Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate expression of many genes. Recent studies suggest roles of miRNAs in carcinogenesis. We and others have shown that expression profiles of miRNAs are different in lung cancer vs. normal lung, although the significance of this aberrant expression is poorly understood. Among the reported down-regulated miRNAs in lung cancer, the miRNA (miR)-29 family (29a, 29b, and 29c) has intriguing complementarities to the 3′-UTRs of DNA methyltransferase (DNMT)3A and -3B (de novo methyltransferases), two key enzymes involved in DNA methylation, that are frequently up-regulated in lung cancer and associated with poor prognosis. We investigated whether miR-29s could target DNMT3A and -B and whether restoration of miR-29s could normalize aberrant patterns of methylation in non-small-cell lung cancer. Here we show that expression of miR-29s is inversely correlated to DNMT3A and -3B in lung cancer tissues, and that miR-29s directly target both DNMT3A and -3B. The enforced expression of miR-29s in lung cancer cell lines restores normal patterns of DNA methylation, induces reexpression of methylation-silenced tumor suppressor genes, such as FHIT and WWOX, and inhibits tumorigenicity in vitro and in vivo. These findings support a role of miR-29s in epigenetic normalization of NSCLC, providing a rationale for the development of miRNA-based strategies for the treatment of lung cancer.
Keywords: epigenetics, tumor-suppressor genes
Lung cancer is the leading cause of cancer mortality in the United States, with an incidence of ≈213,000 new cases per year and a very high mortality (1). Despite new drugs and therapeutic regimens, the prognosis for lung cancer patients has not significantly changed in the last 20 years. Non-small-cell lung cancer (NSCLC) accounts for ≈80% of lung cancers. Surgery remains the main therapy for NSCLC, but a large fraction of patients cannot undergo curative resection. Innovative therapeutic strategies are urgently needed for lung cancer treatment. Hypermethylation is responsible for the silencing of tumor suppressor genes (TSGs) involved in lung carcinogenesis, such as CDKN2A (2), CDH13 (2), FHIT (3), WWOX (3, 4), CDH1 (5), and RASSF1A (5). Specific alterations in DNA methylation patterns are hallmarks of human diseases and therefore could represent specific targets for treatment (6, 7). Methylation changes to the epigenome are controlled by DNA methyltransferases (DNMTs), which catalyze the transfer of a methyl group from the methyl donor S-adenosyl methionine onto the 5′ position on the cytosine ring. Three catalytically active DNMTs have been identified in mammals, Dnmt1, Dnmt3A, and Dnmt3B (8). Dnmt3A and -3B proteins can introduce methyl groups onto CG sites that were unmethylated on the parental template strands of DNA (9). All DNMTs possess de novo methylation activity, but Dnmt1 is inefficient in de novo methylation. Once a DNA methylation pattern has been established, it is maintained during DNA replication by the maintenance Dnmt1 (10). The levels of DNMT1, DNMT3A, and DNMT3B mRNA are reportedly elevated in various malignancies, including hepatomas, prostate, colorectal, and breast tumors (11–14). More recently, the mRNA levels of DNMT1 and DNMT3B have been found to be elevated in 53 and 58% of 102 NSCLCs, respectively (15), with DNMT1 levels independently correlated with prognosis in NSCLC patients (15). Dnmt1, -3A, and -3B protein expression, assessed by immunohistochemical analyses, also have been reportedly highly expressed coordinately in lung tumors of smokers (16). In lung squamous cell carcinomas, elevated Dnmt1 expression has been shown to predict a poorer prognosis, and elevated expression of both Dnmt1 and Dnmt3B have been shown to be correlated with hypermethylation of TSG promoters (16). A specific polymorphism in the DNMT3B promoter, which significantly increases promoter activity, has been associated with an increased risk of lung cancer in a hospital-based, case-control study of 659 patients (17). The inhibition of Dnmt1-mediated DNA methylation and histone deacetylation reduced tobacco carcinogen-induced lung cancer in mice by more than 50% (18).
MicroRNAs (miRNAs) represent a class of naturally occurring small noncoding RNA molecules, distinct from but related to siRNAs. Mature miRNAs are 19- to 25-nucleotide-long molecules cleaved from 70- to 100-nucleotide hairpin pre-miRNA precursors (19). In animals, single-stranded miRNAs bind, through partial sequence homology, to the 3′-UTR of target mRNAs and cause a block of translation or, less frequently, mRNA degradation (19). Deviations from normal miRNA expression patterns play roles in human diseases, including cancer, as reviewed elsewhere (20–23). It has been demonstrated that expression of miRNA (miR)-29a, -29b, and -29c is down-regulated in NSCLCs (24, 25). In this study, we show that expression of miR-29s is inversely correlated to DNMT3A and -3B expression in lung cancer tissues and that miR-29s directly target both DNMT3A and -3B. The enforced expression of miR-29s in lung cancer cell lines restores normal patterns of DNA methylation, induces reexpression of methylation-silenced TSGs, such as FHIT and WWOX, and inhibits tumorigenicity both in vitro and in vivo.
Results and Discussion
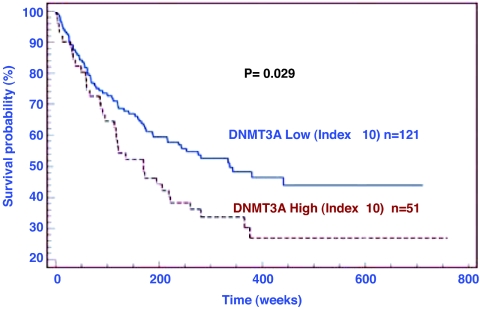
To confirm the relevance of expression of DNMTs in lung cancer, we assessed the expression of Dnmt1, -3A, and -3B proteins by immunohistochemical analyses in 172 matched nonneoplastic/cancerous lung tissues for which clinical features and time of survival were available; as shown in Fig. 1, high-level expression of Dnmt3A protein was significantly associated with lower overall survival (P = 0.029). No statistically significant correlation with survival was observed for Dnmt1 or Dnmt3B in this patient population.
Fig. 1.
Dnmt3A protein expression level in NSCLCs is inversely associated with overall survival. Kaplan–Meier curve showing survival of 172 NSCLC patients with different levels of Dnmt3A expression in tumors relative to adjacent normal lung. Patients with higher expression of Dnmt3A had shorter overall survival (P = 0.029).
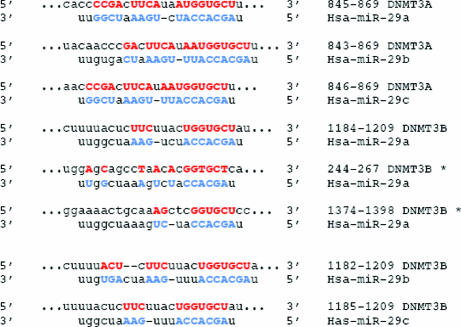
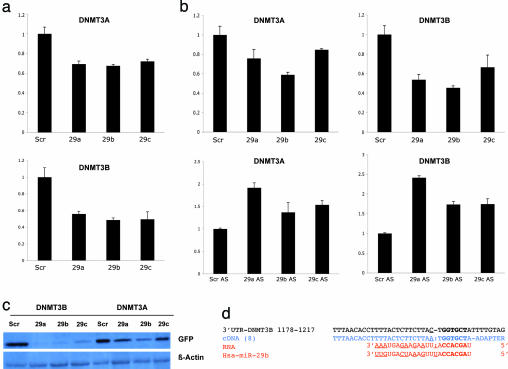
To seek a link between altered miRNA expression profiles and aberrant cancer genome methylation patterns, we chose to study miRNAs that might target DNMT3A and -3B in NSCLC-derived cells and in cancer tissues. We focused on the miR-29 family because miR-29s have been shown previously to be down-regulated in NSCLC (24, 25), and these miRNAs have intriguing complementarity to sites in the 3′-UTRs of DNMT3A and -3B genes, as predicted by several in silico methods for target gene prediction [PicTar (26), TargetScan3.1 (27), MiRanda (28), and miRGen (29)] (Fig. 2). To validate these miRNA–target interactions, the DNMT3A and -B complementary sites were cloned into the 3′-UTR of the firefly luciferase gene and cotransfected with miR-29a, -b, or -c in A459 (NSCLC) cells. As shown in Fig. 3a, all three miRNAs significantly reduced the luciferase activity with respect to the scrambled oligonucleotide. To assess whether ectopic expression of individual miR-29 sequences induces down-regulation of endogenous DNMT3A and -3B mRNA levels, we performed quantitative RT-PCR (qRT-PCR) for RNA from A549 and H1299 cells transfected with scrambled RNA or with miR-29a, -29b, or -29c. Results are shown in Fig. 3b, Upper; similar results were observed for H1299 cells (data not shown). Overexpression of individual miR-29s induced marked reduction of DNMT3A and -3B mRNA levels. On the other hand, silencing of miR-29s with antisense molecules induced up-regulation of DNMT3A and -3B mRNA levels (Fig. 3b, Lower). To demonstrate that overexpression of miR-29s results in reduction of Dnmt3A and -3B at a protein level, we used a GFP reporter vector, QBI–GFP25. Briefly, we cloned the 3′-UTRs of DNMT3A and -B downstream of the GFP-encoding sequence of the QBI–GFP25 vector, allowing expression of a fusion GFP protein containing the 3′-UTR of DNMT3A or -B. A549 cells were cotransfected with the GFP-3A/3B–3′-UTR vector plus miR-29a, -29b, -29c, or scrambled oligonucleotide. Marked reduction in GFP protein expression was observed in cells transfected with miR-29s (Fig. 3c), especially GFP-3B–3′-UTR protein; the protein expression results were in accordance with the qRT-PCR results shown in Fig. 3b, in which endogenous DNMT3B mRNA was more significantly reduced by expression of miR-29s, possibly due to more predicted match “seeds” between miR-29s and the 3B 3′-UTR (three for 29a, one for 29b, one for 29c) than the 3A 3′-UTR (one for each miR-29); also the three different predicted sites for miR-29a differ from -29b/c matches by only one base, possibly allowing partial interaction of the 3B 3′-UTR miR-29a sites with miR-29b and -29c. Thus, the transfection with any member of the miR-29 family may result in more robust silencing of DNMT3B than DNMT3A, according to the “coordinate principle” that miRNAs may act cooperatively through multiple target sites in one gene (28, 30). To show a direct, functional interaction of the DNMT3B 3′-UTR with miR-29b, we used a recently described detection method (31) by which we showed that the endogenous miR-29b, at the PicTar-predicted site of interaction with 3′-UTR (26), was able to function as a “natural” primer to initiate the retrotranscription of DNMT3B mRNA (Fig. 3d).
Fig. 2.
Complementarity sites for miR-29s in the 3′-UTR region of DNMT3A and -3B. The capital and bold letters identify perfect base matches, according to the TARGETSCAN 3.1 software. The PicTar software identifies two additional match regions between miR-29a and DNMT3B, indicated with an asterisk.
Fig. 3.
MiR-29s directly target DNMT3A and -B. (a) Results of the luciferase assay for DNMT3s expression after transfection with miR-29s in A549 cells. (b) (Upper) Assessment of expression of DNMT3A and DNMT3B mRNAs by qRT-PCR, after transfection of A549 cells with miR-29s or a negative control. (Lower) Silencing of miR-29s with antisense molecules (AS) induces increased expression of DNMT3A and DNMT3B mRNA. (c) Western blot of proteins extracted from A549 cells that were cotransfected with the GFP repression vectors for the DNMT3A and -B−3′-UTR plus miR-29s or scrambled (Scr) oligonucleotides. (d) miR-29b acts as an endogenous primer to retrotranscribe its predicted DNMT3B mRNA target. Black, DNMT3B cDNA (GenBank accession no. NM_175848); blue, cloned and sequenced cDNAs experimentally obtained (eight clones analyzed); red, deduced RNA sequences and corresponding miR-29b. The upper underlined black and blue nucleotides have no homology between target and experimental cDNAs. The lower underlined red nucleotides represent RNA sequence complementary to cDNAs that lack homology to the miR-29b sequence. Nucleotides in bold represent the PicTar-predicted match site.
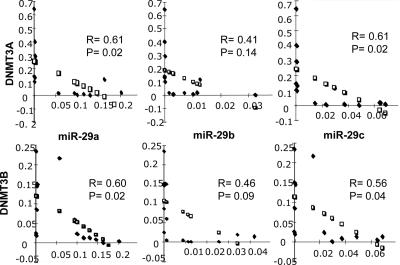
Next we asked whether DNMT3A and -B mRNA expression was inversely correlated with levels of miR-29s in NSCLC tissues; 14 NSCLCs were analyzed for expression levels of DNMT3A and -B mRNAs and for miR-29a, -29b, and -29c expression by qRT-PCR (32). A statistically significant inverse correlation (Fig. 4) was observed between DNMT3A mRNA and miR-29a (P = 0.02) and miR-29c (P = 0.02). A similar inverse correlation was observed for DNMT3B mRNA levels and miR-29a (P = 0.02) and miR-29c (P = 0.04). Although there was a trend toward inverse correlation of DNMT3A and -3B mRNA levels with miR-29b level, the association was not statistically significant; this may be due either to the small number of patients we analyzed or to the fact that whereas miR-29a and -29c are transcribed from only one chromosomal location, on chromosome 7 and 1 respectively, mature miR-29b is transcribed from two different primary transcripts on different chromosomes, the miR-29b-1/miR-29a cluster on 7q32.3 and the miR-29b-2/miR-29c cluster on 1q32.2. The probe used by qRT-PCR to determine the mature product of miR-29b is unable to distinguish between the 29b-1 or 29b-2 gene products.
Fig. 4.
Correlation of endogenous miR-29 levels with DNMT3A/B mRNA levels. Inverse correlation between endogenous mRNA levels of DNMT3A and DNMT3B and endogenous levels of miR-29s determined by qRT-PCR in 14 NSCLCs. R, regression coefficient; □, regression line; ♦, actual sample correlations.
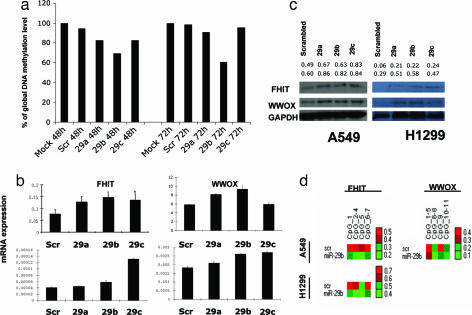
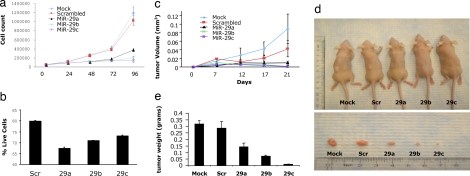
The observation that miR-29s target DNMT3A and -B suggested that expression of these miRNAs contributes to the DNA epigenetic modifications in cancer. To address this issue, we transfected A549 cells with miR-29a, miR-29b, miR-29c, or scrambled oligonucleotides and analyzed global DNA methylation 48 and 72 h later by using an liquid chromatography–tandem MS method (33). As shown in Fig. 5a, all three miR-29s reduced global DNA methylation with respect to the control. The effect was most robust for miR-29b, with a reduction of 30% after 48 h and 40% after 72 h. The percentage of global methylation reduction observed in cells treated with miR-29b is comparable with that observed with Dnmt1 inhibitors, such as decitabine (33), and is partial with either approach. We hypothesize that a more robust global DNA hypomethylation could be achieved combining decitabine (or other nucleoside analogs) with miR-29s, therefore blocking both de novo and maintenance DNMT pathways. To characterize the effects of the methylation changes on gene expression, we analyzed the mRNA expression levels of two TSGs, FHIT and WWOX, which are frequently silenced by promoter methylation in lung cancer (3). As shown in Fig. 5b, Upper, 48 h after transfection of A549 cells, FHIT expression was increased by miR-29a, -29b, and -29c expression by ≈65, 89, and 74%, respectively, and the WWOX mRNA level was increased by ≈40 and 60% by miR-29a and -29b, respectively; a similar trend was observed in H1299 cells (Fig. 5b, Lower). Increased expression of both Fhit and Wwox proteins was also observed in both cell lines (Fig. 5c). To determine whether miR-29s regulated the expression of FHIT and WWOX by altering promoter methylation of these genes, we examined the methylation status of the regulatory region of FHIT and WWOX by using the MassARRAY system (34) (quantitative high-throughput DNA methylation analysis) in A549 and H1299 cells transfected with miR-29b. Two bisulfite reactions (one for each gene CpG island) were designed, which covered 7 CpGs and 11 CpGs for FHIT and WWOX, respectively. In miR-29b-transfected H1299 and A549 cells, the MassARRAY analysis for FHIT showed an average reduction of 19.1 and 54.3% methylation, respectively, whereas for WWOX in H1299 showed an average reduction of 32.1% compared with the scrambled oligonucleotide (Fig. 5d). Finally, we assessed the effects of reexpression of miR-29s on tumorigenicity of A549 cells. The ectopic expression of miR-29s in A549 inhibited in vitro cell growth (Fig. 6a) and induced apoptosis with respect to the scrambled control transfection (Fig. 6b). The inhibitory effect of miR-29s on A549 tumorigenicity was also observed in vivo. Transfection with miR-29s inhibited the growth of A549 engrafted tumors with respect to mock and scrambled oligo transfected cells (Fig. 6 c–e), thus illustrating a likely antineoplastic effect of these miRNAs. Given the high number of mRNAs targeted by miR-29s, which includes well known oncogenes such as TCL1 (35) and MCL1 (36), the reactivation of TSGs by demethylation could represent another mechanism through which miR-29s exert their oncosuppressor function. The present study did not address the relative contribution of these two mechanisms to induce an antitumoral effect but confirmed a role of miR-29s as oncosuppressor genes both in vitro and in vivo.
Fig. 5.
Effect of restoration of miR-29s on the cancer cell epigenome. (a) Global DNA methylation changes induced by miR-29s on A549 cells harvested 48 and 72 h after transfection. The results are compared with nontransfected (mock) cells and cells transfected with a scrambled oligonucleotide. (b) Determination of FHIT and WWOX mRNA levels in A549 and H1299 cells 48 h after transfection with miR-29s or a negative control by qRT PCR; miR-29s induced reexpression of FHIT and WWOX mRNAs. (c) Immunoblot of Fhit and Wwox proteins in A549 and H1299 cells 72 h after transfection with miR-29s or negative control; by 72 h, miR-29s induce increased expression of Fhit and Wwox proteins. The numbers above the immunoblot images represent the intensity of the bands relative to the GAPDH gene (upper row, Fhit; lower row, Wwox). (d) Graphical representation of the quantitative DNA methylation data for FHIT and WWOX promoter region by using the MassARRAY system. Each square represents a single CpG or a group of CpGs analyzed, and each arrow represents a sample. Methylation frequencies are displayed for each experiment in a color code that extends from light green (lower methylation frequencies) to bright red (higher methylation frequencies).
Fig. 6.
Effects of miR-29s on tumorigenicity of A549 cells. (a) Growth curve of A549 cells transfected in vitro with miR-29s or scrambled oligonucleotide or mock-transfected. (b) Percentages of live cells were measured in A549 cells transfected with scrambled oligonucleotide or with miR-29s oligonucleotides (100 nM final concentration). (c) Growth curve of engrafted tumors in nude mice injected with A549 cells pretransfected (48 h before injection) with miR-29s, scrambled oligonucleotides, or mock transfected. (d) Comparison of tumor engraftment sizes of mock-, scrambled-, and miR-29s-transfected A549 cells 21 days after injection in nude mice. The images show average-sized tumors from among five of each category. (e) Tumor weights ± SD in nude mice.
In summary, this study has shown that expression of miR-29 family members is inversely correlated with DNMT3A and -3B expression in lung cancers and that these miRNAs down-modulate expression levels of both enzymes. Furthermore, enforced expression of these miRNAs in lung cancer cells leads to reduced global DNA methylation, restores expression of TSGs, and inhibits tumorigenicity both in vitro and in vivo. Results of this study provide a strong rationale for developing epigenetic therapies that use synthetic miR-29s, alone or in combination with other treatments, to reactivate tumor suppressors and normalize aberrant patterns of methylation in lung cancer. Because loss of expression of miR-29 family members is observed in other common human malignancies, this approach may be extended to the treatment of other human malignancies.
Materials and Methods
Tissues.
We obtained 172 lung cancer samples, including squamous cell, adeno-, large-cell, and neuroendocrine large-cell carcinomas, collectively referred to as NSCLCs, from the Pathology Core Facility at the Ohio State University to perform tissue microarrays (TMAs) for DNMTs expression. Clinical features (histological diagnosis, sex, age, tumor node metastasis status, and survival time) were available for these patients.
Primary lung cancer tissues (eight squamous carcinomas and six adenocarcinomas) for performing qRT-PCR analysis were purchased from the Cooperative Human Tissue Network–Midwestern Division (Columbus, OH). Total RNAs were isolated by TRIzol (Invitrogen, Carlsbad, CA) extraction, according to the instructions of the manufacturer.
TMAs.
Each array contained four samples of each lung cancer, along with multiple appropriate lung and other normal tissue spots. The TMAs, usually two for each antiserum, were stained with antisera against Dnmt1, Dnmt3A, and Dnmt3B proteins, and expression of each of these enzymes in lung cancer was compared with clinical features to seek significant correlations. Dnmt1, Dnmt3A, and Dnmt3B protein expression was assessed on the lung cancer TMAs by using Dnmt1 antiserum (GTX13537; GeneTex, San Antonio, TX) at a dilution of 1:150, Dnmt3A antiserum (ab-4897; Novus Biologicals, Littleton, CO) at a dilution of 1:25, and Dnmt3B antiserum (AP1035a; Abgent, San Diego, CA) at a dilution of 1:32. Sections (4 μm) from TMA blocks were placed on positively charged slides, placed in a 60°C oven for 1 h, cooled, deparaffinized, and rehydrated through xylene and graded ethanol solutions to water. Slides were quenched for 5 min in 3% hydrogen peroxide to block endogenous peroxidase. Antigens were retrieved in TRS (Dako, Carpinteria, CA) solution at 95°C for 25 min. Slides were exposed to primary antisera for 1 h at room temperature and to secondary antisera (1:200) for 20 min at room temperature; secondary antisera were goat anti-mouse for Dnmt1 and goat anti-rabbit for Dnmt3A and -3B. All slides were blocked for endogenous biotin before application of the biotinylated secondary antisera. Chromogen detection was with a Vectastain Elite (catalog no. PK-6100; Vector Laboratories, Burlingame, CA) for 30 min. The substrate chromogen was DAB+ (catalog no. K3468; Dako). Slides were counterstained with hematoxylin, dehydrated through graded ethanol solutions, and coverslipped.
TMAs were read and scored by a pathologist who was blinded to clinical features; expression scores were determined by multiplying the percentage of positive cells in an individual sample by the intensity of staining; the intensity of staining was assessed on a scale from 1 to 3, where 1 was the least intense staining and 3 was the most intense. For example, a sample with 10% positive cells with intensity 3 was assigned a score of 30, the same score as a sample with 30% positive cells with intensity 1.
qRT-PCR.
qRT-PCR analysis for miRNAs was performed in triplicate with the TaqMan MicroRNA assays kit (Applied Biosystems, Foster City, CA) according to the instructions of the manufacturer, and 18S RNA was used for normalization; qRT-PCR analyses for other genes of interest were performed as described previously (32). RNA was reverse transcribed to cDNA with gene-specific primers and IQ SYBR Green Supermix (Bio-Rad, Hercules, CA). GAPDH served as normalization control. For the silencing of miR-29s, A549 and H1299 cells were transfected in six-well plates by using Lipofectamine 2000 reagent (Invitrogen) according to the protocol of the manufacturer, with 100 nM (final) antisense miR-29a, -29b-1, -29c, or scrambled antisense miR (Fidelity Systems, Gaithersburg, MD).
Cell Culture.
A549 and H1299 lung cancer cells from the American Type Culture Collection (Manassas, VA) were maintained in RPMI medium 1640 with 10% FBS and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin).
Luciferase Reporter Assay for Targeting DNMT 3′-UTRs.
For luciferase reporter experiments, a DNMT3A 3′-UTR segment of 979 bp and a DNMT3B 3′-UTR segment of 978 bp were amplified by PCR from human genomic DNA and inserted into the pGL3–control vector with simian virus 40 promoter (Promega, Madison, WI) by using the XbaI site immediately downstream from the stop codon of luciferase. The following sets of primers were used to generate specific fragments: DNMT3A-UTR forward, 5′-GCTCTAGACGAAAAGGGTTGGACATCAT-3′; DNMT3A-UTR reverse, 5′-GCTCTAGAGCCGAGGGAGTCTCCTTTTA-3′; DNMT3B-UTR forward: 5′-GCTCTAGATAGGTAGCAACGTGGCTTTT-3′; DNMT3B-UTR reverse, 5′-GCTCTAGAGCCCCACAAAACTTGTCAAC-3′. Underlined sequences indicate the endonuclease restriction site.
The amplified 3′-UTR of DNMT3A contains an XbaI restriction site in position 583; therefore, we cloned separately the upstream 3′-UTR (DNMT3A 3′-UTR up fragment, 583 bp) and the downstream fragment (DNMT3A 3′-UTR down fragment, 396 bp) into the pGL3 vectors. The predicted match seed of miR-29s is located in the DNMT3A 3′-UTR down fragment, which was used to perform the luciferase assay.
A549 cells were cotransfected in 12-well plates by using Lipofectamine 2000 reagent (Invitrogen) according to the protocol of the manufacturer, with 0.4 μg of the firefly luciferase report vector and 0.08 μg of the control vector containing Renilla luciferase pRL–TK vector (Promega). For each well, 100 nM (final) precursor miR-29a, -29b-1, -29c, or scrambled miRNA (Ambion, Austin, TX) was used. Firefly and Renilla luciferase activities were measured consecutively by using dual-luciferase assays (Promega) 24 h after the transfection. The experiments were performed in triplicate.
GFP Repression Constructs to Assess Effect of DNMT 3′-UTRs on Protein Expression.
For GFP repression, a DNMT3A 3′-UTR segment of 1472 bp and a DNMT3B 3′-UTR segment of 1566 bp (corresponding to the entire length of the 3′-UTRs) were amplified by PCR from human genomic DNA and inserted into the AFP pQBi25F vector (Qbiogene, Irvine, CA) by using the BamHI–Hind III cloning sites located 3′ of the GFP encoding sequence of the vector (which has no stop codon at the end of the GFP coding sequence). The following primer sets were used to generate specific fragments: DNMT3A-GFP forward, 5′-CGGGATCCGCAGGATAGCCAAGTTCAGC-3′; DNMT3A-GFP reverse, 5′-CCCAAGCTTAAGTGAGAAACTGGGCCTGA-3′; DNMT3B-GFP forward, 5′-CGGGATCCCTCGATCAAACAGGGGAAAA-3′; DNMT3B-GFP reverse: 5′-CCCAAGCTTGTTACGTCGTGGCTCCAGTT-3′. Underlined sequences indicate the endonuclease restriction site.
A549 cells were cotransfected in 12-well plates by using Lipofectamine 2000 reagent (Invitrogen) according to the protocol of the manufacturer, with 2 μg of the GFP repression vector containing the 3′-UTR of DNMT3A (QBI–GFP25–DNMT3A) or the 3′-UTR of DNMT3B (QBI–GFP25–DNMT3B) and with 100 nM (final) precursor miR-29a, -29b-1, -29c, or scrambled oligonucleotide (Ambion). As an additional control, a group of cells was also transfected with the GFP vector (no miRNA). Cells were harvested after 24 h. Protein extraction and immunoblot analysis were performed as described previously (4). The following primary antiserum was used: rabbit polyclonal anti-GFP (1:1,000; Novus Biologicals, Littleton, CO).
Detection of miR-29b–DNMT3B RNA Complexes.
To detect miR-29b–DNMT3B RNA complexes, we used the method described by Vatolin et al. (31) to determine whether endogenous miR-29b was able to serve as primer for retrotranscription of DNMT3B mRNA in A549 cells. The cDNAs were cloned in pCR2.1–TOPO vector (Invitrogen). The following sets of primers and adapter sequence were used (GSP indicates gene-specific primer): GSP–DNMT3B, 5′-GAGATGACAGGGAAAACTGC-3′; GSP–DNMT3B 5N, 5′-ACAGGGAAAACTGCAAAGCT-3′; adapter, 5′-CGACTGGAGCACGAGGACACTGACATGGACTGAAGGAGTAGAAA-3′; adapter 5N, 5′-CTGAAGGAGTAGAAA-3′. Primers designated as 5N represent nested primers from the adapter and GSP sequence used to sensitize the detection of the PCR bands.
Global Methylation Studies.
The global methylation status of A549 cells after transfection with scrambled miRNA and with miR-29s was determined as described previously (33). For this assay, 2 × 106 A549 cells were transfected as described above for the luciferase assay and collected 48 and 72 h later.
Quantitative DNA Methylation.
Quantitative DNA methylation analysis of the regulatory regions of FHIT and WWOX was performed by using the EpiTYPER methylation analysis assay (Sequenom, San Diego, CA). Two bisulfite reactions (one for each gene CpG island) were designed, which covered 7 CpGs and 11 CpGs for FHIT and WWOX, respectively. The DNA of scrambled- or miR-29b-transfected A549/H1299 was extracted 48 h after the transfection and 1 μg of DNA was bisulfite treated, in vitro transcribed, cleaved by Rnase A, and subjected to MALDI-TOF MS analysis to determine methylation patterns, as described previously (34). The following primers were used to amplify the regulatory regions of the FHIT and WWOX genes: FHIT forward, 5′-GGGGAGGTAAGTTTAAGTGGAATATTGTT-3′; FHIT reverse, 5′-CACCCCCAAAACCAAAAACTATAAC-3′; WWOX forward, 5′-TTGAAAGAAAGTTTTTTAAAATTAGGAAAT-3′; WWOX reverse, 5′-TCAAAAAAACAAAACCTAAAAAAAA-3′.
The heat map in Fig. 5d was created by using Heatmap builder version 1.0 by Stanford University.
Western Blot Analysis for Fhit and Wwox Proteins.
Protein extraction and immunoblot analysis were performed as described previously (4). The following primary antisera were used: rabbit polyclonal anti-Fhit (1:1000; Zymed, San Francisco, CA); mouse monoclonal anti-Wwox (1:500), as described in ref. 4. Quantification of Fhit, Wwox, and Gapdh signals was performed by densitometry with a Personal Densitometer SI (Molecular Dynamics, Sunnyvale, CA) and IMAGEQUANT 5.2 software (Image Products International, Chantilly, VA).
Cell Growth Curve.
A549 cells (5 × 104) were plated in 6× multiwell plates and transfected, after 24 h, with scrambled oligonucleotides or miR-29s oligonucleotides from Ambion at a final concentration of 100 nM, with Lipofectamine 2000 (Invitrogen), according to the protocol of the manufacturer. Nontransfected (mock) cells were included as a control. Cells were harvested and counted at 24-h intervals by using a ViCell counter (Beckman Coulter, Fullerton, CA). Each sample was run in triplicate.
Apoptosis and Flow Cytometric Studies.
A549 cells (2 × 105) were transfected with scrambled oligonucleotides or miR-29 oligonucleotides (Ambion) at a final concentration of 100 nM, with Lipofectamine 2000 (Invitrogen), according to the protocol of the manufacturer. After 24 h, cells were resuspended in binding buffer containing annexin V–FITC and propidium iodide according to the instructions of the supplier (BD Biosciences, San Diego, CA) and assessed by flow cytometry by using a model EPICS XL cytometer (Beckman Coulter). Each sample was run in triplicate.
In Vivo Studies.
Animal studies were performed according to institutional guidelines. A549 cells were transfected in vitro with 100 nM (final concentration) scrambled oligonucleotides or miR-29a, -29b, or -29c or were mock transfected by using Lipofectamine 2000 reagent (Invitrogen), according to the protocol of the manufacturer. At 48 h after transfection, 3 × 106 viable cells were injected s.c. into the left flanks of 6-wk-old female nude mice (Charles River Breeding Laboratories, Wilmington, MA), five mice per group. Tumor diameters were measured after 7 days from injection and then every 5 days. At 21 days after injection, mice were killed and tumors were weighted after necropsy. Tumor volumes were determined by using the equation V (in mm3) = A × B2/2, where A is the largest diameter and B is the perpendicular diameter.
Statistical Analysis.
P values were two sided and obtained by using the SPSS 10.0 software package (SPSS, Chicago, IL). Overall, survival was calculated from the time of diagnosis until the date of last follow up. Data were censored for patients who were alive at the time of last follow up. To perform the survival analysis and generate a Kaplan–Meier plot, Dnmt1, Dnmt3A, and -B levels measured by immunohistochemical staining were converted to discrete variables by splitting the samples into two classes [high and low expression, according to the DNMT score <10 (low) or >10 (high)]. Survival curves were obtained for each group and compared by using the log-rank test. To assess correlation between miRNA expression and DNMT expression, we used Pearson correlation and linear regression analysis (SPSS package; SPSS). These functions examine each pair of measurements (one from the miRNA and the other from DNMTs) to determine whether the two variables tend to move together or in the opposite direction, that is, whether the larger values from the miRNA (high expression) are associated with the lower values from DNMT expression.
Acknowledgments
This study was supported by a National Cancer Institute grant (to C.M.C.).
Abbreviations
- DNMT
DNA methyltransferase
- miRNA
microRNA
- miR
miRNA
- NSCLC
non-small-cell lung cancer
- qRT-PCR
quantitative RT-PCR
- TMA
tissue microarray
- TSG
tumor suppressor gene.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, Rosetti M, Mengozzi M, Amadori D. J Cell Physiol. 2006;206:611–615. doi: 10.1002/jcp.20503. [DOI] [PubMed] [Google Scholar]
- 3.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 4.Fabbri M, Iliopoulos D, Trapasso F, Aqeilan RI, Cimmino A, Zanesi N, Yendamuri S, Han SY, Amadori D, Huebner K, et al. Proc Natl Acad Sci USA. 2005;102:15611–15616. doi: 10.1073/pnas.0505485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang JC, Jones PA. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 8.Jeltsch A. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Okano M, Bell DW, Haber DA, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Pradhan S, Talbot D, Sha M, Benner J, Hornstra L, Li E, Jaenisch R, Roberts RJ. Nucleic Acids Res. 2003;25:4666–4673. doi: 10.1093/nar/25.22.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girault I, Tozlu S, Lidereau R, Bieche I. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 12.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S. Int J Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 13.Patra SK, Patra A, Zhao H, Dahiya R. Mol Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 14.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 15.Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Cancer. 2006;107:1042–1049. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- 16.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. Cancer Res. 2002;62:4992–4995. [PubMed] [Google Scholar]
- 18.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 19.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Croce CM. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher A, Slack FJ. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 22.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Pasquinelli AE, Hunter S, Bracht J. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 27.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 28.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. Nucleic Acids Res. 2006;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Vatolin S, Navaratne K, Weil RJ. J Mol Biol. 2006;358:983–996. doi: 10.1016/j.jmb.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Liu S, Xie Z, Blum W, Perrotti D, Paschka P, Klisovic R, Byrd J, Chan KK, Marcucci G. Nucleic Acids Res. 2007;35:e31. doi: 10.1093/nar/gkl1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Proc Natl Acad Sci USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, et al. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 36.Mott JL, Kobayashi S, Bronk SF, Gores GJ. Oncogene. 2007 Apr 2; doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]