Abstract
As a tool to explore proinsulin (PI) trafficking, a human PI cDNA has been constructed with GFP fused within the C peptide. In regulated secretory cells containing appropriate prohormone convertases, the hProCpepGFP construct undergoes endoproteolytic processing to CpepGFP and native human insulin, which are specifically detected and cosecreted in parallel with endogenous insulin. Expression of C(A7)Y mutant PI results in autosomal dominant diabetes in Akita mice. We directly identify the misfolded PI in Akita islets and also show that C(A7)Y mutant PI, either in the context of the hProCpepGFP chimera or not, engages directly in protein complexes with nonmutant PI, impairing the trafficking and recovery of nonmutant PI. This trapping mechanism decreases insulin production in β cells. Thereafter we observe a loss of β cell viability. The data imply that PI misfolding leading to impaired endoplasmic reticulum exit of nonmutant PI may be a key early step in a chain reaction of β cell dysfunction and demise leading to onset and progression of diabetes.
Keywords: diabetes mellitus, endoplasmic reticulum storage disease, insulin secretion, proinsulin disulfide isomers
Pancreatic β cell failure is increasingly recognized as central to progression of diabetes mellitus (DM). Different potential β cell insults are implicated in the onset of β cell stress, dysfunction, and death. Of these, a case can be made for proinsulin (PI) misfolding with resultant endoplasmic reticulum (ER) stress as a proximal event in the molecular pathogenesis of DM. In the PI superfamily, a biophysical basis for misfolding both in vitro (1) and in vivo (2) involves predisposition to disulfide mispairing. This may account for susceptibility to ER stress in pancreatic β cells (3) compared with other cell types. Even when the ER stress response is fully activated (4) and ER-associated protein degradation is up-regulated to clear misfolded PI (5), β cells still remain highly susceptible to proteotoxicity (6).
The Akita mouse expresses wild-type PI from three alleles (two Ins1 and one Ins2), which is more than sufficient to avoid DM (7). Nevertheless, DM in male Akita mice results from pancreatic insulin deficiency (8, 9) with loss of β cell mass within weeks postnatally (10). The Akita gene encodes a mouse PI-C(A7)Y mutant, where A7 refers to the seventh residue of the A chain that normally engages in a crucial disulfide bond (11). One recent study reported no difference in extent of PI misfolding in Akita versus normal mouse islets (12), underscoring the technical difficulty in identifying mutant PI in the presence of large quantities of wild-type PI. We recently reported that nonreducing Tris·tricine·urea· SDS/PAGE can identify abnormal disulfide pairing when recombinant C(A7)Y mutant PI was expressed in 293 cells (2). The ability to “see” misfolded PI in live β cells would be an important advance (13), yet this loss of function would not necessarily address the decrease of insulin production derived from coexpressed nonmutant PI.
To help clarify this, we have developed a strategy that incorporates the C(A7)Y mutation into a PI fusion protein bearing the GFP. Such a strategy requires that the nonmutant version of the chimera serves as a suitable model for the endogenous protein. The concept of a GFP reporter fused within the C peptide GFP allows for unique immunoreactivity with anti-GFP while allowing favorable efficiency of intracellular transport of green PI to secretory granules (14). From this concept, we have developed a GFP chimera of human PI designed to serve as a template for introducing the C(A7)Y mutation. We establish physical interactions of the mutant PI with coexisting wild-type PI, retaining the latter in the ER.
Results
Chimeras with GFP as Models for PI Folding and Trafficking.
We wished to design a GFP chimera to serve as a faithful PI reporter, i.e., leading to insulin production. When positioned N-terminally to the PI B chain, the chimera is retained within the ER (15). We therefore compared a chimeric construct containing GFP at the C terminus of human PI (16) to that with GFP fused within the C peptide [supporting information (SI) Data Set 1]. In transfected 293T cells (a robust protein expression system), a portion of the human PI–GFP [i.e., C-terminal GFP (16)] was unexpectedly cleaved to liberate a GFP-immunoprecipitable band comigrating with authentic cytosolic GFP (SI Fig. 7, lanes 2 and 3) as well as an insulin-immunoprecipitable band comigrating with authentic PI (lanes 16 and 17). No established proprotein processing site exists at the junction of PI and the C-terminal GFP, rendering this construct unsuitable as a reporter of PI within the ER. By contrast, in 293T cells hProCpepGFP expression released neither free GFP (SI Fig. 7, lane 1) nor free PI (lanes 14 and 15), although the chimera was well recognized both by anti-insulin (lane 8) and anti-GFP (lane 9).
We next checked whether transport to secretory granules could endoproteolytically liberate CpepGFP and insulin. An initial experiment was performed in transfected AtT20/PC2 cells (regulated secretory cells containing prohormone convertases 1 and 2), selected because they lack any endogenous (pro)insulin expression. Upon expression of hProCpepGFP and metabolic labeling, immunoprecipitation with anti-GFP revealed an initial translation product corresponding to hProCpepGFP (SI Fig. 8). At 2 h after synthesis, ongoing processing could be detected as smaller GFP-immunoprecipitable forms (described further below), each of which could be secreted. Likewise, upon immunoprecipitation with anti-insulin, a band precisely comigrating with authentic insulin, indicating native disulfide bonds (17), was specifically produced within 2 h of chase (SI Fig. 8), despite that only chimera and not PI was expressed. The data indicate production of insulin derived from hProCpepGFP.
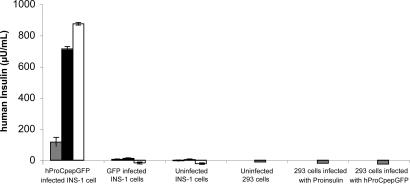
To test hProCpepGFP processing in a β cell context, a human insulin-specific RIA is available that does not recognize uncleaved human PI or fully processed insulin of rodent species. Neither uninfected 293T cells nor those expressing hProCpepGFP or human PI exhibited human insulin-specific immunoreactivity (Fig. 1, fourth, fifth, and sixth sets of bars). Furthermore, neither INS-1 (rat) cells nor those expressing cytosolic GFP (second and third sets of bars) showed significant human insulin-specific immunoreactivity in 2 h of unstimulated secretion, 2 h of secretagogue-stimulated secretion, or final cell lysates (Fig. 1). However, INS-1 cells expressing hProCpepGFP (first set of bars) showed significant human insulin secreted during an initial two unstimulated hours (gray bars), a 7-fold increase after secretagogue stimulation (black bars), and a large remaining intracellular fraction (white bars). This behavior is virtually identical to that of endogenous rat insulin released from INS-1 cells (see below).
Fig. 1.
Detection of processed human insulin in 293T cells and INS-1 cells. Adenoviral expression of constructs was used in preparation for human insulin-specific RIA. Each set of INS-1 cells was incubated first for 2 h under unstimulated conditions (gray bars), then incubated for a further 2 h with a secretagogue mixture (black bars), and finally the cells were lysed and extracted with acid ethanol (white bars). 293T cells were not stimulated. Note the complete absence of human insulin in INS-1 cells that manufacture only rodent insulin or in 293T cells made to express human PI but that cannot endoproteolytically convert to human insulin. By contrast, INS-1 cells expressing hProCpepGFP exhibit stimulated secretion of human insulin and store the remainder intracellularly.
We created recombinant “standards” in 293T cells to identify the molecular masses of expected GFP-containing processing intermediates of hProCpepGFP. The unique mobilities of B chain–GFP, CpepGFP, and GFP–A chain are shown in Fig. 2Top Right. Although untransfected INS-1 cells showed several nonspecific bands (Fig. 2 Top Left), the initial hProCpepGFP translation product was clearly discernible in stably transfected INS-1 cells, and a major form comigrating with CpepGFP accumulated at 4 h of chase. Upon secretagogue stimulation during the last 2 h, CpepGFP release was amplified over that released from unstimulated cells. Only small amounts of bands comigrating with B chain–GFP and GFP–A chain were recovered. An experiment that shows similar stimulus-dependent secretion of CpepGFP was conducted with insulin immunoprecipitation performed in parallel (Fig. 2 Bottom). Similar to CpepGFP, stimulated insulin release to the medium decreased the amount remaining in cells. This behavior appeared similar to that observed from untransfected INS-1 cells (Fig. 2 Bottom Left).
Fig. 2.
Processing and release of hProCpepGFP from INS-1 cells. (Top) Two different portions of the same gel, one for stably transfected INS-1 cells (Left) and another for transiently transfected 293T cells (Right). INS-1 cells were pulse-labeled with 35S-labeled amino acids for 40 min without stimulus (N/A) and then chased for the times indicated. 293T cells transfected with cDNAs encoding B chain CpepGFP (B-chain–GFP), CpepGFP A chain (GFP–A-chain), or CpepGFP were labeled for 60 min without chase. All samples were immunoprecipitated with anti-GFP before SDS/PAGE fluorography. Chase media (M) bathing stably transfected INS-1 cells were collected under either unstimulated or stimulated (Stim +) conditions during a second 2-h period before lysis of cells (C). Note the stimulated secretion of CpepGFP. The experiment shown in Middle and Bottom employs the same protocol, including 293T control cells expressing cytosolic GFP (G) or CpepGFP (far right) and untransfected INS-1 cells (N). [A low-intensity GFP band appearing in cell lysates only at 4 h of chase (arrows) suggests a minor intracellular degradation product that was never recovered in the secretion.] Bottom includes anti-insulin immunoprecipitation from the identical samples and stimulated insulin secretion and final cell lysate from control INS-1 cells.
By confocal microscopy of INS-1 cells stably expressing hProCpepGFP, most INS-1 cells are small and round and lack cytoplasmic processes. Nevertheless, the green fluorescence distribution from hProCpepGFP in ≥75% of cells (n = 105 cells examined) was quite distinct from the ER marker calnexin (in red), instead expressing a peripheral punctate (granule-like) staining pattern that was comparable to the immuofluorescence of insulin (in blue; e.g., Fig. 3A). In occasional INS-1 cells that extend cytoplasmic processes, a granular pattern of green fluorescence was seen to accumulate in the distal tips of processes (Fig. 3C). The data suggest normal intracellular transport of the hProCpepGFP chimera with production of human insulin and CpepGFP that are costored and cosecreted from insulin secretory granules.
Fig. 3.
Expression of hProC(A7)Y-CpepGFP. (A and B) Emerald GFP fluorescence in INS-1 cells expressing hProCpepGFP (A) or hProC(A7Y)-CpepGFP (B) is compared with immunofluorescence of calnexin (anti-Clnx) in red, to which it has been merged. Insulin immunostaining (anti-Ins in blue) is also shown. (C and D) A minority of INS-1 cells extend neurite-like processes, but in such cells the intracellular GFP distribution can be compared in cells expressing hProCpepGFP (C) or hProC(A7Y)-CpepGFP (D). (E) 293T cells were cotransfected, varying the ratio of hProC(A7Y)-CpepGFP plasmid DNA to a fixed amount of wild-type PI cDNA per well. The cells were metabolically labeled for 20 min and either lysed without chase (0 h lanes marked with +) or chased for 1 h in complete medium. Cells (C) and secretion (M) were immunoprecipitated with anti-insulin and analyzed by Tris·tricine·urea· SDS/PAGE. (F) Identical to E, except that the amount of PI cDNA was decreased and a cotransfected plasmid encoding SEAP was included. Cells and media were analyzed for both insulin and SEAP by immunoprecipitation, SDS/PAGE, and autoradiography.
Influence of the C(A7)Y Mutation on PI and hProCpepGFP.
Introduction of the C(A7)Y mutation in the insulin coding sequence dramatically altered the fate of hProCpepGFP. In occasional INS-1 cells that extend cytoplasmic processes, hProC(A7)Y-CpepGFP fluorescence was specifically excluded from these processes, especially their distal tips (Fig. 3D), suggesting failure of the mutant hProC(A7)Y-CpepGFP to be delivered to the distal secretory pathway. In more typical rounded INS-1 cells, none of 115 cells examined revealed hProC(A7)Y-CpepGFP fluorescence in a secretory granule-like distribution; instead GFP fluorescence changed to largely overlap with the ER marker calnexin (e.g., Fig. 3B). Curiously, immunofluorescence reflecting endogenous (pro)insulin distribution changed similarly (Fig. 3B), suggesting a possible effect of the mutant protein on the fate of wild-type PI. Indeed, we found by coexpression in 293 cells that, as the hProC(A7)Y-CpepGFP to wild-type PI ratio was increased, initial synthesis of labeled PI was largely unaffected but secretion of labeled PI at 1 h after synthesis was impaired (Fig. 3E). When using a lower quantity of PI cDNA yet maintaining a high plasmid ratio of hProC(A7)Y-CpepGFP to wild-type PI, pulse–chase studies confirmed that secretion of coexpressed PI was impaired while simultaneous secretion of coexpressed secreted alkaline phosphatase (SEAP) was unimpaired (Fig. 3F). Thus, initial secretory defects caused by hProC(A7)Y-CpepGFP appeared selective for PI.
To understand the extent to which the coexpressed C(A7)Y mutant perturbs insulin production, we performed pulse–chase experiments in islets isolated from young Akita and wild-type mice. Newly synthesized PI from Akita islets exhibited indistinguishable mobility from that of normal islets upon Tris·tricine·urea· SDS/PAGE under reduced conditions, and Akita islets exhibited more labeled PI at the zero chase time than did wild-type islets (Fig. 4A). However, under nonreduced conditions, this increase was no longer apparent (Fig. 4A). In Akita islets under nonreduced conditions (Fig. 4A) there was only a small but reproducible increase in the slower-migrating PI monomer (Fig. 4A, arrowhead) and a portion of immunoprecipitable insulin that was recovered in a disulfide-linked high-molecular-weight complex (Fig. 4A, arrow). By 2 h of chase, the fraction of labeled PI that was converted to insulin (signifying intracellular transport to secretory granules) was clearly diminished in Akita islets.
Fig. 4.
Protein interactions of nonmutant and C(A7)Y mutant versions of PI in Akita islets and INS-1 β cells expressing hProC(A7)Y-CpepGFP. (A) Islets isolated from 5-week-old male wild-type (wt) or Akita (Ak) mice were pulse-labeled with 35S-labeled amino acids for 30 min. Islets without chase or chased for 2 h in the absence of secretagogues were lysed (C), and medium (M) was collected. Samples were immunoprecipitated with anti-insulin and analyzed by Tris·tricine·urea· SDS/PAGE under reduced or nonreduced conditions. The arrowhead denotes a position of aberrant mobility of nonreduced PI with improper disulfide pairing (12). The migration of fully reduced and oxidized (native) PI is indicated. Note that, under nonreduced conditions, newly synthesized PI in Akita islets is less efficiently recovered than under reduced conditions, and there is an increase of a higher-molecular-mass protein complex (open arrow) that is not detected under reduced conditions. (B) Lysates of stably transfected INS-1 cells expressing the constructs shown or pancreatic islets infected with adenovirus driving expression of hProCpepGFP (lane 5) were subjected to Western blotting with anti-GFP after SDS/PAGE under reduced or nonreduced conditions. Lane 4, uninfected control islets; lane 6, a positive control of 293T cells expressing CpepGFP. Note that hProC(A7)Y-CpepGFP is not endoproteolytically processed in β cells, indicating failure to be delivered to immature secretory granules. Also note that pancreatic islets more efficiently process hProCpepGFP to CpepGFP (>90%) than in INS-1 cells (<50%). Finally, note that most hProC(A7)Y-CpepGFP is not recovered in its normal position under nonreduced conditions (Right), with increased higher molecular mass protein complexes (open arrows) that are not detected under reduced conditions. The positions of molecular mass markers are shown on the left. (C) The same cells from B were pulse-labeled with 35S-labeled amino acids for 30 min and then lysed in immunoprecipitation buffer containing 0.1% SDS. GFP-containing peptides were immunoprecipitated, and the samples were analyzed by Tris·tricine·urea· SDS/PAGE under reducing conditions to detect coimmunoprecipitation of endogenous PI. Reduced PI was identified by using direct PI immunoprecipitation from INS-1 control cells (Left).
It is not straightforward to precisely quantify the mutant fraction of initial PI translation product in Akita islets. We therefore examined hProC(A7)Y-CpepGFP expressed in β cells. By Western blotting with anti-GFP, INS-1 cells or primary pancreatic islets exhibited only few nonspecific bands under reduced or nonreduced conditions (Fig. 4B). In INS-1 cells expressing wild-type hProCpepGFP, both this and CpepGFP were detected as specific bands. In primary pancreatic islets expressing wild-type hProCpepGFP, the ratio of processed CpepGFP to unprocessed precursor (Fig. 4B, lane 5) was far greater than in INS-1 cells; this is expected because prohormone processing in islets is much more efficient (18). Upon expression of hProC(A7)Y-CpepGFP (Fig. 4B Left), no processing to CpepGFP was detected, indicating an inability of the mutant to undergo transport to secretory granules.
Under nonreduced conditions, hProC(A7)Y-CpepGFP was largely detected in high-molecular-mass complexes (Fig. 4B, open arrows) that disappeared under reduced conditions. Upon analysis after immunoprecipitation of hProC(A7)Y-CpepGFP with anti-GFP followed by reducing Tris·tricine·urea· SDS/PAGE, coprecipitation of endogenous PI was detected (Fig. 4C). Covalent (but not noncovalent) complexes containing endogenous PI were preserved under our immunoprecipitation conditions (data not shown).
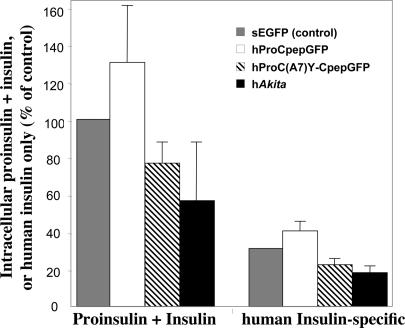
Mutant C(A7)Y PI undergoes ER-associated protein degradation (5). Because the mutant is blocked in intracellular transport (Figs. 3 and 4) and wild-type PI is bound to mutant (Fig. 4C), this could account for loss of β cell insulin. To test this, we transfected INS-832/13 cells with hProCpepGFP or hProC(A7)Y-CpepGFP. At 48 h after transfection, cells underwent FACS. As a control, INS-832/13 were transfected only with secretable GFP (19), which is not expected to augment or decrease intracellular PI or insulin. Transient expression of hProCpepGFP augmented by 30% the total PI plus insulin detectable by RIA that cross-reacts with PI plus insulin of multiple species (Fig. 5); concomitantly, a 30% increase in actual human insulin was observed. By contrast, transient expression of hProC(A7Y)-CpepGFP caused a 28% decrease in the amount of human insulin stored (Fig. 5) but also caused a 23% decrease in total PI plus insulin, most of which is derived from endogenous rat PI. Mutant C(A7)Y human PI (hAkita, with a separately encoded GFP) also markedly lowered the amounts of PI plus insulin (Fig. 5). Thus, with or without the GFP moiety, the C(A7)Y PI mutant exerts dominant negative behavior on endogenous PI and insulin content of pancreatic β cells.
Fig. 5.
Steady-state levels of total PI plus insulin (by rat insulin RIA that cross-reacts with PIs and insulins of multiple species) as well as human insulin specifically (by RIA). INS-832/13 cells were transiently transfected with secretory GFP [sEGFP (31)], hProCpepGFP, hProC(A7)Y-CpepGFP, or hProinsulinC(A7)Y (hAkita in the pCMS vector) as indicated. At 2 d after transfection, fluorescent cells were sorted and total PI plus insulin content as well as processed human insulin were measured in 2 × 105 cells. Total PI plus insulin content in control cells was set at 100%, whereas human insulin specifically contained in control cells (39) was measured at a value that could account for 31% of total. hProCpepGFP expression raises both total PI plus insulin and human insulin specifically. Expression of hProC(A7)Y-CpepGFP (hatched bars) or hAkita (black bars) not only lowers human insulin, but lowers total PI plus insulin derived primarily from endogenous rat PI, indicating dominant negative suppression. The mean and standard deviation from three independent experiments are shown.
β Cell Toxicity of the C(A7)Y PI Mutation.
By restricting analysis to cells exhibiting GFP fluorescence, we could follow the fate of β cells expressing the hProC(A7)Y-CpepGFP mutant. INS-1 cells are nonmotile, allowing daily imaging in culture. In cells expressing hProCpepGFP at 24 h after transfection and followed until 3 d after transfection (Fig. 6A Upper), GFP fluorescence intensity changes over time because of secretion and because gene expression is transient; nevertheless, the identical cells were clearly identified each day. By contrast, upon expression of hProC(A7)Y-CpepGFP (Fig. 6A Lower), cells began to detach from the plate at 2–3 d after transfection. Quantified over three independent experiments (Fig. 6B), the data indicate β cell toxicity of hProC(A7)Y-CpepGFP compared with the nonmutant protein. Taken together, studies of the C(A7)Y PI mutation highlight a sequence of events, including aberrant PI folding, protein complex formation with impairment of nonmutant PI transport, decreased PI and insulin content, and, ultimately, β cell death.
Fig. 6.
Fate of INS-1 β cells expressing hProCpepGFP or hProC(A7)Y-CpepGFP. (A) INS-1 cells plated on gridded glass coverslips were transfected, and the cells were imaged daily in a tissue culture microscope equipped with epifluorescence. Three transfected INS-1 cells are highlighted in each field. The cells are essentially nonmotile, and the relative positions of the cells each day are indicated. Cells expressing hProC(A7)Y-CpepGFP (Lower, yellow arrow) were predisposed to be lost from the culture over time. (B) Quantitation of survival of hProC(A7)Y-CpepGFP-expressing INS-1 cells from three independent experiments like that shown in A. The trend of these data was even more dramatic at 5 d after transfection, although the data were not included here because the 5-d time point was not performed in all experiments.
Discussion
ER stress derived largely from secretory protein misfolding has been implicated in pancreatic β cell failure. In one example, transgenic mice and rats develop type 2 DM that can be specifically attributed to transgenic deposition of human islet amyloid polypeptide (20), as occurs in human type 2 DM (21). Rodent islet amyloid polypeptide is nonamyloidogenic (22), yet there is reason to believe that PI in all species may be predisposed to misfold (2). A primary reason to date for limited investigation of PI folding in living cells is the analytical difficulty in distinguishing well folded from misfolded PI. Indeed, this report demonstrates the misfolding of C(A7)Y mutant PI that coexists with native disulfide-paired PI in islets from Akita mice.
Early work suggested the presence of higher-molecular-weight forms of PI in Akita islets (9), although a recent study from the same laboratory failed to demonstrate PI misfolding beyond that found in normal islets (12). On the other hand, another Cys mutant of PI (disrupting the intra-A chain disulfide bond) has been shown to confer similar dominant negative insulin-deficient DM (23). Indeed, each of the three disulfide bonds of PI impacts not only final PI structure but also the PI folding pathway, and S–S disruption creates potential to block PI folding intermediates. Even for nonmutant PI there is significant potential for nonnative disulfide isomer formation, either as on-pathway or off-pathway products (2).
We have wanted to understand why, in Akita islets, there is less immunoreactive insulin per β cell with fewer insulin secretory granules (8, 24). ER stress response in islets and β cells expressing the C(A7)Y mutant PI (4, 5) [e.g., increased BiP mRNA (13) and protein (9)], general deterioration of the secretory pathway (12), and β cell death have each been proposed to explain disease pathogenesis in male Akita mice, yet most of these phenotypes can be predicted to occur secondary to the deleterious effects of DM on islet function. By contrast, up-regulated PI biosynthesis reflects a relatively early stage of disease progression before β cell failure and death, yet at this time insulin production (Fig. 5A) is already decreased. Moreover, we have clearly established covalent complex formation involving the C(A7)Y mutant PI, in conjunction with a significant decrease in wild-type PI-to-insulin maturation.
Employing an approach in which GFP tags the C-peptide (SI Fig. 7), we report that such a PI construct is efficiently processed in islets (Fig. 4B) and liberates authentic insulin and CpepGFP that is costored and cosecreted upon secretagogue exposure (Fig. 2) along with the human insulin (Fig. 1) from secretory granules (Fig. 3 A and C). We understand that not every aspect of PI biosynthesis will be identical in hProCpepGFP; e.g., the translation time and that required for native PI disulfide bond formation are likely to be substantially longer, whereas folding of GFP will consume additional time. Nevertheless, native PI disulfide pairing is remarkably tolerant of substitutions within the C peptide (17). Altogether, our observations support that hProCpepGFP is a functional model of PI that can serve as a template on which to study effects of the C(A7)Y mutation. The mutation totally eliminated the transport of hProCpepGFP to immature secretory granules as measured by loss of processing (Fig. 4B) and intracellular fluorescence distribution (Fig. 3 B and D) indicating ER retention of hProC(A7)Y-CpepGFP. This is not surprising because the mutant also clearly forms high-molecular-weight protein complexes (Fig. 4B). Most importantly, we directly show that the intermolecular protein complexes containing C(A7)Y mutant also include wild-type endogenous PI, involving covalent (Fig. 4C) as well as possible hydrophobic interactions (25).
ER retention of hProC(A7)Y-CpepGFP appears to confer impaired transport on nonmutant PI with which it is associated (Fig. 3E) but not on SEAP coexpressed from the same cells (Fig. 3F). However, as cells get sicker, secretion of other proteins is likely to be adversely affected (12). Another potential consequence that requires further investigation is possible ER-associated protein degradation of nonmutant PI. Indeed, in Akita islets at 2 h after synthesis, there was a clear loss of labeled PI without a commensurate increase in labeled insulin (Fig. 4A). Consistent with the possibility of ER-associated protein degradation, we establish a dominant negative decrease in steady-state levels of nonmutant PI plus insulin induced by expression of the C(A7)Y mutant (Fig. 5). These data begin to explain insulin deficiency by a mechanism that sets in motion a chain reaction of additional untoward outcomes. Indeed, the data in Fig. 6, similar to a previous report (13), indicate that expression of hProC(A7)Y-CpepGFP is ultimately cytotoxic to β cells (Fig. 6), which can only further compromise net insulin production. However, even before β cell death with resultant loss of β cell mass, we propose that the simplest interpretation is that DM caused by Akita PI is initially a consequence of misfolded PI causing decreased insulin production from nonmutant PI that is blocked in the proximal secretory pathway.
Materials and Methods
Materials.
We used guinea pig anti-rat insulin RIA (catalog no. RI-13K; Linco/Millipore); human insulin-specific RIA (catalog no. HI-14K; Linco/Millipore); rabbit anti-GFP (Immunology Consultants Labs); Zysorbin (Zymed); [35S]methionine/cysteine (ICN); Met/Cys-deficient DMEM, DTT, and RIA-grade BSA (Sigma, St. Louis, MO); Hanks's balanced salt solution Gibco); collagenase P and proteinase inhibitor mixture (Roche); nitrocellulose membranes (Bio-Rad); and ECL solution (Amersham). AtT20/PC2 cells were a gift from R. Mains (University of Connecticut, Farmington, CT).
Construction of hProCpepGFP and Processing Standards.
The cDNA encoding emerald GFP surrounded by flanking ApaI sites (14) was amplified by PCR (forward primer, 5′-GGGCCCTATGGTGAGCAAGGGCGAGGAGCTG-3′; reverse primer, 5′-gggccccgcagcagcagccttgtatagctc-3′) and ligated into pTarget (Promega, Madison, WI) that already contained ApaI-digested human PI cDNA or that bearing the Akita mutation. Constructs encoding B chain CpepGFP (forward, 5′-ggtaccatggccctgtggatgcgcctcctgcc-3′; reverse, 5′-gaattcctactgcagggacccctccagggccaa-3′), CpepGFP (forward, 5′-ggtaccATGGAGGCAGAGGACCTGCAGGTGGG-3′; reverse, 5′-gaattcctactgcagggacccctccagggccaa-3′), and CpepGFP A chain (forward, 5′-ggtaccATGGAGGCAGAGGACCTGCAGGTGGG-3′; reverse, 5′-cctaagctagttgcagtagttctccagctggta-3′) were created by PCR using hProCpepGFP as template. Each PCR product was ligated into pTarget and confirmed by DNA sequencing.
Transfection and Labeling of Cells in Culture.
293T cells were cultured in high-glucose DMEM plus 10% FBS. Cells were plated into six-well plates 1 d before transfection. A total of 2 μg of plasmid DNA was transfected per well using Lipofectamine. At 48 h after transfection, cells were pulse-labeled with 35S-labeled amino acids and chased for the times indicated. For Fig. 7, INS832/13 cells (26) were plated onto 35-mm gridded glass-bottom culture dishes (MatTek) at 30% confluence 1 d before transfection. Each day, live GFP-positive cells were specifically identified by their coordinates on the gridded dish, with fluorescence and phase-contrast images captured at low power in a tissue culture microscope before returning cells to the incubator. Statistical significance was calculated by ANOVA as analyzed by Prism software (GraphPad).
Construction and Use of hProCpepGFP Adenovirus.
A replication-defective adenoviral construct expressing hProCpepGFP was generated by established protocol, purified by cesium chloride ultracentrifugation, and used with another replication-deficient adenovirus encoding GFP alone as a control. One hundred isolated islets were infected (24 h at 37°C in RPMI medium 1640 plus 10% FBS) at 1 × 108 pfu per islet and then transferred into fresh medium for another 48 h before lysis and detection of hProCpepGFP and its processed products. INS-1 and AtT20/PC2 cells were infected for 6 h with adenovirus encoding hProCpepGFP at 150 pfu per cell. Infected cells were washed and returned to regular cell culture growth medium for another 40 h before experiments.
Stimulation of Insulin Secretion.
INS-1 cells infected with hProCpepGFP adenovirus were washed with prewarmed RPMI medium 1640 lacking glucose and then incubated for 2 h at 37°C in KRBH [129 mM NaCl, 5 mM NaHCO3, 4.8 mM KCl, 1.2 mM KH2PO4, 2.5 mM CaCl2, 1.2 mM MgSO4, and 10 mM Hepes (pH 7.4) containing 2.8 mM glucose plus 0.2% RIA-grade BSA]. A second 2-h collection was then made in KRBH plus secretagogue (16.7 mM glucose, 10 mM glutamine, 1 mM tolbutamide, 1 mM isobutylmethylxanthine, and 1 μM phorbol 12-myristate 13-acetate) (27). In Fig. 1, the media were collected and cells were lysed in acid ethanol (28) in preparation for human insulin-specific RIA.
Immunoprecipitation.
Cell lysates and chase media (plus proteinase inhibitor mixture) were precleared with Zysorbin and subjected to immunoprecipitation with antibodies described in the text. Anti-insulin immunoprecipitates were boiled for 5 min in gel sample buffer [1% SDS, 12% glycerol, and 0.0025% Serva Blue in 50 mM Tris (pH 6.8) with or without 100 mM DTT as indicated]. Anti-GFP immunoprecipitates were boiled for 5 min in SDS sample buffer at a final concentration of 2% SDS, 25% glycerol, 0.01% bromophenol blue, and 62.5 mM Tris (pH 6.8), with or with 100 mM DTT as indicated.
SDS/PAGE and Western Blot.
Tris·tricine·urea· SDS/PAGE was used for analysis of PI and insulin, as described previously (2). Conventional SDS/12% PAGE was used for analysis of GFP-containing proteins. For Western blotting, 30 μg of total protein (or 100 infected islets) were boiled in SDS sample buffer with or without 100 mM DTT. Proteins were then resolved by SDS/PAGE, electrotransferred to nitrocellulose, and blotted with rabbit anti-GFP followed by an anti-rabbit secondary antibody conjugated to HRP, with development of the blot by ECL.
Isolation of Mouse Pancreatic Islets.
Pancreata removed from 5-week-old male Akita mice (blood glucose at time of euthanasia: 11 mM), and nonmutant male littermates were digested in 2 mg/ml collagenase-P in Hanks's balanced salt solution containing calcium and magnesium in a shaking water bath for 30 min at 37°C. After 500-μm filtration, sedimentation (1,000 × g for 5 min), and washing twice in ice-cold buffer, islets were hand-picked for overnight recovery in RPMI medium 1640 containing 11.1 mM glucose plus 10% FBS and 1% penicillin–streptomycin.
Metabolic Labeling of Mouse Pancreatic Islets.
One hundred fifty wild-type and Akita islets were washed twice in prewarmed Met/Cys-deficient medium plus 1% BSA and 10 mM Hepes (pH 7.35). Islets were then metabolically labeled with 35S-labeled amino acids in the same medium for 20 min. In Fig. 4A, labeled islets were briefly washed once with RPMI medium 1640 containing 10% FBS and then split into three equal portions: two were directly immersed in lysis buffer containing a proteinase inhibitor mixture for subsequent analysis under nonreduced or reduced conditions, and the third was chased for 2 h at 37°C in RPMI medium 1640 (11.1 mM glucose plus 10% FBS). The media were collected, and the islets were lysed as above. Lysate aliquots were evaluated for trichloroacetic acid-precipitable radioactivity to normalize the immunoprecipitations.
Confocal Imaging of INS-1 Cells.
INS-1 cells expressing hProCpepGFP or hProC(A7)Y-CpepGFP were seeded at low confluency on polylysine-treated coverslips and grown in complete media for 1 d. Cells were fixed with 2% formaldehyde at room temperature for 10 min, washed three times with PBS, and then mounted onto glass microscope slides for imaging of GFP by confocal fluorescence microscopy. For immunofluorescence, fixed cells were permeabilized with 0.1% Nonidet P-40 and then incubated with guinea pig anti-insulin (1:1,000) and rabbit anti-calnexin (1:1,000) and appropriate secondary antibodies (1:1,000) conjugated to aminomethylcoumarin acetate and Alexa Fluor 555, respectively.
Steady-State PI and Insulin Content of Transiently Transected INS-832/13 Cells Expressing hProCpepGFP, hProC(A7)Y-CpepGFP, or a Cytosolic GFP.
INS-832/13 cells were transfected 1 d after plating using Lipofectamine (3 μg of plasmid DNA per well of a six-well plate). After 48 h, trypsinized cells were washed, pelleted, resuspended in PBS, and counted/sorted by green fluorescence. A total of 100 μl of acid ethanol was used to extract each 105 sorted INS-832/13 cells. Total PI plus insulin was measured by rat insulin RIA that cross-reacts with PI and insulins of all species. Processed human insulin was measured by specific RIA.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant DK48280 with help from both the Molecular Biology Core and the Morphology/Image Analysis Core of the University of Michigan Diabetes Research and Training Center (National Institutes of Health Grant DK20572).
Abbreviations
- ER
endoplasmic reticulum
- PI
proinsulin
- DM
diabetes mellitus
- SEAP
secreted alkaline phosphatase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702697104/DC1.
References
- 1.Min CY, Qiao ZS, Feng YM. Eur J Biochem. 2004;271:1737–1747. doi: 10.1111/j.1432-1033.2004.04079.x. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, Li Y, Cavener D, Arvan P. J Biol Chem. 2005;280:13209–13212. doi: 10.1074/jbc.C400475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Kaufman RJ. Neurology. 2006;66:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 4.Nozaki J, Kubota H, Yoshida H, Naitoh M, Goji J, Yoshinaga T, Mori K, Koizumi A, Nagata K. Genes Cells. 2004;9:261–270. doi: 10.1111/j.1356-9597.2004.00721.x. [DOI] [PubMed] [Google Scholar]
- 5.Allen JR, Nguyen LX, Sargent KEG, Lipson KL, Hackett A, Urano F. Biochem Biophys Res Commun. 2004;324:166–170. doi: 10.1016/j.bbrc.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 6.Ron D. J Clin Invest. 2002;109:443–445. doi: 10.1172/JCI15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroux L, Desbois P, Lamotte L, Duvillie B, Cordonnier N, Jackerott M, Jami J, Bucchini D, Joshi RL. Diabetes. 2001;50:S150–S153. doi: 10.2337/diabetes.50.2007.s150. [DOI] [PubMed] [Google Scholar]
- 8.Yoshioka M, Kayo T, Ikeda T, Koizumi A. Diabetes. 1997;46:887–894. doi: 10.2337/diab.46.5.887. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, Lu D, Takata K, Koizumi A, Izumi T. J Clin Invest. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayo T, Koizumi A. J Clin Invest. 1998;101:2112–2118. doi: 10.1172/JCI1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo ZY, Feng YM. Biol Chem. 2001;382:443–448. doi: 10.1515/BC.2001.054. [DOI] [PubMed] [Google Scholar]
- 12.Izumi T, Yokota-Hashimoto H, Zhao S, Wang J, Halban PA, Takeuchi T. Diabetes. 2003;52:409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- 13.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins S, Geng X, Li L, Papworth G, Robbins PD, Drain P. Traffic. 2002;3:461–471. doi: 10.1034/j.1600-0854.2002.30703.x. [DOI] [PubMed] [Google Scholar]
- 15.Pouli AE, Kennedy HJ, Schofield JG, Rutter GA. Biochem J. 1998;331:669–675. doi: 10.1042/bj3310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohara-Imaizumi M, Nakamichi Y, Tanaka T, Ishida H, Nagamatsu S. J Biol Chem. 2002;277:3805–3808. doi: 10.1074/jbc.C100712200. [DOI] [PubMed] [Google Scholar]
- 17.Liu M, Ramos-Castañeda J, Arvan P. J Biol Chem. 2003;278:14798–14805. doi: 10.1074/jbc.M212070200. [DOI] [PubMed] [Google Scholar]
- 18.Irminger JC, Meyer K, Halban P. Biochem J. 1996;320:11–15. doi: 10.1042/bj3200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molinete M, Lilla V, Jain R, Joyce PBM, Gorr S-U, Ravazzola M, Halban PA. Diabetologia. 2000;43:1157–1164. doi: 10.1007/s001250051507. [DOI] [PubMed] [Google Scholar]
- 20.Matveyenko AV, Butler PC. Diabetes. 2006;55:2106–2114. doi: 10.2337/db05-1672. [DOI] [PubMed] [Google Scholar]
- 21.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 22.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. Proc Natl Acad Sci USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbach N, Rathkolb B, Kemter E, Pichl L, Klaften M, Hrabé de Angelis M, Wolf E, Aigner B, Wanke R. Diabetes. 2007;56:1268–1276. doi: 10.2337/db06-0658. [DOI] [PubMed] [Google Scholar]
- 24.Zuber C, Fan JY, Guhl B, Roth J. FASEB J. 2004;18:917–919. doi: 10.1096/fj.03-1210fje. [DOI] [PubMed] [Google Scholar]
- 25.Yoshinaga T, Nakatome K, Nozaki J, Naitoh M, Hoseki J, Kubota H, Nagata K, Koizumi A. Biol Chem. 2005;386:1077–1085. doi: 10.1515/BC.2005.124. [DOI] [PubMed] [Google Scholar]
- 26.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 27.Neerman-Arbez M, Halban PA. J Biol Chem. 1993;268:16248–16252. [PubMed] [Google Scholar]
- 28.Arakawa K, Ishihara T, Oku A, Nawano M, Ueta K, Kitamura K, Matsumoto M, Saito A. Br J Pharmacol. 2001;132:578–586. doi: 10.1038/sj.bjp.0703829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.