Abstract
Integration of patterning cues via transcriptional networks to coordinate gene expression is critical during morphogenesis and misregulated in cancer. Using DNA adenine methyltransferase (Dam)ID chromatin profiling, we identified a protein–protein interaction between the Drosophila Myc oncogene and the Groucho corepressor that regulates a subset of direct dMyc targets. Most of these shared targets affect fate or mitosis particularly during neurogenesis, suggesting the dMyc–Groucho complex may coordinate fate acquisition with mitotic capacity during development. We find an antagonistic relationship between dMyc and Groucho that mimics the antagonistic interactions found for EGF and Notch signaling: dMyc is required to specify neuronal fate and enhance neuroblast mitosis, whereas Groucho is required to maintain epithelial fate and inhibit mitosis. Our results suggest that the dMyc–Groucho complex defines a previously undescribed mechanism of Myc function and may serve as the transcriptional unit that integrates EGF and Notch inputs to regulate early neuronal development.
Keywords: neurogenesis, Drosophila, stem cell, cell fate, mitosis
The Myc family of oncogenes is intimately involved in the genesis of cancer (1). Myc proteins function within the context of a highly conserved basic helix–loop–helix zipper (bHLH) Myc/Max/Mxd(Mad-Mnt) transcriptional network that is essential for normal development (2). Loss-of-function mammalian Myc mutants exhibit embryonic lethality probably because of Myc's role in organogenesis (1, 2). Myc proteins dimerize with Max, and the resulting Myc-Max heterodimers bind to CACGTG (E-box) sequences, where they are associated with gene activation. Max can also heterodimerize with Myc antagonist proteins belonging to the Mxd family. Max-Mxd heterodimers bind to the same E-box sequences; however, this binding results in repression of many Myc-Max target genes. The transcriptional antagonism between Myc and Mxd proteins is well established biologically in the regulation of cell size and cellular growth (1, 2).
The Myc/Max/Mxd network is highly conserved and, in Drosophila, is represented by single dMyc, dMax, and dMnt genes (3, 4). Similar to the vertebrate Myc genes, dmyc is an essential gene involved in cell growth, affecting endoreplication, regulation of cell size, cell competition, and apoptosis (reviewed in ref. 5), whereas dmnt is a nonessential gene that is associated with differentiation, where it functions to limit cell growth (4).
A major challenge in understanding Myc function has been to identify the number and nature of the direct targets that it regulates (6). To this end, we previously used a microarray-based genomic chromatin profiling method termed DNA adenine methyltransferase (Dam)ID to identify the direct binding sites of the Drosophila Myc network (7). DamID, similar to the ChIP-chip chromatin profiling technique, is a powerful tool that allows systematic and global identification of in vivo direct targets of transcriptional networks (8).
We have also used the DamID approach to map the direct binding sites of the bHLH repressor Hairy and its associated cofactors Sir2, CtBP, and Groucho (9). Strikingly, a comparison of the two networks revealed a group of dMyc target genes that overlaps with targets recruiting the Groucho corepressor. Groucho (Gro) and its mammalian orthologs, collectively called transducin-like Enhancer of split (TLE) (TLE1–4), are developmentally regulated corepressors. Groucho was the first cofactor shown to be required for Hairy-mediated repression and was subsequently shown to mediate repression through several other classes of DNA-binding transcriptional regulators, including Engrailed, Dorsal, Tcf, and Runt (10).
Here we show that dMyc and Gro antagonistically coregulate a subset of cell fate and mitotic targets, defining a previously undescribed mechanism of dMyc function. Consistent with this, our phenotypic analyses show that dMyc and Gro are required for neuronal fate and mitosis and phenocopy EGF and Notch signaling, respectively. We also demonstrate a genetic link between dMyc, Gro, and the EGF/Notch pathways and propose that dMyc and Gro integrate EGF/Notch signaling during neuroectoderm development.
Results
dMyc and Gro Share Many Direct Targets.
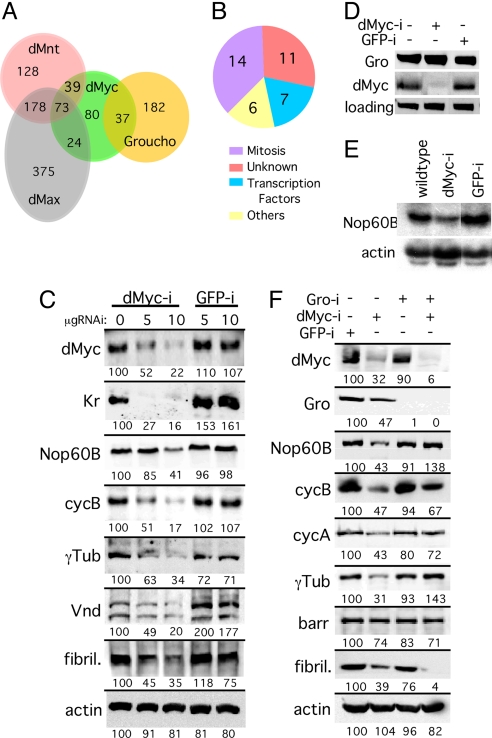
We identified 37 transcriptional direct targets shared between dMyc and Gro in Kc cells [Fig. 1A; supporting information (SI) Table 2; chance probability of overlap is 3 × 10 e-14 (hypergenometric distribution); SI Fig. 5A]. These dMyc-Gro shared targets are not bound by other network proteins (dMax, dMnt, Hairy, dSir2, or dCtBP). Consistent with this, we find that the Myc-Max canonical binding consensus CACGTG (E-box) and its derivatives are absent from most (75%) of these dMyc-Gro target gene promoters. Functional classification of the dMyc-Gro targets show that 37% (14/38) of these genes play roles in mitosis. In addition, these shared targets also include genes encoding fate-determining transcription factors such as ventral nerve defective (vnd); intermediate nerve defective (ind); dMyc's heterodimerization partner, dMax; and other genes involved in early neurogenesis (Fig. 1B; SI Table 2; data not shown). We hypothesized that dMyc and Gro coregulate this subset of transcriptional targets to determine mitotic potential and cell fate during development.
Fig. 1.
Regulation of dMyc and Gro shared targets. (A) Venn diagram of dMyc/dMax/dMnt and Gro targets (see SI Text). (B) Functional distribution of the 37 dMyc and Gro shared targets identified by DamID (plus one target identified experimentally; SI Table 2). (C) dMyc is required for expression of dMyc/Gro targets. Western blot analyses indicating that dMyc/Gro target gene expression is reduced in S2 cells treated with dMyc RNAi (dMyc-i). Kr, Krüppel; γTub, γTubulin; vnd, ventral nerve defective; cycB, cyclin B; fibril, fibrillarin. (D and E) dMyc is required for dMyc/Gro target gene transcription. (D) Western blot analysis of dMyc and Gro in untreated S2 cells or in cells treated with either dMyc or control (GFP) RNAi. (E) Northern blot analysis of Nop60B expression using the same extracts depicted in D. (F) Decline in target gene expression because of reduced dMyc levels can be partially restored by the simultaneous reduction of Gro levels. Western blot analysis of S2 cells treated with RNAi to the indicated gene(s) and assayed for the level of dMyc, Gro, or the shared target proteins Nop60B, CycA, CycB, γTub, Barren (Barr), and the direct Myc-Max target, fibrillarin. Relative quantification of the proteins is given below the lanes with the first 0 μg of dMyc-i RNAi lane (wild type) being set to 100 (see Materials and Methods).
dMyc and Gro Antagonistically Regulate the Expression of Their Shared Targets.
Gro/TLE proteins are corepressors that are recruited by dedicated repressors like Hairy/E(spl) (HES) proteins (10) or by activators such as Dorsal, thereby converting them into repressors (11). We envision two basic models by which dMyc–Gro complexes could regulate transcription: (i) dMyc and Gro cooperate to form a dedicated repressor complex on their shared targets; or (ii) dMyc activates shared target expression but is antagonized by Gro, leading to inhibition of target expression. In model i, reduction of dMyc would be expected to increase target gene expression, whereas in model ii, reduction of dMyc should decrease target gene expression. To distinguish between these models, we reduced dMyc levels in Drosophila S2 cells using RNAi and monitored target gene expression. Reduction of dMyc levels resulted in reduced target gene expression at both the protein and transcriptional levels (i.e., Nop60B; Fig. 1 C–F; SI Fig. 6A). Furthermore, simultaneous RNAi-mediated silencing of both dMyc and Gro partially or fully restored expression of all shared targets tested with the exception of Barren (Fig. 1F), establishing an antagonistic relationship between dMyc and Gro, as predicted by the second model. In contrast, cosilencing of dMyc and Gro did not restore the expression of fibrillarin, a known Myc target (12) that is not shared with Gro, indicating that Gro repression is limited to the dMyc-Gro subset of dMyc targets (Fig. 1F). Reduction of Gro levels alone had a small effect on dMyc-Gro target gene expression, but this is likely indirect, because this effect was also observed with classical Myc target gene (fibrillarin; Fig. 1F). We also find that, although both dMyc and Gro are expressed in proliferating Kc and S2 cells, the dMyc antagonist dMnt is not detected under these conditions, and RNAi to dMnt had no effect on expression of these targets (data not shown). Thus, our data support the second model; dMyc is positively required for target gene expression, whereas Gro inhibits target expression independent of dMnt.
dMyc-Gro Antagonistic Interactions Regulate Fate and Mitosis in the Developing Nervous System and Phenocopy EGF/Notch Antagonistic Signaling.
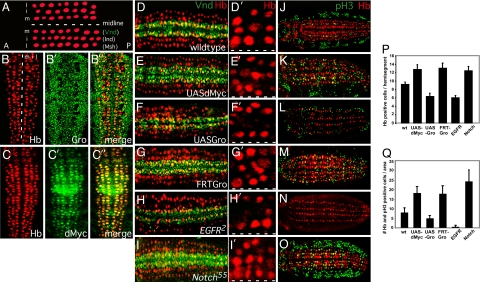
Gro is a downstream transducer of several signaling pathways and was placed at the crossroads of the Notch and EGF signaling pathways during patterning of the Drosophila nervous system, where EGF-induced site-specific phosphorylation of Gro attenuates it repression activity (13–15). During embryonic stage 9, the CNS matures in three bilaterally symmetrical longitudinal rows of neuroblasts (16), with the homeobox transcription factors, Vnd, Ind, and Msh, specifying the medial (ventral), intermediate, and lateral rows, respectively (Fig. 2A; ref. 17). EGF regulates the expression of both Vnd and Ind and is thus required for the formation of the ventral and intermediate rows (13, 17). Interestingly, we find that both Vnd and Ind are among the 38 dMyc-Gro shared targets we identified (SI Table 2), and that Gro and dMyc, but not dMnt, are expressed in neuroblasts of stage 9 embryos (Fig. 2 B–C′′; data not shown). Because dMyc-Gro targets are associated with both neuroblast fate and mitosis, we hypothesized that EGF and Notch coregulate cell fate and mitosis within the developing neuroectoderm via dMyc-Gro antagonism. We compared Vnd expression (a shared Myc-Gro target whose expression overlaps with and is required for establishment of S1 neuroblasts), the overall number of neuroblasts, and mitotic activity in wild-type embryos to groe47 loss-of-function (LOF) mutants (in which the maternal contribution of Gro is removed), Egfr2, or Notch55e11 [note that dMyc LOF embryos cannot be generated (18); Fig. 2 D and D′, G–I′, J, and M–O)]. We also evaluated these parameters in embryos overexpressing either dMyc or Gro using the conditional Gal4/upstream activating sequence (UAS) expression system (Fig. 2 E, F, K, and L). Vnd expression is stronger and expanded in both Notch and gro LOF embryos, as well as in embryos overexpressing dMyc (Fig. 2 I, G, and E, respectively) when compared with wild type (Fig. 2D). These mutants also show neuroblast hyperplasia (Fig. 2 I′, G′, and E′, respectively; Fig. 2P) and elevated mitotic activity (Fig. 2 O, M, and K, respectively; Fig. 2Q). Furthermore, Egfr LOF or Gro-overexpressing embryos show reduced Vnd expression (Fig. 2 H and F), neuronal hypoplasia (Fig. 2 H′, F′, and P), and reduced mitotic activity (Fig. 2 N, L, and Q), consistent with the molecular nature of the dMyc-Gro common targets.
Fig. 2.
Regulation of cell fate and mitosis in the developing Drosophila neuroectoderm (CNS). (A) Diagram depicting the three rows of neuroblasts (red) that are observed on each side of the midline (white dashed line) of a stage 9 embryo and the homeobox transcription factors (Vnd, Ind, and Msh) that specify them: m, middle (ventral); i, intermediate; l, lateral. Anterior (A) is to the left. (B–C′′) Gro and dMyc are expressed during Drosophila neurogenesis. (B) Stage 9 embryos were immunostained with Gro (B′ and B′′) or dMyc (C′ and C′′). Neuroblasts are identified by Hunchback (Hb) staining (red; B and C). Anterior is up. (D–O) Opposing regulation by dMyc/EGF and Gro/Notch of neuroectoderm cell fate, neuroblast number, and mitosis in the developing Drosophila CNS/neuroectoderm. Stage 9 embryos were immunostained with Hb (red) to identify neurons (D–O) and either Vnd (green), a dMyc-Gro direct target whose expression overlaps with and is required for S1 neuroblasts (D–I′), or the mitosis marker phosphohistone3 (pH3; green) (J–O). Anterior is to the left. Wild-type embryos are depicted in D and J. Rho-Gal4 was used to drive the expression of dMyc (two copies; UAS-dMyc) (E and K) or Gro (one copy; UAS-Gro) (F and L) in the neuroectoderm. Embryos lacking Gro function (FRT-Gro) were generated via germ-line clones (G and M). Egfr2 (H and N) and N55e11 (I and O) are LOF alleles for EGF receptor and Notch, respectively. (D′–I′) Higher magnification of one hemisegment from the embryos in D–I, respectively, depicting neuroblasts (red). The dashed white line indicates the position of the midline. (P) Quantification of the number of neuroblasts as identified by Hb-positive staining per hemisegment (n = 40 hemisegments). (Q) Quantification of the number of neuroblast mitoses as identified by cells dually positive (yellow) for Hb (red) and phospho-H3 (green) staining per 10 hemisegment area (n = 10 embryos).
Another patterning/fate determination process governed by EGF and Notch signaling is the specification of mesothoracic sensory bristles in the peripheral nervous system (PNS) (13–15). Similar to our findings during neuroblast development, we find that loss of Gro or dMyc overexpression phenocopies activation of the EGF pathway, whereas loss of dMyc expression or overexpression of Gro phenocopies activated Notch signaling (SI Fig. 7). Importantly, cooverexpression of dMyc along with Gro results in a dose-dependent partial rescue of the Gro phenotype (SI Fig. 7 and SI Text).
dMyc Interacts Genetically with EGF Receptor and Notch During Neurogenesis.
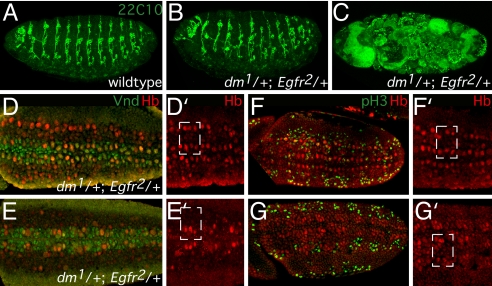
The genetic interaction between dMyc and Gro fits well with previously reported genetic interactions between Notch-Gro and EGF-Gro (15). We find that dMyc also interacts genetically with EGF and Notch, leading to disruptions in neurogenesis. We tested for interactions between an EGF receptor mutant (Egfr2; ref. 19) and two dMyc mutants (dm1 and dm2; refs. 4 and 18) and observed a dose-sensitive genetic interaction in which a reduced number of transheterozygous progeny survive (i.e., synthetic lethality; Table 1). Embryos from mothers heterozygous for either Egfr or dMyc alone (or hemizygous for dm* in males; dm*/Y) are viable. In contrast, simultaneously reducing the dose of both dMyc and Egfr (dm*/+; Egfr2/+ or dm*/Y; Egfr2/+) results in inappropriate development and subsequent increased embryo lethality. Analysis of dm1/Egfr2 doubly heterozygous mutant embryos using the panneuronal marker 22C10 revealed severe patterning phenotypes and aberrant neurogenesis due in part to improper development of neuroblasts (Fig. 3).
Table 1.
dMyc(dm) exhibits genetic interaction with Egfr and N
| Female | Male | N | Percent flies with ectopic bristles* | Percent lethality† |
|---|---|---|---|---|
| +/+; Egfr2/+ | +/Y; +/+ | 2,969 | 3 | |
| dm1/+; +/+ | +/Y; +/+ | 2,575 | 3 | |
| dm2/+; +/+ | +/Y; +/+ | 2,423 | 6 | |
| dm1/+; Egfr2/+ | +/Y; +/+ | 2,254 | 20 | |
| dm2/+; Egfr2/+ | +/Y; +/+ | 2,028 | 17 | |
| dm1/+ | 98 | 0 | ||
| N55e11/+ | 91 | 89 | ||
| N55e11 +/+ dm1 | 116 | 38 |
*dm1 suppresses ectopic bristle formation associated with N55e11.
†dm/Egfr doubly heterozygous embryos exhibit synthetic lethality.
Fig. 3.
Phenotypic analysis of the genetic interaction between dMyc-EGF receptor. (A–G′) Analysis of dm1/Egfr2 doubly heterozygous mutant embryos. Wild-type (A) or doubly heterozygous dm1/Egfr2 (B and C) stage 17 embryos stained with the 22C10 panneuronal marker outlining the embryonic PNS. At this late stage, the majority of mutant embryos (>90%) exhibit a severely defective morphology and staining pattern (C), whereas the minority exhibits moderate neuronal abnormalities and patterning defects. (D–G′) dm1/Egfr2 doubly heterozygous stage 9 embryos show reduced Vnd protein expression, neuroblast hypoplasia, and reduced mitotic activity. Moderately defective (D and D′, F and F′) and severely defective (E and E′, G and G′) doubly heterozygous dm1/Egfr2 embryos immunostained with Hb (red) to identify neuroblasts (D–G′) and either Vnd (green), a dMyc-Gro direct target whose expression overlaps with and is required for S1 neuroblasts (D and E), or the mitosis marker phosphohistone3 (pH3; green) (F and G). Hemisegments are indicated by a white rectangle. All other abbreviations are as in Fig. 2. Anterior is to the left.
Because Gro is a downstream transducer of the Notch pathway during neurogenesis (14, 15), our results suggest that an antagonistic relationship will exist between dMyc and Notch during neurogenesis. Because dMyc and Notch are located in close proximity on the X chromosome, we tested for dominant genetic interaction between a LOF mutant in Notch (N55e11; ref. 20) and dm1. Although homozygous N55e11 is lethal, heterozygous N55e11 females are viable (Table 1) and display a greatly increased number of mesothoracic bristles (SI Fig. 7J). We hypothesize that, if dMyc antagonizes Notch and is required to promote neuronal fate, partial loss of dMyc will result in suppression of the ectopic bristle formation observed in N55e11 heterozygous females. Indeed, dm1 exhibits a dominant genetic interaction with N55e11: ectopic bristle formation is partially rescued (>50%) in doubly heterozygous N55e11/dm1 females (Table 1; SI Fig. 7L). Thus, our genetic results suggest that EGF and Notch are the upstream regulators of dMyc-Gro function: dMyc is required for establishing the neurogenic program, in addition to antagonizing the Notch signaling pathway.
dMyc and Gro Form a Complex in Vivo and in Vitro.
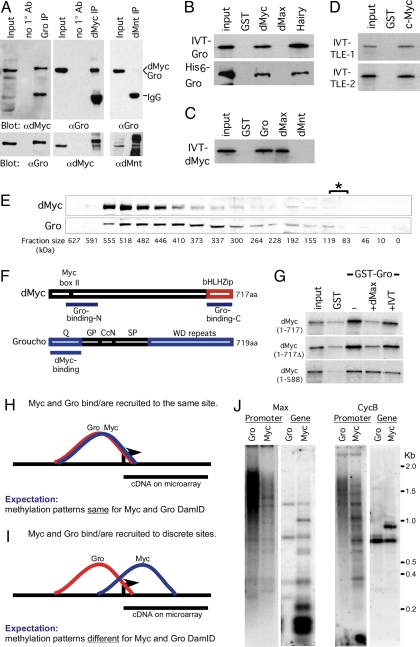
Our observations suggest a coordinated role for dMyc and Gro during neuroectoderm development, likely via antagonistic regulation of a distinct group of shared direct targets. Although this regulation may be mediated via independent dMyc and Gro complexes, it is also possible that it is mediated via a direct dMyc-Gro protein–protein interaction (Fig. 4). To test this, we immunoprecipitated endogenous dMyc or Gro from 3.5- to 4.5-h Drosophila embryo lysate (Fig. 4A) or Kc cell lysate (SI Fig. 5B) and tested for the presence of the other protein in the immunoprecipitated complex. We find that dMyc and Gro associate with each other in vivo, whereas the dMyc antagonist dMnt (4) (and the unrelated proteins GFP and cadherin) could not be detected in association with Gro (Fig. 4A and SI Fig. 5C).
Fig. 4.
dMyc and Gro form a complex in vivo and in vitro. (A) Immunoprecipitation of dMyc–Gro complexes from stage 8 and 9 Drosophila embryos. Embryo extracts were immunoprecipitated with the indicated antibodies and analyzed by Western blot. Note that the dMyc and Gro proteins are 717 and 719 aa, respectively. (B) IVT-35S-Met-labeled full-length Gro or purified full-length His6-Gro interacts with GST-Myc (81–504 aa) and GST-Hairy. (C) IVT-35S-dMyc interacts with GST-Gro and GST-dMax. (D) Full-length IVT-35S-Met-labeled mammalian TLE-1 and -2 proteins bind to GST-c-Myc (1–262 aa). (E) dMyc and Gro protein colocalize to a ≈520-kDa fraction upon gel filtration. The asterisk denotes the fractions (83–110 kDa) where dMyc and Gro monomers would be expected to localize. (F) Diagram depicting the regions mediating dMyc–Gro interaction. (G) IVT-dMax outcompetes binding of full-length 35S-dMyc to GST-Gro (at the top). This displacement requires dMyc's bHLHZ domain. For all cases, 5% input is shown. (H–J) dMyc and Gro generate differential Dam-methylation patterns within a shared target gene locus and are likely recruited to independent sites at their shared targets. (H and I) Graphic representation of a theoretical binding and methylation pattern generated by DamID fragments from Dam-dMyc and Dam-Gro if they bind or are recruited to a single site (H) or discrete sites (I) within their shared target locus. (J) Southern blot analysis of dMyc and Gro DamID generated fragments hybridized with probes to the promoter (“promoter”) or coding regions (“gene”) of two representative dMyc-Gro shared targets, CycB and dMax (see also SI Text).
We also find that dMyc and Gro form a complex in vitro using GST pulldown assays (Fig. 4 B and C; data not shown). Similarly, the mammalian Gro orthologs, TLE-1 and -2, interact with c-Myc, suggesting this interaction is evolutionarily conserved (Fig. 4D). To exclude the possibility that this interaction is mediated by bridging proteins within the in vitro translated (IVT) lysate, we purified bacterially expressed His6-tagged Gro. His6-Gro binds directly to GST-dMyc and to the GST-Hairy control (Fig. 4B). Furthermore, separation of embryo extract over a gel-filtration column suggests that dMyc and Gro may be part of a larger protein complex, because they colocalize in ≈520-kDa fraction, which is larger than the ≈80- to 110-kDa fraction, where dMyc and Gro monomers would be expected (Fig. 4E).
Because dMyc lacks motifs previously known to facilitate Gro recruitment by other transcription factors (10), we mapped the domains within dMyc and Gro that are required for their interaction. We find that Gro binds to two regions within the dMyc protein: an N-terminal region containing the conserved Myc BoxI and BoxII motifs (50–150 aa) and its C-terminal bHLH leucine zipper motif (bHLHZip) region (624–717 aa) (Fig. 4F and SI Fig. 5 F and G). We find that both Myc's N- and C-terminal Gro-interacting regions bind strongly to Gro's N-terminal glutamine-rich Q domain (1–133 aa; Fig. 4F; SI Fig. 5 H and I). The requirement for two dMyc-binding sites with Gro is unlike the single-domain protein interactions reported for Engrailed-Gro and Hairy-Gro through Gro's WD domain (21, 22).
Because the bHLHZip region of Myc is required for its dimerization with Max (1, 3), we tested whether exogenous dMax modulates dMyc binding to Gro. Adding dMax to a dMyc-Gro binding assay competed the binding of full length Myc protein and requires an intact C-terminal dMyc bHLHZip region. However, excess Gro was not able to compete dMax binding to dMyc in a similar setting (Fig. 4G; SI Fig. 5 D and E). The ability of dMax to outcompete for Gro binding to dMyc could be important, because we identified dMax as a dMyc-Gro shared target that, once expressed, could shift Myc molecules from dMyc-Gro complexes into canonical dMyc–dMax complexes.
To further understand the relationship between dMyc and Gro on DNA in vivo, we used DamID Southern mapping (Fig. 4 H–J; see also SI Text). We find that dMyc binding occurs mainly in the vicinity of the coding region, whereas Gro recruitment is mostly associated with the 5′ upstream regions of the promoter (Fig. 4J). Taken together with our dMyc-Gro protein–protein interaction data, Gro's ability to multimerize via its Q domain, and Gro's ability to function as a long-range repressor (10), our results support the notion that a dMyc–Gro complex is associated with more than one site within the target gene locus.
Discussion
Identification of a Gro–dMyc Interaction Outside the Canonical dMyc/dMax/dMnt Network.
Myc proteins are required for both cell growth/size and cell proliferation. The model in which Myc functions are mediated by heterodimerization with Max and antagonized by Mxd (Mad/Mnt) proteins has been well established (1, 2). However, recent studies suggest that a set of interactions outside the canonical Myc/Max/Mxd network also regulate some of Myc's functions (2, 7). Interestingly, our studies point to a subset of dMyc direct targets that are not shared with either dMax or dMnt (Fig. 1A; ref. 7). Furthermore, dMnt-Dam and dMax-Dam were not recruited to these dMyc targets even in experiments where the Dam fusions were coexpressed in the presence of high levels of dMax or dMyc, respectively (A.O. and R.N.E., unpublished data), suggesting that previously uncharacterized mechanisms may mediate Myc's recruitment to DNA, and proteins other than dMnt may antagonize its transcriptional activity on this set of targets. Here we report the identification of Gro as the first component in a pathway that antagonizes dMyc function independent of dMnt and operates during Drosophila neurogenesis.
Gro Antagonizes dMyc Function.
Transcriptionally, we find that dMyc is positively required for the expression of dMyc-Gro targets, activity that is antagonized by Gro. Importantly, dMyc is not a Gro target, and reducing Gro levels does not affect dMyc protein levels (Fig. 1D; ref. 9). Furthermore, Gro antagonism is limited only to the dMyc-Gro subset of shared targets and does not involve dMnt: there is no overlap between genes bound by dMnt or Gro, dMnt is not expressed in cells where the dMyc–Gro interaction is observed, RNAi to dMnt does not affect Myc-Gro shared target expression (not shown), and overexpression of dMnt does affect PNS development (SI Fig. 6).
Although we cannot exclude the possibility that dMyc-Gro targets are coregulated by individual dMyc and Gro complexes, our results suggest that dMyc and Gro are part of a single larger protein complex. First, the observation that RNAi to dMyc results in reduction of target expression and is restored by coreducing Gro suggests that other activators coregulate shared target expression along with dMyc. Second, our biochemical purification, binding data, and DamID Southern analyses support the idea that both proteins physically interact with one another yet associate with DNA through distinct binding sites. Third, Gro does not bind directly to DNA but must be recruited to targets by sequence-specific DNA-binding transcription factors. Fourth, most of the dMyc-Gro targets lack E-box sequences associated with canonical Myc network targets, suggesting that dMyc and Gro may be recruited to shared targets via a novel mechanism or by other protein(s) yet to be identified. Candidates for recruiting Gro may be the E(spl) proteins that convey the Notch signal, antagonize the EGF pathway, interact with Gro, and exhibit similar phenotypes (14). Thus, the identification of the entire dMyc–Gro complex and its regulation will be an important next step.
Biological Role of dMyc-Gro During Neuroectoderm Development.
Gro's role as a downstream transducer of Notch signaling during neurogenesis is well documented (15), and mounting evidence supports Myc as a key player in progenitor cell proliferation (23, 24). Here we have identified a previously undescribed role for dMyc, together with Gro, during Drosophila early neuronal development. dMyc and Gro are required to directly regulate key fate controlling genes such as the homeodomain proteins vnd and ind that are downstream targets of EGF signaling (SI Table 2; Fig. 3). Because Vnd was identified as a regulator of the proneural gene complex (25, 26), the differential regulation of vnd by dMyc and Gro implicates them as antagonistic regulators upstream of proneural genes. Thus, we propose that dMyc is transiently required within the neuroectoderm, where it promotes specific fate acquisition and allows mitotic expansion of committed neuronal cells.
Phenotypically, we observe that, similar to EGF, dMyc promotes neurogenesis both in the PNS and CNS, whereas Gro and Notch inhibit neuroblast formation and mitosis (Figs. 3 and 4; SI Fig. 6). This is a different role than that previously ascribed to dMyc, because it is usually associated with regulation of cell size and organismal growth (5), functions that are antagonized by dMnt. Consistent with this, a recent study identified EGF-induced phosphorylation of c-Myc, Max, and TLE proteins in mammalian cells (27). The antagonistic relationship of Myc/EGF to Gro/Notch is likely to be highly dependent on the developmental context and the specific progenitor niche (28). For example, in cellular contexts in which Notch promotes proliferation, such as during the development of T cells in acute leukemia, Myc is a direct target of mutated Notch1 and is required for T cell proliferation and development (29). Our findings also fit well with observations that N-Myc is required during mouse progenitor development, and that the fly tumor suppressor Brat regulates dMyc levels posttranscriptionally in larval neuroblasts resulting in a “tumorous” phenotype (30, 31).
A Proposed Pathway for Transducing Signaling Cues into Transcriptional Outputs.
Taken together, the snapshot provided by our DamID data leads us to suggest a model in which changes in neuronal progenitor fate and mitosis are determined by the balance between EGF and Notch signaling that is likely transcriptionally mediated by the dMyc–Gro complex. During epithelial development, Notch, like Gro, is required to specify and maintain epithelial fate. We propose that Gro sequesters dMyc in an inactive multiprotein complex formed by associating with dMyc, preventing the activation of dMyc-Gro shared targets. Upon EGF signaling, a molecular switch takes place whereby Gro is phosphorylated, and its repression is attenuated (14). dMyc, as part of an as-yet-to-be-identified activation complex, is then liberated to activate zygotic transcription of a subset of targets that determines neuronal fate and enhances mitosis. One of these targets is dMax, which is specifically expressed in the neuroectoderm (32). Activation of dMax would be expected to establish a feed-forward loop required for the subsequent activation of (E box-containing) Myc targets to promote cell growth. As development progresses, the dMnt gene would be induced, and dMnt-dMax complexes would replace dMyc–Max complexes, thereby promoting cellular differentiation.
Finally, both EGF/dMyc and Notch/Gro misregulation and mutation are intimately involved in hematological, epithelial, and neuroectodermal cancers (1, 10). Thus, our identification of a dMyc–Gro complex that could serve as a molecular junction to integrate EGF and Notch signaling inputs is highly relevant for both developmental biology and cancer.
Materials and Methods
Flies and Genetics.
Flies were cultured on yeast/cornmeal/molasses/malt extract medium at 25°C. Alleles used in this study: y w dm2/FM70, Kr-GFP was from D. Stein; y w1118; y w dm1/FM7; wa N55e11/FM7; lac-Z; cn Egfrf2 bw sp/CyO; w ovoD1 v24 P{w[+mW.hs] = FRT(w[hs])}101/C (1)DX, y f/Y; P{ry[+t7.2] = hsFLP}38, FRT82B ovoD1/TM3; and y w hs-flp122; Gl/TM3 were all obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN). The transgenic strains UAS-λTOP3; w; UAS-dMyc UAS-dMyc/TM3, Sb; w; UAS-dMyc/CyO; UAS-gro/TM6b, and UAS-dMnt/TM3, UAS-dMax/TM3 were expressed by using the Gal4/UAS system with the following drivers: w1118; P{w[+mW.hs] = GawB}C253 (performed at 29°C), or Rho-Gal4 (performed at 25°C). To generate dMyc and gro LOF mosaic thoraces, we used the flippase recombination target (FRT)-DFS system as described (33) using w, dm4 FRT14AB/Bin and groe47 FRT82B/TM3, Ser Sb. Scoring of the embryonic lethality was performed as described (9).
Plasmids and Constructs.
A detailed list of constructs used in this study is provided in SI Text.
Binding Assays.
In vitro binding experiments were preformed by using GST-pulldown assays similar to those described (4, 34, 35).
Immunoprecipitations and Immunofluorescence.
Embryos (3.5–4.5 h) or Kc cells were lysed on ice for 15 min in buffer L (1× PBS/1% BSA/0.5% Nonidet P-40) with 10 mM EDTA and protease and phosphatase inhibitors, and then sonicated, followed by centrifugation at 4°C. Protein (750 μg) was used for each immunoprecipitation with the indicated antibody and then subjected to SDS/PAGE and Western blot analysis. Embryos and cells were immunostained as described (ref. 35; see also SI Text).
Drosophila Cell Culture and RNAi Experiments.
S2 cells were maintained at 25°C Schneider's media (Invitrogen, Carlsbad, CA), 10% FBS/10 mM glutamine. RNAi to Drosophila cells was performed by using the MegaScript RNAi Kit (Ambion, Austin, TX). One hundred fifty micrograms of cell extract per lane was separated via SDS/PAGE.
Supplementary Material
Acknowledgments
We thank Bas van Steensel and David Ish-Horowicz for sharing personal communications and Christos Delidakis, Mark Groudine, Adi Salzberg, Phil Soriano, and Valera Vasioukhin for discussions and comments. We are grateful to R. Basom, D. Bianchi-Frias, L. Carlos, M. Conacci-Sorrell, D. Elzi, F. Remington, and B. Schneider for help at different stages of this research. We also thank H. Bellen, C. Delidakis, C. Doe, D. Glover, C. Grandori, S. Henikoff, J. Reinitz, H. Ruohola-Baker, D. Mellerick, S. Poole, T. Tsukiyama, and the Bloomington Stock Center for antibodies, DNAs, flies, and reagents used in this study. This work was supported by National Institutes of Health Grants CA57138 (to R.N.E.) and GM073021 (to S.M.P.). R.N.E. is an American Cancer Society Research Professor. Z.P. is a Braun Lecturer and was supported by Israeli Science Foundation Grant 501/04 and the Krol Foundation. A.O. is supported by Human Frontiers Science Program Grant CDA 0048/2004 and is a Special Fellow of the Lymphoma and Leukemia Society and a Horev-Taub-Shalom Fellow.
Abbreviations
- bHLH
basis helix–loop–helix
- bHLHZip
bHLH leucine zipper motif
- Dam
DNA adenine methyltransferase
- dm
diminutive
- IVT
in vitro translation
- LOF
loss-of-function
- PNS
peripheral nervous system
- UAS
upstream activating sequences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707418104/DC1.
References
- 1.Adhikary S, Eilers M. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Hooker CW, Hurlin PJ. J Cell Sci. 2006;119:208–216. doi: 10.1242/jcs.02815. [DOI] [PubMed] [Google Scholar]
- 3.Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 4.Loo LW, Secombe J, Little JT, Carlos LS, Yost C, Cheng PF, Flynn EM, Edgar BA, Eisenman RN. Mol Cell Biol. 2005;25:7078–7091. doi: 10.1128/MCB.25.16.7078-7091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Cova C, Johnston LA. Semin Cancer Biol. 2006;4:303–312. doi: 10.1016/j.semcancer.2006.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levens DL. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- 7.Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greil F, Moorman C, van Steensel B. Methods Enzymol. 2006;410:342–359. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buscarlet M, Stifani S. Trends Cell Biol. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collar HH, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culi J, Martin-Blanco E, Modolell J. Development (Cambridge, UK) 2001;128:299–308. doi: 10.1242/dev.128.2.299. [DOI] [PubMed] [Google Scholar]
- 14.Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, Dinur T, Volk T, Courey AJ, Paroush Z. Nat Genet. 2005;37:101–105. doi: 10.1038/ng1486. [DOI] [PubMed] [Google Scholar]
- 15.Hasson P, Paroush Z. Br J Cancer. 2006;94:771–775. doi: 10.1038/sj.bjc.6603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman CS, Doe CQ. In: The Development of Drosophila melanogaster 2. Bates M, Martinez-Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. pp. 1131–1206. [Google Scholar]
- 17.Cornell RA, Von Ohlen T. Curr Opin Neurobiol. 2000;10:63–71. doi: 10.1016/s0959-4388(99)00049-5. [DOI] [PubMed] [Google Scholar]
- 18.Maines JZ, Stevens LM, Tong X, Stein D. Development (Cambridge, UK) 2004;14:775–786. doi: 10.1242/dev.00932. [DOI] [PubMed] [Google Scholar]
- 19.Schejter ED, Shilo BZ. Cell. 1989;56:1093–1094. doi: 10.1016/0092-8674(89)90642-9. [DOI] [PubMed] [Google Scholar]
- 20.Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 21.Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Howoricz D. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein RE, Cook O, Dinur T, Pisante A, Karandikar UC, Bidwai A, Paroush Z. Mol Cell Biol. 2005;25:10711–10720. doi: 10.1128/MCB.25.24.10711-10720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy MJ, Wilson A, Trumpp A. Trends Cell Biol. 2005;15:128–137. doi: 10.1016/j.tcb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Skeath JB, Panganiban GF, Carroll SB. Development (Cambridge, UK) 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- 26.Chu H, Parras C, White K, Jiménez F. Genes Dev. 1998;12:3613–3624. doi: 10.1101/gad.12.22.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Shilo BZ. Development (Cambridge, UK) 2005;132:4017–4027. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- 29.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoepfler PS, Kenney AM. Cell Cycle. 2006;5:47–52. doi: 10.4161/cc.5.1.2292. [DOI] [PubMed] [Google Scholar]
- 31.Betschinger J, Mechtler K, Knoblich JA. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. research0088.1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TB, Perrimon N. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poortinga G, Watanabe M, Parkhurst SM. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini D, Parkhurst SM. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.