Abstract
The product of the ataxia-telangiectasia gene (ATM) was identified by using an antiserum developed to a peptide corresponding to the deduced amino acid sequence. The ATM protein is a single, high-molecular weight protein predominantly confined to the nucleus of human fibroblasts, but is present in both nuclear and microsomal fractions from human lymphoblast cells and peripheral blood lymphocytes. ATM protein levels and localization remain constant throughout all stages of the cell cycle. Truncated ATM protein was not detected in lymphoblasts from ataxia-telangiectasia patients homozygous for mutations leading to premature protein termination. Exposure of normal human cells to γ-irradiation and the radiomimetic drug neocarzinostatin had no effect on ATM protein levels, in contrast to a noted rise in p53 levels over the same time interval. These findings are consistent with a role for the ATM protein in ensuring the fidelity of DNA repair and cell cycle regulation following genome damage.
Ataxia-telangiectasia (A-T) is an autosomal recessive disorder marked by progressive cerebellar ataxia and oculocutaneous telangiectases. Other clinical features include extreme cellular and humoral immune deficiencies and an increased predisposition to lymphoreticular malignancies (1, 2). Furthermore, A-T carriers were reported to be at increased risk for cancer (3, 4). At the cellular level, homozygous A-T cells are hypersensitive to ionizing radiation and resistant to inhibition of DNA synthesis following irradiation (5).
In response to DNA damage, the cell cycle is blocked at the G1/S and G2/M transitions, allowing the repair of DNA damage before initiating DNA replication and entry into mitosis (6). Recent studies have shown that a signal transduction pathway involving p53 mediates the G1-S checkpoint after exposure to ionizing radiation (7–9) by acting as a positive transcriptional regulator of, among others, the cell growth suppressor p21 (10–12). A series of studies (7, 13, 14) have shown that A-T cells do not show an increase in radiation-induced levels of p53 as seen in normal cells.
The A-T gene (designated ATM) was recently identified (15, 16) and encodes a protein with a predicted mass of ≈350 kDa. The C terminus of the ATM protein is similar to the catalytic domain of phosphatidylinositol 3 kinase and is found in several yeast, Drosophila, and mammalian proteins known to be involved in mitogenic signal transduction, meiotic recombination, and cell cycle control (17). Despite these homologies, the function of the ATM gene product remains unknown.
To characterize the ATM protein, an antiserum was generated and utilized to characterize the human ATM protein by immunoblot analysis and to determine subcellular location by cell fractionation and indirect immunofluorescence. ATM protein localization at all stages of the cell cycle was examined as well as ATM protein and p53 levels following γ-irradiation exposure and treatment with the radiomimetic drug neocarzinostatin (NCS).
MATERIALS AND METHODS
Cell Culture.
The human lymphoblast cell line B-310 and A-T patient lymphoblasts were cultured in RPMI 1640 medium with 15% fetal calf serum (FCS). Low-passage human foreskin fibroblast line Hs-68 and lung fibroblast line MRC-5 were obtained from the American Type Culture Collection and cultured as indicated. The human fibroblast line F-2054 was grown in DMEM containing 15% FCS. The A-T fibroblast line F-169 was cultured in a customized media formulation (M-99094; Sigma) containing 20% FCS. Peripheral blood mononuclear cells were fractionated from whole blood by centrifugation over Ficoll-Paque PLUS (Pharmacia) cushions and further fractionated from monocytes and macrophages by adherence to tissue culture flasks. When indicated, fibroblasts, lymphoblasts, or lymphocytes were fractionated using established protocols (18).
Construction and Expression of ATM Fusion Proteins.
One segment of ATM cDNA was amplified from a full-length ATM cDNA via PCR using the primers 5′-CCAGTATTGGATCCCTCTGTC-3′ (ATM ORF nt 2026–2045) and 5′-CATTCCTTCCTGAG AATTCAAGTATGC-3′ (nt 3392–3371). The ≈1.3-kb PCR product that encodes ATM protein residues 679-1126 was digested at the primer-encoded BamHI and EcoRI sites and cloned into digested pGEX-2T in a carboxyl-terminal fusion to the protein glutathione S-transferase (GST). Similarly, another region of the ATM ORF was amplified using the primer 5′-GCTGTGGATCCTCTGAGTGGCAGCTGG-3′ (nt 6853–6879) and 5′-GGTTTAGTCAGGAATTCATCTCTGTTTCG-3′ (nt 7723–7695) and the ≈0.9-kb PCR product that encodes ATM protein residues 2287–2572, was similarly cloned into pGEX2T. The two plasmids constructed in this scheme, pGEX AT 2026–3392 (pGEX AT-6) and pGEX AT 6853–7723 (pGEX AT-4), were authenticated by restriction and sequence analysis. pGEX AT-6 and pGEX AT-4 were transformed into the Escherichia coli strain DH5α and the encoded fusion proteins were induced by adding isopropyl β-d-thiogalactoside (final concentration, 1.0 mM) to a culture of logarithmically growing cells cultured in Luria–Bertani broth containing 50 μg/ml ampicillin.
Antibody Preparation and Affinity Purification.
The peptide NH2-CKSLASFIKKPFDRGEVESMEDDTNG-COOH, which corresponds to amino acid residues 819–844 of the predicted ATM gene product primary structure, was synthesized by automated methodologies. (Computer searches indicated that the sequence was unique to ATM when searched against currently available protein sequence databases.) This peptide (designated AT peptide 4) was coupled to keyhole limpet hemocyanin and injected into two adult rabbits at three week intervals. (Peptide synthesis, coupling, and immunizations were performed by Phoenix Pharmaceuticals, St. Joseph, MO). The serum from one of these rabbits (designated pAb 132) detected the ATM protein following the third boost.
Anti-ATM protein immunoreactivity was purified from pAb 132 in the following manner: E. coli transformed with pGEX AT-6 were induced to express the encoded fusion protein by addition of isopropyl β-d-thiogalactoside and lysates were subjected to SDS/PAGE followed by transfer to a nitrocellulose sheet. The sheet was stained with Ponceau-S and a thin strip of nitrocellulose containing the GST–AT6 was cut from the sheet. The strip was then blocked by incubation (30 min) in Tris-buffered Saline (TBST; 10 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1% Tween-20) containing 5% nonfat dry milk. Following this, the strip was rinsed in TBST, incubated in 1 ml of pAb 132 for 4 h, rinsed extensively in TBST, and the antibody was eluted from the nitrocellulose by a 1-min incubation in 1 ml of 100 mM Na-citrate (pH 2.2). The sample was then neutralized by addition of 100 μl of 1 M Tris·HCl (pH 8.0) and dialyzed for 16 h at 4°C against 500 times volume of PBS. Antibody was stored at 4°C following the addition of Na-azide to 0.02%.
Electrophoresis and Immunoblotting.
Cells were harvested and rinsed twice in cold PBS, and SDS lysates were formed and protein assays were conducted as previously outlined (19). SDS/PAGE and electrotransfer to nitrocellulose was carried out using standard procedures (20, 21). Protein molecular weight standards were purchased from BRL and Pharmacia. Immunoblotting was conducted as outlined (19) using chemiluminescent substrate and recorded on Kodak XAR x-ray film. Quantitation of immunoblot signals was performed using a Molecular Dynamics densitometer and software. Where indicated, blots were stripped by incubation (80°C for 30 min) in 100 mM Tris·HCl (pH 7.5), 1% SDS, and 50 mM 2-mercaptoethanol followed by TBST rinses.
For immunoblotting, pAb 132 serum was used at a dilution of 1:2000 and affinity purified pAb 132 at a final concentration of 2 μg/ml. Additionally, immunoblotting was performed using the anti-α-tubulin mAb DM1A (22), the anti-p53 mAb DO-1 (Santa Cruz Biotechnologies), or a rabbit anti-NuMA antisera (23).
Immunofluorescence Microscopy.
Fibroblasts were cultured on sterilized glass coverslips to ≈75% confluency before processing. The cells were fixed by immersion in PBS containing 3.0% formaldehyde for 30 min (room temperature). Immunofluorescent staining was done as described (19) using fluorescein isothiocyanate-labeled goat anti-rabbit secondary antibody. Images were captured with a Zeiss Axioplan microscope equipped with a Photometrics (Tucson, AZ) charged-coupled device (CCD) camera.
Cell Synchronization.
Hs-68 fibroblasts were synchronized in G0 by culturing cells for 96 h under reduced (0.5% FCS) serum conditions in DMEM. To synchronize cells in G1, cultures of serum deprived cells were recultured in DMEM plus 10% FCS for 6 h [cell cycle exit and re-entry was confirmed by proliferating cell nuclear antigen (PCNA) staining]. Cells were synchronized at the G1/S border by culturing in the presence of 2 mM hydroxyurea for 44 h, and mitotic cells were obtained by culturing in colcemid (0.1 μg/ml) for 24 h.
γ-Irradiation and Radiomimetic Drug Treatment of Cells.
Exponentially growing cells were plated into tissue culture dishes and irradiated with a 137Cs source at a dose rate of 2 Gy/5 sec. Postirradiation, cells were incubated in a 37°C, 5% CO2 atmosphere for indicated times, and extracts from ≈106 cells were analyzed for ATM protein levels. Approximately 105 cells were analyzed for p53 levels.
NCS was obtained from Kayaku (Tokyo) and diluted with PBS before each experiment and applied directly to the medium of cultures (80 ng/ml) of the normal fibroblast line F-2054. Following NCS addition, the cultures were incubated in a 5% CO2, 37°C atmosphere for the indicated period of time, and subsequently cell extracts were formed, and immunoblotting was performed as outlined above.
RESULTS
Characterization of ATM Protein Antiserum pAb 132.
A rabbit antiserum was prepared to a synthetic peptide antigen, designated AT peptide 4, which corresponds to ATM protein amino acid residues 819–844 (16). In addition, two segments of the ATM ORF were expressed in bacteria as carboxyl-terminal GST fusion proteins. These two ATM segments, termed GST–AT6 and GST–AT4, comprise amino acid residues 679-1126 and 2287–2572, respectively, of the ATM protein ORF. Hence, AT peptide 4 is encoded within the GST–AT6 fusion protein.
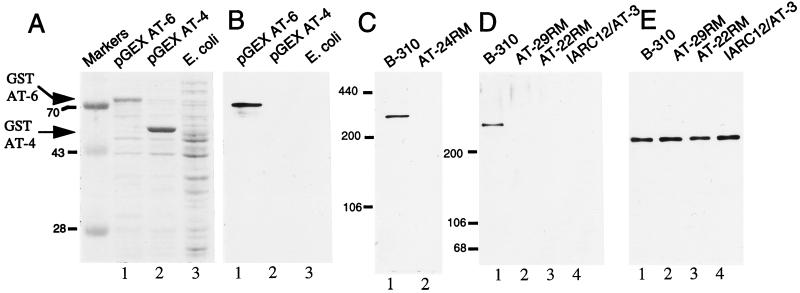
When E. coli transformed with these GST fusion protein expression constructs were induced to express their respective fusion proteins, accumulation of a ≈75-kDa protein in the bacteria transformed with pGEX-AT6 (Fig. 1A, lane 1) and an ≈55-kDa protein in the pGEX AT-4 transformed bacteria (Fig. 1A, lane 2) were observed when compared with untransformed E. coli (Fig. 1A, lane 3). pGEX AT-4 encodes GST-AT4, an ≈33-kDa segment of the ATM polypeptide in a carboxyl-terminal fusion to GST (molecular weight ≈ 25 kDa) and pGEX AT-6 encodes GST-AT6, an ≈50-kDa segment of the ATM polypeptide in a similar fusion to GST. Thus, the relative masses of these accumulated proteins are in close agreement with the predicted sizes of the encoded fusion proteins. At high dilution (1:2500), serum from a rabbit immunized with AT peptide 4 (designated pAb 132) detected an ≈75-kDa protein present in the extract from bacteria expressing GST–AT6 (Fig. 1B lane 1). Nothing was detected in extracts from either E. coli expressing GST AT-4 or untransformed E. coli (Fig. 1B, lanes 2 and 3, respectively), indicating that pAb 132 detected the ATM portion of the GST AT-6 fusion protein. Additionally, preimmune serum displayed no reactivity to GST–AT6 (data not shown).
Figure 1.
Characterization of ATM protein antiserum pAb 132. (A) Lysates from E. coli transformed with the plasmids pGEX AT-6 (lane 1) and pGEX AT-4 (lane 2) following fusion protein induction and untransformed E. coli (lane 3) were subjected to SDS/PAGE and Coomassie blue staining. Migration of the specific GST–AT fusion proteins are indicated by arrowheads. (B) Extracts shown in A were immunoblotted with pAb 132. (C) A total of 50 μg of B-310 (lane 1, wild type for ATM) and AT24RM (lane 2, homozygous mutant for ATM) cell extracts were immunoblotted with pAb 132. (D) A total of 50 μg of SDS-lysate from B-310 lymphoblasts (lane 1) and A-T patient lines AT29RM (lane 2), AT22RM (lane 3), and IARC12/AT3 (lane 4) were immunoblotted with pAb 132. (E) Blot displayed in D was stripped and reprobed with an antisera to NuMA.
To test whether pAb 132 was capable of detecting the native ATM protein in extracts from human cells, SDS extracts were made from the human lymphoblast cell line B-310 and the A-T lymphoblast line AT24RM. [Cells derived from the A-T patient AT24RM possess a homozygous frameshift mutation in the ATM gene at codon 252, leading to premature termination of the ATM protein before the pAb 132 epitope (24)]. Immunoblot analysis of these extracts showed that pAb 132 detected a high-molecular weight protein present exclusively in B-310 cell extract (Fig. 1C). Furthermore, pAb 132 immunoreactivity with this protein was ablated by preincubation of the antiserum with 100 ng/ml of AT peptide 4 (data not shown). Molecular weight estimates using appropriate electrophoretic standards determined that the protein detected by pAb 132 possessed a mass of ≈320 kDa, in close agreement with the predicted molecular weight of the ATM gene product (≈350 kDa) (16).
Close to 90% of ATM mutations identified to date consist of frameshifts resulting from out of frame insertion or deletion events predicted to result in truncations throughout the protein (24, 25). Lysates were prepared from three lymphoblastoid cell lines derived from homozygous A-T patients whose mutations have been previously determined (24) and lead to premature termination of the ATM polypeptide. IARC12/AT3 cells are homozygous for a frameshift mutation causing premature protein termination at codon 2714, which would predict a mutant ATM protein of ≈305 kDa. AT22RM and AT29RM cells possess similar frameshifts expected to yield truncated products of 281 and 163 kDa, respectively. Immunoblot analysis revealed that while the ATM protein was detectable in the B-310 extract (Fig. 1D, lane 1), no bands were detected at the expected molecular weight of the mutated ATM protein products from these patient cell lines (or elsewhere in their respective lanes) (Fig. 1D, lanes 2–4). This blot was stripped and reprobed with an antiserum to the 235-kDa nuclear protein NuMA (23) to demonstrate that equal amounts of extracts were assayed (Fig. 1E). Hence, these findings indicate that, in general, truncated ATM protein products are unstable.
ATM Protein Localization in Normal Human Cells.
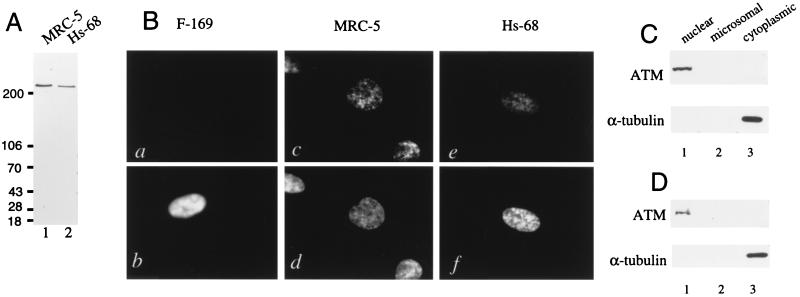
pAb 132 was affinity purified and subsequently used for immunoblot analysis of the normal human fibroblast lines MRC-5 and Hs-68. Extracts from these cells electrophoresed on steep (5–15%) acrylamide gradient gels showed that affinity-purified pAb 132 was monospecifically reactive with the ATM protein present in these lines (Fig. 2A).
Figure 2.
Localization of ATM protein in normal human fibroblasts. (A) A total of 50 μg of SDS lysate from the normal human fibroblast lines MRC-5 (lane 1) and Hs-68 (lane 2) was immunoblotted with affinity purified pAb 132. (B) F-169 (a and b), MRC-5 (c and d), and Hs-68 (e and f) cells were stained with affinity purified pAb 132 (a, c, and e), and Hoechst 22358 (b, d, and f). (C) MRC-5 fibroblasts (1 × 107) were fractionated and 10% of the nuclear (lane 1), microsomal (lane 2), and cytoplasmic (lane 3) fractions were immunoblotted with pAb 132 (Upper) or 1% of each fraction were immunoblotted with antitubulin (Lower). (D) Hs-68 fibroblasts (1 × 107) were fractionated and analyzed as in C.
Indirect immunofluorescence was conducted on MRC-5 and Hs-68 fibroblasts, and the A-T fibroblast line F-169, which is homozygous for an ATM frameshift mutation (24) and which possesses no pAb 132 immunoreactivity as judged by immunoblotting (data not shown). F-169 (Fig. 2B a and b), MRC-5 (Fig. 2B c and d), and Hs-68 (Fig. 2B e and f) cells were stained with affinity-purified pAb 132 (Fig. 2B a, c, and e), and counterstained with the DNA stain Hoechst 33258 (Fig. 2B b, d, and f). No significant pAb 132 staining was observed in the A-T cell line F-169, but punctate nuclear staining was observed in both MRC-5 and Hs-68 cells. The lack of definite staining in the cytoplasm of both normal fibroblast cell lines indicates that the presence of the ATM protein is largely confined to the nucleus in these cells.
Nuclear, microsomal, and cytoplasmic fractions were obtained from both MRC-5 (Fig. 2C) and Hs-68 (Fig. 2D) cells. Immunoblot analysis of equal percentages of these fractions with pAb 132 (Fig. 2 C and D Upper) showed that the ATM protein was confined to the nuclear fractions in these cells. α-Tubulin was solely detected in the cytoplasmic fractions, indicating that lysis was complete (Fig. 2 C and D Lower).
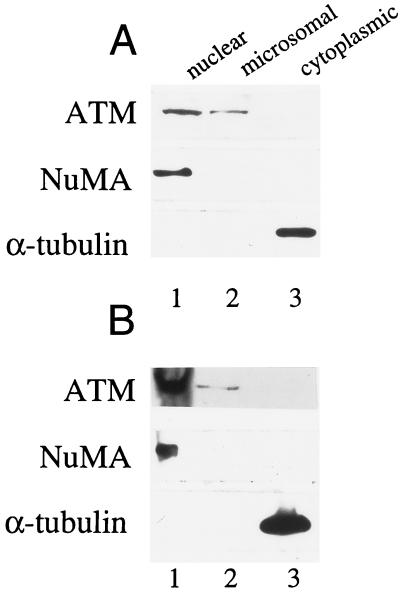
When normal human lymphoblasts and lymphocytes were fractionated and analyzed for the presence of the ATM protein, this protein was found to cofractionate with both nuclei and microsomes in both transformed and peripheral blood lymphocytes (Fig. 3 A and B Top). Immunoblot analysis of these fractions with anti-NuMA showed minimal nuclear contamination of the microsome fraction (Fig. 3 A and B Middle), and analysis with anti-tubulin determined lysis was complete (Fig. 3 A and B Bottom).
Figure 3.
Analysis of ATM protein in fractionated human lymphoblasts and peripheral blood lymphocytes. (A) B-310 lymphoblasts (1 × 107) were fractionated. Ten percent of the resultant nuclear (lane 1), microsomal (lane 2), and cytoplasmic (lane 3) fractions were immunoblotted with pAb 132 (Top). To evaluate purity of the preparations, 2% of each fraction was analyzed with NuMA antisera (Middle), or 1% of each fraction was analyzed with anti-tubulin (Bottom). (B) Normal peripheral blood lymphocytes (2 × 108) were fractionated and analyzed as in A.
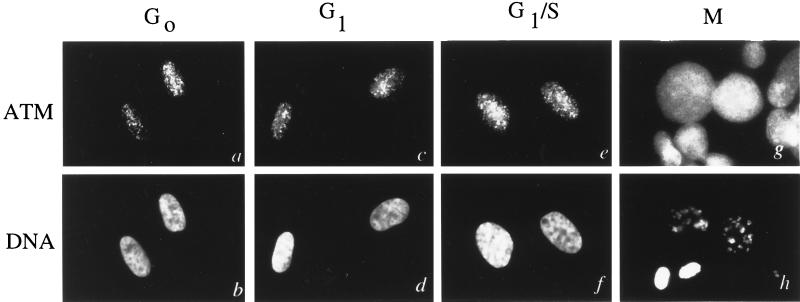
The location of the ATM protein at various stages of the cell cycle was evaluated by indirect immunofluorescence microscopy on synchronized Hs-68 fibroblasts. pAb 132 stains only the nucleus during the G0, G1, and G1/S-phases of the cell cycle (Fig. 4 a–f). Following breakdown of the nuclear envelope at the onset of mitosis, ATM protein staining appears to be diffuse throughout the cytoplasm and does not appear to be associated with any particular structures in the cell (Fig. 4 g and h). Immunoblot analysis of extracts from synchronized cell populations reveal that ATM protein levels are relatively constant throughout the cell cycle (data not shown).
Figure 4.
Localization of ATM protein at various cell cycle phases. Cells were synchronized at various cell cycle phases and processed for immunofluorescence with affinity-purified pAb 132 (a, c, e, and g) and Hoechst 22358 (b, d, f, and h). (a and b) Cells synchronized at G0. (c and d) Cells synchronized at G1. (e and f) Cells synchronized at G1/S. (g and h) Cells synchronized in mitosis.
Effects of Genome-Damaging Agents on ATM Protein Abundance.
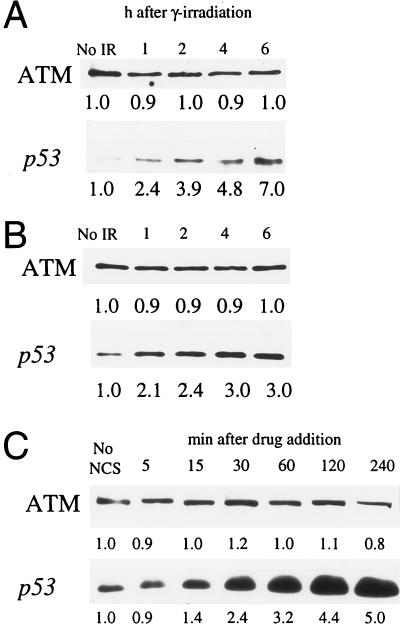
Normal ATM protein function appears to play a role in arresting cell cycle advance following genome damage stemming from events such as exposure to ionizing radiation or radiomimetic drugs (2, 26). To assess the effects of γ-ray exposure on ATM protein abundance, both Hs-68 fibroblasts (Fig. 5A) and B-310 lymphoblasts (Fig. 5B) were exposed to 5 Gy of ionizing radiation. Quantitative analysis of extracts from unirradiated cells as well as cells at 1, 2, 4, and 6 h postirradiation showed that, in each cell line, the cellular levels and electrophoretic mobility of the ATM protein remained relatively unchanged following exposure to ionizing radiation (Fig. 5 A and B Upper). In contrast to the ATM protein, and as previously observed (7), a quantitative increase in p53 was observed following ionizing radiation exposure in both cell lines (Fig. 5 A and B Lower). Additionally, we obtained similar results at other irradiation doses (2 and 10 Gy) in both cell lines, and immunofluorescence of irradiated Hs-68 fibroblasts showed no change in localization of the ATM protein following irradiation (data not shown).
Figure 5.
Effects of genome damaging agents on cellular ATM levels. (A) Hs-68 fibroblasts were exposed to 5 Gy of ionizing radiation. Unirradiated cells (No IR) and cells 1, 2, 4, and 6 h after exposure were analyzed for ATM protein (Upper) and p53 (Lower) levels by immunoblotting. (B) B-310 lymphoblasts were exposed to 5 Gy of IR and analyzed as in A. (C) The normal human fibroblast cell line F-2054 was treated with NCS and extracts from treated cells at 5, 15, 30, 60, 120, and 240 min following drug addition, and untreated cells (No NCS) were analyzed for ATM protein (Upper) and p53 (Lower). Relative immunoblot signal intensity (determined by densitometric scanning of exposed films) is displayed below each band. Minor differences in recorded ATM protein levels are likely due to slight inconsistencies in protein loading and/or protein transfer.
ATM protein levels following exposure to the radiomimetic drug NCS (27) were also assayed. Similar to what was observed following γ-irradiation, NCS induced a 5-fold increase in p53 levels over the course of 4 h (Fig. 5C Lower), but quantitative analysis of ATM protein levels determined that this protein remained steady during this time interval (Fig. 5C Upper).
DISCUSSION
An initial characterization of the ATM gene product was conducted following the generation of a specfic antiserum. This antiserum detects a single distinct high-molecular weight protein which was primarily, but not exclusively, located in the nucleus of normal human cells. Transport of many proteins to the nucleus is facilitated by the presence of a short contiguous stretch of predominantly basic residues termed a nuclear localization sequence (NLS) (28). However, sequence inspection of the predicted ATM gene product failed to detect the presence of a NLS within the primary structure of this protein (16). Perhaps a currently undefined NLS exists within the ATM protein or, alternatively, nuclear import is facilitated by the interaction of the ATM protein with cofactor(s). Interestingly, a punctate staining pattern was observed within the nucleus of fibroblasts stained with affinity purified pAb 132 (Figs. 2 and 4). This is unlikely to be a fixation artifact because alternative fixation yielded similar results and suggests that the ATM protein may be limited to certain domain(s) or associated with particular structure(s) within the nucleus.
Immunoblot analysis of three lymphoblast cell lines derived from homozygous A-T patients with mutations predicted to result in premature termination of the ATM protein failed to detect the presence of truncated ATM protein within these cells. Similarly, expression of other mutant genes predicted to encode prematurely terminated proteins often results in greatly diminished or subdetectable accumulation of the mutant protein (29–31), a feature that is usually attributed to disruption of proper protein folding (32). The failure to detect mutant ATM protein levels in these cells indicates that these patients suffer from greatly diminished, if not total, loss of ATM protein function(s). Furthermore, these findings suggest that A-T carriers who are heterozygous for similar alleles may possess 2-fold reduced cellular ATM protein levels and that this may account for the phenotype reported in these individuals (3).
The observation that detectable levels of microsome-associated ATM protein were found in lymphocytes and lymphoblasts but not in fibroblasts suggests this protein may perform function(s) in lymphoblasts and lymphocytes not utilized in fibroblasts. Furthermore, this difference in ATM protein distribution may be related to the striking differences in chromosomal rearrangements seen between A-T lymphoblasts and fibroblasts (33). While we cannot rule out the hypothesis that this cytoplasmic component represents a storage pool, irradiated lymphoblasts showed no increase in the nuclear ATM protein fraction (data not shown).
Although A-T cells do not appear to be grossly defective in the kinetics of DNA double-strand break (DSB) repair (34), their radiosensitivity and chromosomal instability indicate a defect in appropriate response to genomic damage. This is supported by studies which demonstrate the presence of elevated levels of both unrepaired DSBs and chromosomal breaks in A-T cells (35, 36). In addition to these defects, A-T cells fail to respond to DNA damage by activating cell cycle checkpoints which normally function during G1, S, and G2 (2, 26). This is thought to stem, in part, from faulty up-regulation of cellular p53 levels following genome damage (7, 13, 14). These observations have led to the view that the ATM gene product participates in either sensing DSBs and/or triggering a signal transduction pathway that insures the fidelity of DSB repair and halting cell cycle advance following genome damage.
Given the rapidity of cellular response to genome damage [i.e., mitotic fractions are generally reduced by 80–90% in normal fibroblasts within 1 h after irradiation (37)] one would expect that both the damage sensing mechanisms and immediate downstream elements of the signal transduction pathway which respond to DSB would be constitutively present within the cell. Our finding that the ATM protein is located in the nucleus during all phases of the cell cycle indicates that it is appropriately located to perform an omnipresent genome surveillance function. Furthermore, the observation that p53 levels are up-regulated following γ-irradiation without a coordinate increase in ATM protein indicates that the cell does not require the time consuming constraint of synthesizing nascent ATM protein to elicit an effective response to radiation-induced genomic damage. Thus, it seems likely that the ATM protein undergoes posttranslational modification(s) and/or forms damage-specific complex(es) required to activate p53-dependent checkpoints. Similarly, G2 delay by RAD9 does not require protein synthesis but is mediated through a rapid posttranslational control (38).
Genomic damage does not solely stem from external forces such as radiation exposure, but may also arise from gene rearrangements, recombination and replication errors during the course of normal DNA metabolism. Furthermore, insufficient monitoring of genome fidelity would be predicted to result in high rates of chromosomal aberrations. Indeed, high levels of genomic instability have been noted in A-T patient lymphocytes, a cell type that undergoes routine recombination as a function of normal cell physiology and is likely to be a major contributing factor to the high incidence of leukemias and lymphomas observed in A-T patients (39). Moreover, elevated rates of thymic lymphomas that contain chromosomal anomalies have been observed in ATM deficient mice (40, 41). These mice also lack germ cells, consistent with an essential role of the ATM protein in the proper development of cells that, in this case, routinely undergo recombination during the process of meiosis.
The pathway involved in the recognition of DNA DSBs is not well characterized. Recent studies, however, have shown that the Ku autoantigen recognizes and binds to ends of double-stranded DNA and is able to recruit DNA-PKcs to form a DNA-dependent protein kinase complex important in the repair of DNA damage (42). DNA-PKcs, just like the ATM protein, belongs to the family of phosphatidylinositol 3 kinase-like proteins, and mice that lack functional DNA-PKcs show phenotypes that partially overlap those of ATM-deficient mice (43). It is plausible, since the ATM protein is located in the nucleus, that a potential direct interaction of the ATM protein with damaged DNA is modulated by a yet to be identified Ku-like molecule. In support of this possibility, the ATM protein was recently found to colocalize with meiotic chromosomes (44).
Another unresolved issue is the substrate specificity of the ATM protein’s putative kinase function. For example, the phosphatidylinositol 3 kinase-like family member DNA-PKcs has been found to have protein kinase activity but undemonstrable lipid kinase activity (45), and the ATM yeast homologs TEL1 and MEC1 were recently found to phosphorylate RAD53 (46). Whether the putative kinase activity of the ATM protein is to act as a protein and/or as a phospholipid kinase in a signaling mechanism remains unanswered.
In summary, our findings of constitutive expression and nuclear localization of the ATM protein are consistent with its potential role in choreographing appropriate cellular responses to genomic damage. Identification of proteins that functionally interact with the ATM protein and that are substrates for the putative kinase function following DNA damage should lead to a greater understanding of the role of the ATM protein in the maintenance of genome integrity.
Acknowledgments
We are grateful to Dr. Gilbert Lenoir (International Agency for Research on Cancer, Lyon, France) for providing us with the IARC12/AT3 cell line. We are also grateful to Dr. Duane Compton (Dartmouth Medical School, Hanover, NH) for providing us anti-NuMA antisera. Special thanks go to Dr. Anthony Wynshaw-Boris (National Human Genome Research Institute, National Institutes of Health) for critical reading of the manuscript and to Deborah Mosbrook for her technical support. Work in the laboratory of Y.S. was supported by research grants from the A-T Medical Research Foundation, The A-T Children’s Project, The U.S.–Israel Binational Science Foundation, and the National Institute of Neurological Disorders and Stroke (NS31763). This work is dedicated to A-T patients and their families.
ABBREVIATIONS
- A-T
ataxia-telangiectasia
- FCS
fetal calf serum
- GST
glutathione S-transferase
- NCS
neocarzinostatin
- DSB
double-strand break
References
- 1.Sedgwick R P, Boder E. In: Handbook of Clinical Neurology. Vinken P J, Bruyn G W, editors. Amsterdam: North-Holland; 1972. pp. 267–339. [Google Scholar]
- 2.Shiloh Y. Eur J Hum Genet. 1995;3:116–138. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- 3.Swift M, Morrell D, Massey R B, Chase C L. New Eng J Med. 1991;325:1831–1836. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 4.Easton D F. Int J Radiat Biol. 1994;66:S187–S182. doi: 10.1080/09553009414552011. [DOI] [PubMed] [Google Scholar]
- 5.Painter R B, Young B R. Proc Natl Acad Sci USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell L M, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 7.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 8.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 9.Livingston L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 10.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1024. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 13.Khanna K K, Lavin M F. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 14.Canman C E, Wolff A C, Chen C-Y, Fornace A J, Kastan M B. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 15.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 16.Savitsky K, Sfez S, Tagle D A, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 17.Zakian V A. Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 18.Abrams H D, Rohrschneider L R, Eisenmann R N. Cell. 1982;29:427–439. doi: 10.1016/0092-8674(82)90159-3. [DOI] [PubMed] [Google Scholar]
- 19.Brown K D, Coulson R M R, Yen T J, Cleveland D W. J Cell Biol. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Nature (London) 1970;337:650–655. [Google Scholar]
- 21.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blose D H, Meltzer D I, Feramisco J R. J Cell Biol. 1984;98:847–858. doi: 10.1083/jcb.98.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaglio T, Saredi A, Compton D A. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilad S, Khosravi R, Shkedy D, Uziel T, Ziv Y, et al. Hum Mol Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 25.Byrd P J, McConville C M, Cooper P, Parkhill J, Stankovic T, McGuire G M, Thick J A, Taylor A M R. Hum Mol Genet. 1996;5:145–149. doi: 10.1093/hmg/5.1.145. [DOI] [PubMed] [Google Scholar]
- 26.Meyn M S. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- 27.Shiloh Y, Tabor E, Becker Y. Carcinogenesis. 1982;3:815–820. doi: 10.1093/carcin/3.7.815. [DOI] [PubMed] [Google Scholar]
- 28.Silver P A. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 29.Baumann B, Potash M J, Kohler G. EMBO J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehrman M A, Schneider W J, Brown M S, Davis C G, Elhammer A, Russell D W, Goldstein J L. J Biol Chem. 1987;262:401–410. [PubMed] [Google Scholar]
- 31.Brody L C, Mitchell G A, Obie C, Michaud J, Steel G, Fontaine G, Robert M-F, Sipila I, Kaiser-Kupfer M, Valle D. J Biol Chem. 1992;267:3302–3307. [PubMed] [Google Scholar]
- 32.Pakula A A, Sauer R T. Annu Rev Genet. 1989;23:289–310. doi: 10.1146/annurev.ge.23.120189.001445. [DOI] [PubMed] [Google Scholar]
- 33.Kojis T L, Schreck R R, Gatti R A, Sparkes R S. Hum Genet. 1989;83:347–352. doi: 10.1007/BF00291379. [DOI] [PubMed] [Google Scholar]
- 34.Ganesh A, North P, Thacker J. Mutat Res. 1993;299:251–259. doi: 10.1016/0165-1218(93)90101-i. [DOI] [PubMed] [Google Scholar]
- 35.Cornforth M N, Bedford J S. Radiat Res. 1985;111:385–405. [PubMed] [Google Scholar]
- 36.Pandita T K, Hittleman W N. Radiat Res. 1992;131:214–223. [PubMed] [Google Scholar]
- 37.Zampetti-Bosseler F, Scott D. Int J Radiat Biol. 1981;39:547–558. doi: 10.1080/09553008114550651. [DOI] [PubMed] [Google Scholar]
- 38.Weinert T A, Hartwell L H. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor A M R, Metcalfe J A, Thick J, Mak Y-F. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 40.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J, Reid T, Tagle D, Wynshaw-Boris A. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Ashley T, Brainerd E E, Bronson R T, Meyn M S, Baltimore D. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 42.Anderson C W, Lees-Miller S P. Crit Rev Eukaryotic Gene Expression. 1992;2:283–314. [PubMed] [Google Scholar]
- 43.Blunt T, Finnie N J, Taccioli G E, Smith GCM, Demengeot J, Gottleib T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 44.Keegan K S, Holtzman D A, Plu A W, Christenson E R, Brainerd E E, Flaggs G, Bentley N J, Taylor E M, Meyn M S, Moss S B, Carr A M, Ashley T, Hoekstra M F. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 45.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]