure of PilX reveals the α/β roll fold shared by all pilins, and we show that this protein colocalizes with Tfp. These observations suggest that PilX is a minor, or low abundance, pilin that assembles within the filaments in a similar way to pilin. Deletion of a PilX distinctive structural element, which is predicted to be exposed on the filament surface, abolishes aggregation and adhesion. Our results support a model in which surface-exposed motifs in PilX subunits stabilize bacterial aggregates against the disruptive force of pilus retraction and illustrate how a minor pilus component can enhance the functional properties of pili of rather simple composition and structure.
Keywords: adhesion, aggregation, pilus retraction, protein crystallography
Type IV pili (Tfp) are hair-like bacterial surface filaments that have been detected in dozens of Gram-negative and Gram-positive species (1–3). These organelles mediate bacterial adhesion to surfaces as diverse as stainless steel (4) and host cells (1), a property that is tightly linked to their ability to promote the formation of adhesive 3D microcolonies by mediating interbacterial interactions (5, 6). Tfp are therefore one of the most widespread virulence factors in bacteria and play a key role in infection in several human pathogens, including our working model Neisseria meningitidis. A remarkable property of Tfp is that they can be retracted into the bacterium from which they originate, via the action of the force-generating ATPase PilT (7, 8), which promotes other phenotypes including bacterial motility, competence for DNA transformation, and signaling to host cells.
Across divergent species, there is striking homogeneity among Tfp, long thin fibers that interact laterally to form bundles. Tfp are substantially polymers of one protein, the pilin PilE, that is produced by cleavage of a distinctive N-terminal sequence pattern from prepilin by the prepilin peptidase PilD (9). 3D structures of type IV pilins from several species reveal a core protein fold consisting of a ≈54-aa-long N-terminal α-helix (α1), the C-terminal half of which (α1C) is cradled against a four-stranded antiparallel β-sheet (10, 11). These similarities suggest that the molecular mechanisms underlying biogenesis and functions of these filaments are conserved (1, 10) but leave open the question of how Tfp, which have rather simple structure and composition, can mediate such a vast array of functions. In all bacteria expressing Tfp, there are additional proteins with prepilin-like N-terminal sequences that play important roles in Tfp biology (12). In pathogenic Neisseria species, there are seven such proteins cleaved by PilD (PilH, PilI, PilJ, PilK, ComP, PilV, and PilX also known as PilL in Neisseria gonorrhoeae). ComP, PilV and PilX, which have canonical PilD cleavage motifs and mature lengths similar to PilE, are particularly interesting because they modulate various Tfp-linked functions and yet are dispensable for Tfp biogenesis (although pilL and unencapsulated pilX mutants exhibit reduced piliation) (13, 14). ComP and PilV in N. gonorrhoeae are necessary for competence for DNA transformation (15) and adhesion to human cells (16) respectively, whereas N. meningitidis PilX is essential for bacterial aggregation and adhesion (17). These three proteins thus provide excellent models to study the relationship between the structure and function of pilin-like proteins because the phenotypes associated with the corresponding mutants are not obscured by the absence of pili.
Fundamental questions regarding proteins with prepilin-like N termini remain unanswered. Are they minor components of pilus filaments? Do they adopt the same fold as pilin and hence qualify as “minor pilins”? How do they modulate the diverse functions associated with Tfp? Providing answers to these questions would not only be desirable because of the widespread nature of Tfp as virulence factors, but also because of the important roles played by similar proteins in type II secretion (18) or assembly of flagella in archaebacteria (19). In this study we have investigated PilX, which we identified previously in N. meningitidis strain 8013 as a preprotein that is processed by PilD and copurifies with Tfp (17). We reported that a pilX mutant, despite having quantitatively and qualitatively unaltered fibers, displays a selective loss of Tfp-linked phenotypes. Although motile and naturally competent, this mutant is unable to form bacterial aggregates and to adhere to human cells (17) because interbacterial interactions are essential for Tfp-facilitated adhesion (5, 6). Interestingly, because aggregation is restored in a pilX/T double mutant, it seemed that PilX participates in the formation of aggregates by somehow counterbalancing PilT-mediated Tfp retraction. To understand the molecular basis for PilX's properties, we have carried out a 3D structure/function analysis of this protein.
Results
PilX Has Structural Features of a Type IV Pilin.
The sequence similarity between the mature PilX and N. gonorrhoeae PilE proteins, after processing, by PilD, of 10 and 7 aa, respectively, is restricted to the highly hydrophobic first 27 aa, 96% of which are identical or conserved (Fig. 1). This region in PilX is therefore likely to adopt the same α-helical structure as the corresponding α1N filament-forming domain in PilE. Otherwise, PilX exhibits no sequence similarity with pilin, or any other protein, over the remaining 125 residues. To avoid solubility problems and facilitate crystallization, we produced a soluble truncated form of PilX missing the first 28 residues of the mature protein similarly to what was done for the Pseudomonas aeruginosa PAK pilin (20). We solved the x-ray crystal structure of six independent noncrystallographically related PilX monomers, represented in two space groups [supporting information (SI) Table 1]. The six compare with one another with a Cα root mean square deviation (rmsd) of <1 Å for all possible pairs.
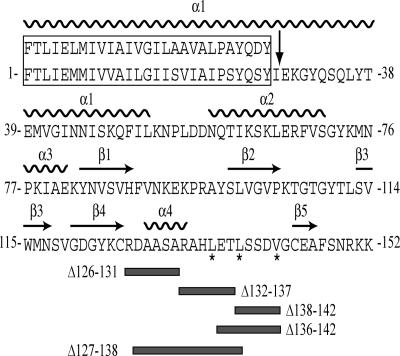
Fig. 1.
Relevant features of the PilX protein and the PilX variants. Sequence of the mature PilX protein is from N. meningitidis strain 8013. The boxed N-terminal 27 residues represent the hydrophobic part of PilX highly conserved in N. gonorrhoeae pilin (upper sequence). A 28-aa region of PilX was truncated, as depicted by arrow, to facilitate crystallization. The surface-exposed hydrophobic residues in the D-region that were exchanged for serines are identified by *, whereas those that were deleted are indicated by thick gray lines.
Truncated PilX adopts the classic pilin fold with an α1C helix cradled against a four-stranded anti-parallel β-sheet (21) (Fig. 2A). Seventy-nine residues from the conserved core of PilX can be aligned with analogous residues of the secondary structural elements from PilE with a rmsd of 3 Å (Fig. 2B). Notable differences are the lack of curvature in α1C in the truncated form of PilX, although we cannot rule out that curvature might be present in the full-length molecule, and the longer insertion between β1 and β2, which is even longer in some pilX alleles (14). Like in PilE, hydrophobic packing at the core of the conserved fold serves to stabilize the interaction of α1C against the interior surface of the β-sheet. The three methionines (Met40, Met75, Met116) are arranged in notable proximity within the densely packed hydrophobic interior between sheet and helix (SI Fig. 7A). A C-terminal disulfide bond, an invariant feature of type IV pilins, is also found in PilX (Fig. 2A). Hovever, although in both PilX and PilE the first of the cysteines is at the C-terminal end of β4 and both disulfide bonds staple β4 to the D-region, the covalent links are on opposite faces of the central β-sheet.
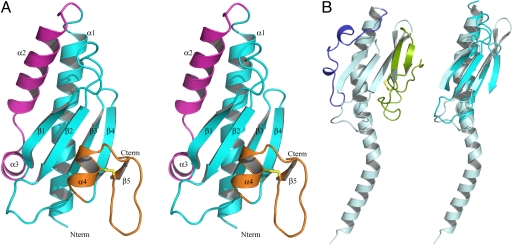
Fig. 2.
The 3D structure of PilX reveals an archetypal type IV pilin. (A) The PilX fold, represented as a stereo ribbon drawing, includes the N-terminal α1C helix and 4-stranded antiparallel β-sheet (cyan) that characterize type IV pilins, as well as distinctive antiparallel α2-α3 helices (magenta) and D-region delimited by a disulfide bond (yellow) consisting of the α4 pigtail and the α4-β5 connection hook (orange). (B) Full-length PilE with its species-variable edges α1/β- (blue) and D-regions (green) highlighted (Left). The superposition of PilX (cyan) and PilE (gray blue) without α1/β- and D-regions emphasizes the conserved core architecture (Right).
In pilins, the two regions flanking the central β-sheet show substantial sequence, length and structural variation and therefore define structurally variable “edges” (11). These edges are the α1/β-region found between α1 and the first β-strand, and the D-region delimited by an invariant disulfide bond near the C terminus (Fig. 2B). In PilX, two α-helices form the α1/β-region (Fig. 2A). α2 is a 4-turn helix nearly parallel to α1, whereas α3 is a two-turn helix perpendicular to α1. A stripe of hydrophobic residues from α2 (Ile62, Leu66, Phe69, Val70) packs between α1 (Ile43, Ile46, Phe50) and the β1-β2 loop (Val85, Val87, Phe89, Pro95) so that the helix participates in the hydrophobic core of PilX. An intriguing observation is that among the six independently determined structures, the orientations of Phe50 and Phe69 vary on either side of the α2-α3 turn (SI Fig. 7B). Although this variation may be promoted by crystal packing interactions, the apparent unraveling of α3 by one residue seen in two monomers (P43 chains A and B) is interestingly compensated by a re adjustment of other residues in this region (SI Fig. 7B). The D-region of PilX, which forms a protrusion, consists of a short “pigtail” helix α4 and a “hook” (Fig. 2A). The pigtail occupies the volume filled with the sequence variable β-hairpin in the PilE subunit (21). The D-region of PilX is packed against the central β-sheet by the stacking of β3 Trp115 between β4 Tyr123 and α4 His134, covered from the front by hook residue Glu136.
Our x-ray crystallographic analysis of truncated PilX shows that it has the structure of a type IV pilin with a conserved α/β roll fold, whereas the α1/β- and D-regions form structural edges unlike those seen in any pilin.
PilX Is a Minor Pilin.
By appending the closely homologous α1N from the PilE structure (Fig. 1) onto the N terminus of the truncated PilX structure, we have created a full-length 3D representation of PilX. This model supports the obvious hypothesis that PilX assembles within the pilus filament as pilin itself does and that its previously reported copurification with Tfp is not merely due to a loose association (17). We modeled the possible packing of this protein within a recently reported 12.5 Å resolution structural model for Tfp (22), which is based on a N. gonorrhoeae pilin with 93% identical or conserved residues with the pilin in strain 8013. We found that PilX fits readily into this model (Fig. 3). The straight α1C helix in PilX does not preclude its assembly into filaments and leads to better presentation of surface-exposed structural edges of PilX that jut out from the filament surface. Residues that make up the D-region and the top of α2 are fully or partially exposed, respectively, on the fiber surface (Fig. 3).
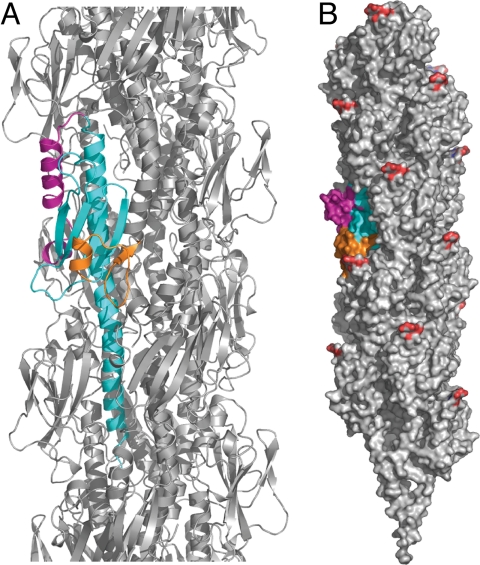
Fig. 3.
Model for packing of full length PilX (colored as in Fig. 2A) into the current Tfp model (22) (gray protein atoms and red posttranslational PilE disaccharide modifications). To create this model, a least-squares superposition of the backbone atoms of the core α/β fold elements in PilE and PilX was calculated. Because of the different curvatures of α1 in PilX and PilE, the best junction was judged by eye to be between residues 30 and 31 and thus the globular domain of PilE was replaced beginning with residue 30 with the truncated PilX structure. (A) The ribbon representation highlights the fit of the PilX monomer into the filament. (B) Within this packing arrangement, displayed as a solvent-accessible surface, the D-region α4 pigtail and hook protrude from the filament surface.
We next sought experimental evidence that PilX indeed colocalizes with Tfp. In immuno-fluorescence (IF) experiments, strains were doubly stained by using a monoclonal antibody (20D9) that specifically recognizes the filaments of N. meningitidis strain 8013 (23) and a polyclonal antibody generated against the native PilX used for crystallization. Merged images revealed that PilX colocalizes with Tfp in the wild-type (WT) strain and in the pilX/pilXWT complemented mutant (SI Fig. 8) in which a WT copy of the pilX gene is expressed ectopically under the transcriptional control of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter. To analyze this colocalization in more detail, we examined bacteria by immuno-transmission electron microscopy (TEM) using the same anti-PilX antibody. The number of gold particles is dramatically smaller than in previous experiments in which pilin was detected by using the 20D9 antibody (24), which is consistent with our immunoblot estimate that PilX represents 3% of total pilus proteins (data not shown), or ≈30 PilX monomers per 1,000-subunit filament. No labeling was seen for the pilX mutant (Fig. 4). Several gold particles corresponding to PilX were consistently observed on each pilus bundle in the WT and pilX/pilXWT strains, with no apparent periodicity or preference for the tip of the pilus, suggesting that PilX is distributed along the filaments. In the pilX/pilXWT strain, in which PilX is overproduced, a mean 4.8-fold increase in gold particles was observed associated with pilus bundles. In this strain, up to 30% of PilX was observed apparently outside of bundles, most likely associated with poorly stained individual pilus filaments or pieces of pili, or alternatively secreted or released by lysis.
Fig. 4.
PilX colocalizes with the pilus filaments. By using immuno-transmission electron microscopy, gold particles that correspond to PilX molecules were seen on Tfp bundles in the WT and pilX/pilXWT strains. (Scale bars: 100 nm.) Images at 27,000-fold magnification show on average 8.6 gold particles for the WT and 41.3 gold particles for the pilX/pilXWT strain, 87% and 78% of which, respectively, are associated with visualizable filaments.
Taken together, PilX's structure, 3D modeling of its assembly within the filament and its colocalization with pili on the surface of bacteria strongly support the view that PilX is a minor component of Tfp that assembles into filaments as the major subunit PilE does, with the consequence that its distinctive structural edges are surface-exposed.
PilX's D-Region Is Key to Its Function in Aggregation and Adhesion.
Our filament incorporation model highlights the surface-protruding D-region of PilX as potentially important in its role in aggregation and adhesion. This possibility is particularly appealing because the D-region in pilin plays a pivotal role in some Tfp-linked functions (22). We therefore created a series of substitutions and deletions within this region of PilX to test this hypothesis (Fig. 1). To complement the pilX mutant, variant pilX alleles constructed by site-directed mutagenesis were placed under the control of an IPTG-inducible promoter and integrated ectopically in the genome of the pilX mutant. We previously reported that low level expression of PilX in the pilX/pilXWT strain, in the absence of induction, restored bacterial aggregation and adhesion to human cells at levels slightly lower than those of the WT strain (17) (SI Fig. 9). In the presence of 0.1 mM IPTG, the aggregative and adhesive abilities of the pilX/pilXWT strain are higher (SI Fig. 9). The finding that in these conditions the pilX/pilXWT strain aggregates slightly more than the WT strain demonstrates that the aggregative ability of a strain correlates with PilX levels and reinforces the notion that this minor pilin is directly involved in this phenotypic trait (17).
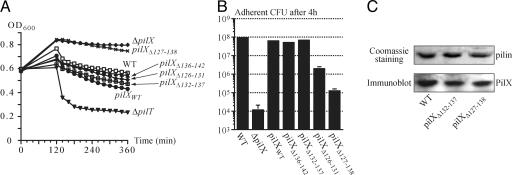
We targeted the D-region of PilX with a series of single residue substitutions and 5- to 12-aa deletions (Fig. 1). The change of three hydrophobic residues (Leu135, Leu138, Val142) into serines did not affect aggregation (data not shown), indicating that these somewhat surprisingly exposed hydrophobic residues within the hook are not critical for PilX's function. Similarly, strains pilX/pilXΔ132–137, pilX/pilXΔ136–142 and pilX/pilXΔ138–142 displayed levels of aggregation (Fig. 5A) and adhesion (Fig. 5B) comparable with those of the pilX/pilXWT and WT strains, indicating that the deleted residues are not required for PilX's function. In contrast, strains harboring deletions covering most or all of the α4 pigtail presented striking phenotypes. The pilX/pilXΔ127–138 strain in which the α4 pigtail and the neighboring region of the hook were deleted was the most affected, with abolished aggregation (Fig. 5A) and dramatically reduced adhesion to human cells (Fig. 5B). The adherence was reduced 1,000-fold when compared with the WT strain (Fig. 5B), but was 10-fold higher than for the pilX mutant (17). For pilX/pilXΔ126–131, the kinetics of aggregation were similar to those of pilX/pilXWT, but movement of the cultures led to a slight decrease in the number of aggregates as visualized by phase-contrast microscopy (data not shown), which suggested that these aggregates were more fragile. This observation may explain why the pilX/pilXΔ126–131 strain displays a 50-fold lower adhesive ability than the WT strain (Fig. 5B). Importantly, immunoblot analysis of pilus preparations confirmed that an unimpaired PilX variant such as PilXΔ132–137 and the inactive PilXΔ127–138 both copurify with Tfp in amounts comparable with the WT strain (Fig. 5C). This control rules out the possibility that the observed phenotypic defects are due to protein instability or mislocalization. Taken together, our results indicate that PilX's protruding D-region is key to its function in aggregation and adhesion.
Fig. 5.
Analysis of strains expressing variant PilX proteins (pilX/pilXvariant). (A) Quantification of aggregation by measuring the decrease in OD600 upon sedimentation of bacterial aggregates in static liquid cultures. Values are means of 2–3 independent experiments. (B) Quantification of the adhesive abilities of the various strains compared with WT and pilX strains. After a 30-min contact during which bacteria adhered to HUVEC, cells were incubated with hourly washes and the number of adherent CFU after 4 h of infection were recovered and counted. Values are the means ± standard deviations of three to four independent assays. (C) Expression and copurification with Tfp of the PilXΔ127–138 variant. Equal amounts of pilus preparations as shown by Coomassie-blue staining (Upper) were separated by SDS/PAGE. Immunoblot detection of PilX (Lower) confirms that the PilXΔ127–138 variant copurifies with Tfp as well as PilXWT and PilXΔ132–137, which displays no obvious phenotypic defects and was included as a control.
Discussion
Because of the widespread importance of Tfp in host-pathogen interactions, a better understanding of the molecular mechanisms underlying Tfp functions is of general interest. Whatever aspect of Tfp biology is to be studied, pilus retraction must be taken into account because it generates remarkable mechanical forces (25), likely promoted by domain movements within PilT (26). PilT-powered retraction is indispensable for intimate adhesion (27, 28), twitching motility (7, 8), competence for DNA transformation (29) and signaling to host cells (30), yet it must be counteracted to achieve efficient biogenesis of pilus filaments. Accordingly, almost half of the 15 proteins essential for Tfp biogenesis in N. meningitidis are required to stabilize the fibers by counterbalancing the action of PilT (31). Bacterial aggregation also is negatively affected by retraction because a N. meningitidis pilT mutant displays dramatically increased aggregation (17). This finding is readily envisioned: fibers retracting in opposite directions generate disruptive forces on the antiparallel pilus–pilus interactions that hold bacteria together within aggregates. Therefore, bacteria must have evolved mechanisms to interfere with PilT-mediated retraction to enable formation of stable bacterial aggregates, which is a crucial property for adhesion to host cells. Our previous work in N. meningitidis identified PilX, one of the proteins with a prepilin-like N-terminal signature, as a candidate for this property (17).
Our crystal structure of PilX sheds light on the molecular nature of a prepilin peptidase substrate other than pilin. Truncated PilX closely resembles type IV pilins, with a canonical α/β roll fold. A structural model of full length PilX suggests that it is an elongated ladle-shaped molecule like PilE, which fits well within the recently proposed high-resolution Tfp model (22). By analogy to PilE, it is likely that the N terminus of PilX provides the major assembly interface that is buried within the filament core, with the α1 helix axis nearly parallel to the filament long axis. Together with the confirmation that it colocalizes with Tfp, these findings experimentally validate PilX as a minor pilin. One important finding in this study is that the α1/β- and D-regions flanking the β-sheet in PilX are surface-exposed on the pilus filament like the corresponding regions in pilin subunits. This observation is of particular interest because the D-region in pilins mediates some of the diverse functions of Tfp (22). For example, the 12-residue D-region of the P. aeruginosa PAK pilin has been proposed to form the binding domain for specific receptors present on epithelial cells of the lungs and retina (20). Mutational analysis of PilX's D-region demonstrates that it is pivotal in the role of this protein in bacterial aggregation and adhesion. Deletion of the α4 pigtail and the neighboring region of the hook (PilXΔ127–138) does not affect the ability of the protein to be incorporated in Tfp but abrogates bacterial aggregation and, as a result, dramatically decreases adhesion. The α4 pigtail is particularly important for PilX function because the PilXΔ126–131 variant, despite presenting normal aggregation kinetics, forms fragile aggregates and displays impaired adhesion.
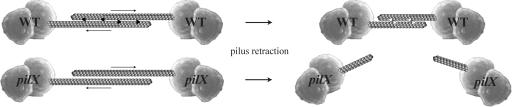
It is now possible to propose an integrated molecular model for a Tfp-mediated phenotype crucial for adhesion to host cells, i.e., the formation of bacterial aggregates (Fig. 6). When pili interact they establish electrostatic interactions (11). Anti-parallel interactions between the pili of neighboring bacteria play a role in aggregation, whereas parallel interactions between pili from one cell account for Tfp's characteristic ability to form bundles. Bundling and bacterial aggregation are nevertheless distinct properties because a N. meningitidis pilX mutant displays bundles of pili but fails to aggregate (17) and a pilT mutant in N. gonorrhoeae displays dramatically increased aggregation but no obvious change in pilus bundling (29). This latter finding is important because it indicates that whereas PilT-mediated retraction works against formation of bacterial aggregates, it does not preclude formation of bundles of pili. Pilus-pilus interactions are probably strong enough to promote pilus bundling because the fibers emanate from a single bacterium and are likely to be retracted in the same direction by PilT. In contrast, these interactions are insufficient to maintain lasting contacts between neighboring bacteria in the face of active retraction because fibers are pulled in opposite directions, hence the necessity of a protein such as PilX to counterbalance retraction. How could PilX function? We previously reported that aggregation occurs only when PilX is present in both interacting partners, which suggested that is likely to result from PilX-PilX interactions (17). Whereas alternative scenarios cannot be excluded, it is now reasonable to propose that PilX subunits within the fibers of interacting meningococci counterbalance Tfp-mediated retraction by bracing against each other through their protruding D-regions (Fig. 6). Multiple PilX-PilX contacts of this nature, occuring along the length of interacting Tfp, could prevent slippage of pili against each other, pull the bacteria closer together and thus explain the role of this minor pilin in bacterial aggregation and adhesion. The variants in which the D-region protrusion has been erased do not present an obstacle to slippage of pili. As a result, pilus-pilus electrostatic interactions are not sufficient to maintain bacterial aggregates and the pili slide past each other (Fig. 6). It is possible to envision that PilX senses and modulates the tension along filaments, thereby controlling the switch between pilus extension and retraction (32) and fine-tuning the regular ordered aggregates of Neisseria on human cells (28). In this context, the slight unraveling of α2 observed in two monomers leads us to speculate that PilX could accommodate some of the retraction force through conformational changes (Fig. 6), as the type I pilus adhesin FimH does (33). Although it remains to be tested, a similar molecular mechanism may account for aggregation in other Tfp-harboring bacteria through another minor pilin or the pilin itself.
Fig. 6.
Conceptual model for PilX's molecular role in the formation of bacterial aggregates. In WT cells, retraction of pili by PilT slides PilX molecules (black) close to one another. When interacting with another PilX, and under tension, PilX subunits may adopt a different conformation (white) and prevent further slippage of pili (Upper). In the absence of PilX, pilus retraction disrupts the interactions between cells and no aggregation is observed (Lower).
In conclusion, experimental evidence that PilX is a minor pilin suggests an enormous potential for these proteins in enhancing Tfp functional versatility without affecting the conserved structural or biochemical properties of Tfp. Moreover, because immunization with a minor component of type I pili of uropathogenic Escherichia coli can prevent infection (34), minor type IV pilins that exhibit a relatively small repertoire of sequence variants, as shown for PilX (14), may be suitable targets for the development of vaccines and drugs against N. meningitidis and other human pathogens.
Materials and Methods
Detailed methods are described in SI Materials and Methods.
Bacterial Strains and Culture Conditions.
E. coli TOP10 (Invitrogen, Carlsbad, CA) was used for cloning experiments, whereas BL21(DE3) was used for protein expression and purification. A methionine auxotroph of the latter strain was used for selenomethionine labeling as described (26, 35). To facilitate crystallization, we engineered a strain producing a PilX protein missing the first 28 residues of the mature protein. The portion of the pilX gene encoding residues 29–152 was cloned in the pMAL-p2X vector (New England Biolabs, Ipswich, MA). The resulting plasmid, pYU15, contains the malE gene, which encodes maltose-binding protein (MBP), followed by the Factor Xa protease recognition site and the truncated pilX gene, all under the control of the Ptac promoter. Because of the signal peptide of the MBP, the fusion protein is directed to the periplasm, which is desirable for disulfide bond formation.
Derivatives of the serogroup C N. meningitidis 8013 isolate were grown as described (17). Isogenic mutants and the pilX/pilXWT complemented strain were described elsewhere (17). To test their functionality, pilX alleles altered in vitro by site-directed mutagenesis were ectopically expressed in the pilX mutant. The variant alleles were generated by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
PilX Purification and Crystallization.
The fusion protein was purified on amylose resin (New England Biolabs) by using the affinity of MBP for amylose, then cleaved with Factor Xa (New England Biolabs). PilX was separated from MBP and uncut fusion on HiTrap SP Sepharose resin (Amersham Pharmacia Biotech, Pittsburgh, PA). Pooled PilX fractions were concentrated to 7 mg/ml and dialyzed against 25 mM KCl, 50 mM Hepes (pH 7.5). The final truncated PilX product contains 4 vector-derived residues (ISEF) followed by residues 29–152 of the mature PilX protein.
Crystallization conditions were found by using vapor diffusion and commercial (Nextal) or in-house sparse matrix screens. Optimized crystal form I (P43) was obtained with mother liquor consisting of 22% PEG 8000, 120 mM LiSO4, and 100 mM Bis-Tris (pH 6.5). Crystal form II (C2221) was obtained with 4 M sodium formate. To obtain selenomethionine-containing PilX crystals, 15 mg/ml purified selenomethionine-containing protein were streak-seeded with a horse hair by using previously shattered native crystals.
Data Collection and Structure Determination.
Multiwavelength anomalous dispersion (MAD) data were collected on a single vitrified form I (P43) crystal at beamline 32-ID-B at the Advanced Photon Source (APS). The data were used for structure determination with available crystallographic software as described (26, 35) (SI Materials and Methods). Radiation damage was apparent, so one monomer from this model was used as the molecular replacement search model for 2.5 Å resolution data from crystal form II (C2221) collected in-house (SI Table 1). Refinement of the C2221 structure led to a final model with R/Rfree values of 20.4/23.6% (SI Table 1). Finally, this model was used to phase 2.4 Å resolution in-house data for crystal form I. The overall geometry of both final structures is excellent, with 98% and 98.7% of residues in favored regions of the Ramachandran plot for the P43 and C2221 structures, respectively, and no Ramachandran outliers.
Aggregation and Adhesion Assays.
N. meningitidis' ability to form aggregates and to adhere to human umbilical vein endothelial cells (HUVEC) was monitored and quantified as described (17).
Pilus Purification.
Pilus preparations were obtained as described with slight modifications (17)
SDS/PAGE and Immunoblotting.
Preparation of protein samples, protein quantification, SDS/PAGE separation, transfer to membranes, and immunoblotting were done as described (17).
Localization of PilX.
To detect and localize native PilX protein, polyclonal antibodies were raised in New Zealand White rabbits (Centre LAGO, Vonnas, France) against the protein that was used for crystallization. The serum was immunopurified twice against this protein and used to detect PilX by immunogold labeling and IF microscopy as described (24) with slight modifications (SI Materials and Methods).
Supplementary Material
Acknowledgments
We acknowledge the excellent technical assistance of J. Brunzelle for x-ray data collection and M. Garfa for confocal microscopy. We thank H. Winther-Larsen for sharing the Tfp purification method. We are grateful to G. Duménil, D. Holden, and C. Tang for critical reading of the manuscript. S.H. was supported by the Université Paris 5 and the Fondation pour la Recherche Médicale. This project was funded by National Institutes of Health Grant GM5972 (to K.T.F.), the W. M. Keck Foundation (K.T.F.), and Agence Nationale de Recherche Grant JC05-44953 (to V.P.).
Abbreviations
- Tfp
type IV pili
- IPTG
β-d-thiogalactoside
- MBP
maltose-binding protein.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for PilX have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2OPD and 2OPE for the C2221 and P43 crystal forms, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707581104/DC1.
References
- 1.Mattick JS. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 2.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, Melville SB. Mol Microbiol. 2006;62:680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- 3.Rakotoarivonina H, Jubelin G, Hebraud M, Gaillard-Martinie B, Forano E, Mosoni P. Microbiology. 2002;148:1871–1880. doi: 10.1099/00221287-148-6-1871. [DOI] [PubMed] [Google Scholar]
- 4.Giltner CL, van Schaik EJ, Audette GF, Kao D, Hodges RS, Hassett DJ, Irvin RT. Mol Microbiol. 2006;59:1083–1096. doi: 10.1111/j.1365-2958.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- 5.Kirn TJ, Lafferty MJ, Sandoe CMP, Taylor RK. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 6.Marceau M, Beretti JL, Nassif X. Mol Microbiol. 1995;17:855–863. doi: 10.1111/j.1365-2958.1995.mmi_17050855.x. [DOI] [PubMed] [Google Scholar]
- 7.Skerker JM, Berg HC. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merz AJ, So M, Sheetz MP. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 9.Lory S, Strom MS. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 10.Hansen JK, Forest KT. J Mol Microbiol Biotechnol. 2006;1:192–207. doi: 10.1159/000094054. [DOI] [PubMed] [Google Scholar]
- 11.Craig L, Pique ME, Tainer JA. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 12.Koomey M. Trends Microbiol. 1995;3:409–411. doi: 10.1016/s0966-842x(00)88989-1. [DOI] [PubMed] [Google Scholar]
- 13.Winther-Larsen HC, Wolfgang M, Dunham S, van Putten JP, Dorward D, Lovold C, Aas FE, Koomey M. Mol Microbiol. 2005;56:903–917. doi: 10.1111/j.1365-2958.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- 14.Lappann M, Haagensen JA, Claus H, Vogel U, Molin S. Mol Microbiol. 2006;62:1292–1309. doi: 10.1111/j.1365-2958.2006.05448.x. [DOI] [PubMed] [Google Scholar]
- 15.Wolfgang M, van Putten JP, Hayes SF, Koomey M. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]
- 16.Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Proc Natl Acad Sci USA. 2001;98:15276–15281. doi: 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. Mol Microbiol. 2005;55:65–77. doi: 10.1111/j.1365-2958.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- 18.Nunn D. Trends Cell Biol. 1999;9:402–408. doi: 10.1016/s0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- 19.Ng SY, Chaban B, Jarrell KF. J Mol Microbiol Biotechnol. 2006;1:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 20.Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT. J Mol Biol. 2000;299:1005–1017. doi: 10.1006/jmbi.2000.3801. [DOI] [PubMed] [Google Scholar]
- 21.Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 22.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Pujol C, Eugène E, de Saint Martin L, Nassif X. Infect Immun. 1997;65:4836–4842. doi: 10.1128/iai.65.11.4836-4842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Mol Microbiol. 2005;55:54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- 25.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. Proc Natl Acad Sci USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Structure (London) 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 28.Pujol C, Eugène E, Marceau M, Nassif X. Proc Natl Acad Sci USA. 1999;96:4017–4022. doi: 10.1073/pnas.96.7.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfgang M, Lauer P, Park HS, Brossay L, Hébert J, Koomey M. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 30.Howie HL, Glogauer M, So M. PLoS Biol. 2005;3:e100. doi: 10.1371/journal.pbio.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbonnelle E, Helaine S, Nassif X, Pelicic V. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- 32.Maier B, Koomey M, Sheetz MP. Proc Natl Acad Sci USA. 2004;101:10961–10966. doi: 10.1073/pnas.0402305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 34.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, et al. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 35.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.