Abstract
Potassium (K+) is an essential nutrient for plant growth and development. Plants often adapt to low K+ conditions by increasing their K+ uptake capability. Recent studies have led to the identification of a calcium signaling pathway that enables plants to act in this capacity. Calcium is linked to two calcineurin B-like calcium sensors (CBLs) and a target kinase (CBL-interacting protein kinase 23 or CIPK23) that, in turn, appears to phosphorylate and activate the potassium channel, Arabidopsis K+ transporter 1 (AKT1), responsible for K+ uptake in roots. Here, we report evidence that this regulatory mechanism is more elaborate than earlier envisaged. The recently described pathway is part of an extensive network whereby several CBLs interact with multiple CIPKs in the activation of the potassium channel, AKT1. The physical interactions among the CBL, CIPK, and AKT1 components provide a mechanism for specifying the members of the CBL and CIPK families functional in AKT1 regulation. The interaction between the CIPKs and AKT1 was found to involve the kinase domain of the CIPK component and the ankyrin repeat domain of the channel. Furthermore, we identified a 2C-type protein phosphatase that physically interacts and inactivates the AKT1 channel. These findings provide evidence that the calcium-sensitive CBL and CIPK families together with 2C-type protein phosphatases form a protein phoshporylation/dephosphorylation network that regulates the AKT1 channel for K+ transport in plants.
Keywords: calcium sensor, protein kinase, protein phosphatase, signal transduction
As the most abundant cation in a living plant cell, K+ is central to the fundamental processes of growth, development, maintenance of turgor pressure, and plasma membrane polarization (1–4). Several different families of transporters function in the uptake, tissue distribution, and intracellular storage of K+ (5, 6). The Shaker-like K+ channels, which function in K+ nutrition in Arabidopsis, consist of nine members that are divided into several groups based on their biophysical properties and structure (5, 6). The Arabidopsis K+ transporter (AKT) family (AKT1, AKT2/3, AKT5, AKT6/SPIK) and the K+ transporters of Arabidopsis thaliana (KAT)-type (KAT1, KAT2, KC1) channels are believed to be inward rectifying K+ channels active in the uptake of K+ (7–12). The outward K+ channels include the stellar K+-outward rectifying (SKOR) and guard cell outward rectifying K+ (GORK) channels that display high sequence homology but are expressed in different cell types in Arabidopsis (6, 13, 14). Shaker-type channels of plants are tetrameric structures composed of identical or related subunits that display a typical six-transmembrane segment and a pore domain together with putative cyclic nucleotide-binding and KHA domains at the C terminus (15, 16). The AKT, SKOR, and GORK channels all contain an ankyrin repeat domain that is absent in KAT1, KAT2, and KC1 (5, 17, 18).
Shaker-like channels appear to play different roles in K+ nutrition. Thus, although AKT1 has been shown to function in plant growth under low K+, the SKOR channel is responsible for xylem loading and long-distance root-shoot transport (7, 13). The AKT2 channel, on the other hand, seems to participate in K+ loading in the phloem (9), and AKT6 (also named SPIK) acts in K+ uptake by germinating pollen grains (10).
Despite extensive studies on the biophysical properties of the channels, relatively little is known about the mechanisms that govern their function and regulation under physiological conditions. According to recent studies (19, 20), one Shaker-type channel (AKT1) is regulated by a calcium-dependent signaling pathway involving calcineurin B-like calcium sensors (CBLs) and a target kinase. The CBL–CBL-interacting protein kinase (CIPK) network has been recently established as a major signaling mechanism in plant response to environmental cues (21). Genetic analysis combined with electrophysiological studies demonstrated that CBL1 and CBL9 and their common target kinase (CIPK23) form alternative complexes located at the plasma membrane where they jointly activate the AKT1 channel, thereby increasing K+ uptake capacity. Current results suggest that a complex network involving multiple CBLs and CIPKs may cooperate in the regulation of AKT1 activity. Such a network is difficult to study in planta with a genetic approach. Consequently, we have taken advantage of heterologous model systems, namely, yeast two-hybrid and Xenopus oocyte patch-clamping, to illustrate principles of AKT1 regulation by the CBL–CIPK network. We now report results that give insight into how CBL–CIPK regulates the AKT1 potassium channel. In particular, we have found that the physical interaction among the CBL–CIPK–AKT components determines the specificity of CBL–CIPK complexes that function in AKT1 regulation. In addition, a 2C-type protein phosphatase (PP2C) was identified to counteract the CIPKs, forming a kinase/phosphatase partnership that enables AKT1 activity to be switched on and off. The finding that individual members of the protein families functional in the network interact specifically with one another would seem to provide flexibility that enables the plant to adjust and survive in response to changing environmental conditions.
Results and Discussion
Multiple CBLs Associate with Multiple CIPKs That then Interact with AKT1.
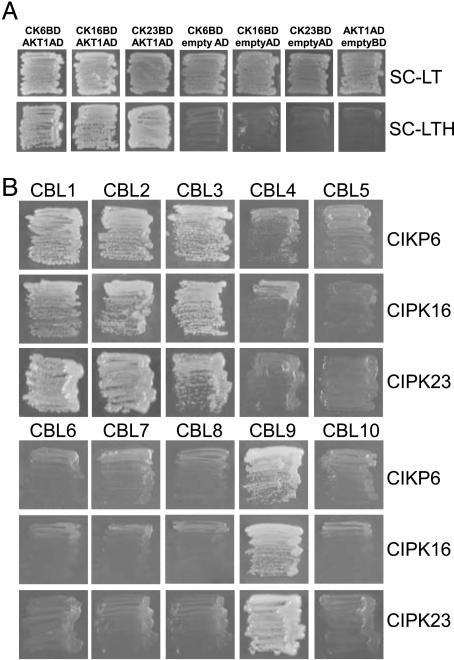
The finding that AKT1 is a target for a CBL–CIPK pathway has uncovered a mechanism for the regulation of a plant K+ channel by protein phosphorylation (19, 20). As both CBL and CIPK families consist of multiple members that can interact to form an integrated CBL–CIPK network, we suspected that the regulation of AKT1 by the pathway recently identified may be more complex than thought earlier (19, 20). The finding that AKT1 activity was not completely eliminated in either the cipk23 single mutant or the cbl1cbl9 double mutant was consistent with this possibility (19). To examine this point further, we applied a yeast two-hybrid assay to probe the interaction between AKT1 and each of the 25 CIPKs. These experiments revealed that, in addition to CIPK23, at least two other CIPKs, CIPK6 and CIPK16, interacted with the C terminus of AKT1 (Fig. 1A). However, among CIPK family members, CIPK6, CIPK16, and CIPK23 are not the most related in overall protein sequence. For instance, CIPK3 and CIPK9 are the closest homologues to CIPK23, yet these two CIPKs failed to interact with AKT1 (data not shown).
Fig. 1.
Yeast two-hybrid assay of interactions among CBLs, CIPKs, and AKT1. (A) CIPK6, CIPK16, and CIPK23 interacted with AKT1 C-terminal domain. Yeast strain PJ69-4A was transformed with 25 CIPKs cloned into pGBT9 and ATK1 cytoplasmic C-terminal domain cloned into pGADGH. (Lower) Growth on the selection medium [SC-leucine-tryptophan-histidine (SC-LTH) including 2 mM 3′-AT] was used as indicator for interaction. (Upper) Growth in SC-leucin-tryptophan (SC-LT) was used as control. (B) CBL1, CBL2, CBL3, and CBL9 interacted with CIPK6, CIPK16, and CIPK23. CBLs were cloned into pGADGH, and CIPKs were cloned into pBGKT9. These clones were transformed into yeast PJ69-4A strain and selected on SC-LTH medium supplemented with 2 mM 3′-AT.
The three CIPKs physically associated with AKT1 should interact with a set of CBLs to provide CBL–CIPK complexes for regulating AKT1. To identify these partners, we performed yeast two-hybrid assays using the three CIPKs and all CBLs. Interestingly, despite the low overall sequence similarity in the three CIPK proteins, they interacted with the same members of the CBL family, including CBL1, CBL2, CBL3, and CBL9 (Fig. 1B). CBL1 and CBL9 share high sequence identity, while CBL2 and CBL3 are also highly homologous to each other. This result possibly suggests that, after activation by the four calcium-sensing proteins, these three CIPKs function in the same signal transduction pathway.
CBL–CIPK–AKT1 Physical Interactions Define the Specificity of the CBL-CIPK Complexes That Activate AKT1 Channel.
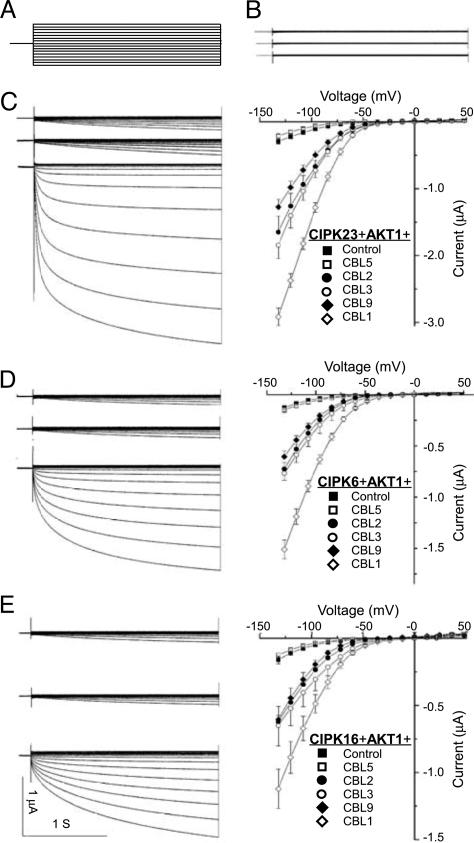
The assays described above led to the identification of four CBLs and three CIPKs forming multiple complexes that interacted with AKT1. Such an interaction network may be functionally relevant to the regulation of AKT1 activity. To test this idea, we coexpressed AKT1 with various combinations of the CBL–CIPK complexes in Xenopus oocytes and measured AKT1 activity. Consistent with the earlier studies (19, 20), the CBL1–CIPK23 duplex strongly activated AKT1 (Fig. 2C). Like CIPK23, CIPK6 and CIPK16 both activated AKT1 in a CBL1-dependent manner, although the magnitude of AKT1 activity was lower than with CIPK23 (Fig. 2 D and E). In this experiment, we also tested the effect of CIPK1 and found that AKT1 was not activated by the CIPK1–CBL1 combination (Fig. 2B). We noted that CIPK1 interacted with CBL1 but did not physically interact with AKT1 (data not shown) (22).
Fig. 2.
AKT1 channel is activated by multiple CIPKs interacting with multiple CBLs in the Xenopus oocytes. (A) Two-electrode voltage-clamp protocols. Voltage steps of 48 to −132 mV (in 12-mV decrements, 2.25-s duration) were applied from a 0.25-s prepulse to −40 mV. (B) CIPK1 that does not interact with AKT1 did not activate AKT1 after coexpression with CBL1 and AKT1. Typical whole-cell current traces were recorded in cells injected with cRNA of, from top to bottom, AKT1, AKT1+CIPK1, and AKT1+CIPK1+CBL1. (C) CIPK23 interacts with multiple CBLs to activate AKT1 channel activity. (Left) Currents were recorded from oocytes injected with cRNA of, from top to bottom, AKT1+CIPK23, AKT1+CIPK23+CBL5, and AKT1+CIPK23+CBL1. (Right) The current–voltage relationship is shown for AKT1 (Control) or AKT1+CIPK23 and each of the CBLs (including CBL5, CBL2, CBL3, CBL9, and CBL1). (D) CIPK6 interacts with multiple CBLs to activate AKT1 channel. The presentation is the same as in B except that the CIPK is now CIPK6. (E) CIPK16 interacts with multiple CBLs to activate AKT1 channel. The presentation is the same as in B except that the CIPK is now CIPK16. The current value data shown in C–E Right were at 0.9 s of each voltage-clamp episode and presented as means ± SE, with n > 6 in each case.
In addition to CBL1, we tested whether the two other CBLs (CBL2 and CBL3) that interacted with the three CIPKs (CIPK23, CIPK6, and CIPK16) enhanced the activation of AKT1 by these CIPKs. To this end, we coexpressed AKT1 in oocytes with each of the possible combinations formed by CBL1, CBL2, CBL3, CBL5, or CBL9 and the three CIPKs found to interact with AKT1. As shown in Fig. 2, CBL5, a component that does not interact with these CIPKs, had no effect on the level of AKT1 activation (Fig. 2 C–E). However, CBL1, CBL2, CBL3, and CBL9 all dramatically enhanced the magnitude of AKT1 activation by CIPK23, CIPK6, and CIPK16 (Fig. 2 C–E). Among the CBLs tested, CBL1 produced the strongest activation, and CBL2, CBL3, and CBL9 showed similar, but lower, effects (Fig. 2 C–E). Among the three CIPKs interacting with AKT1, CIPK23 was more effective than CIPK6 or CIPK16 (Fig. 2 C–E). These results show that physical interaction with CBLs is required for the activation of CIPKs and that the interaction between CIPKs and AKT1 defines the specificity of CIPK members for AKT1 channel activation.
Ankyrin Repeat of AKT1 Serves as a “Docking” Domain for Interaction with the Kinase Domain of CIPKs.
The cytoplasmic region of AKT1 available for interaction with CIPKs consists of cAMP binding, ankyrin repeat, and KHA domains. It has been shown that these C-terminal cytosolic domains are important for the assembly of the AKT1 tetrameric channel (19), but little is known about their function in the regulation of channel activity. The finding of an AKT1–CIPK interaction indicates that at least part of the cytosolic domain of AKT1 mediates regulation of the channel by CIPKs.
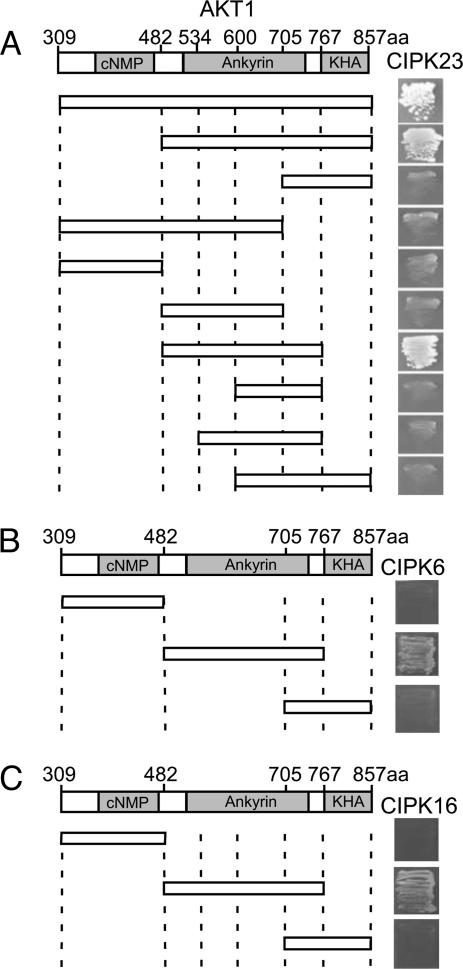
To investigate which AKT1 domain interacts with CIPK23, we made several truncated forms of the AKT1 C-terminal region and tested their interaction with CIPK23 in the yeast two-hybrid system. The analysis revealed that the ankyrin domain, which consists of six repeats, is sufficient for interaction with the kinase (Fig. 3A). Further analysis indicated that all six ankyrin repeats are required for the interaction and that deletion of any one abolished the ability to interact with CIPK23 (Fig. 3A). We conducted similar yeast two-hybrid assays with two other AKT1-interacting CIPKs (CIPK6 and CIPK16) and found that, like CIPK23, both interacted with the ankyrin repeat domain of AKT1 (Fig. 3 B and C). As the ankyrin repeat domain is a common feature of several Shaker-like K+-channel proteins in Arabidopsis, this finding suggests that other K+ channels with this domain may interact with CIPKs, thereby forming an even larger network for CBL–CIPK ion channel regulation in plants.
Fig. 3.
Comparative yeast two-hybrid interaction analysis of CIPKs with various domains of AKT1. (A) Ankyrin repeat domain of AKT1 is required and sufficient for interaction with CIPK23. (B) CIPK6 interacts with ankyrin repeat domain of AKT1. (C) CIPK16 interacts with ankyrin repeat domain of AKT1. Each of the domains of the AKT1 cytoplamic C-terminal region was cloned into the pGADGH and transformed into yeast PJ69-4A strain with each of the CIPKs cloned into pGBT9. Growth in selection media (SC-leucine-tryptophan-histidine supplemented with 2 mM 3′-AT) indicates the interaction between CIPKs and domains of AKT1.
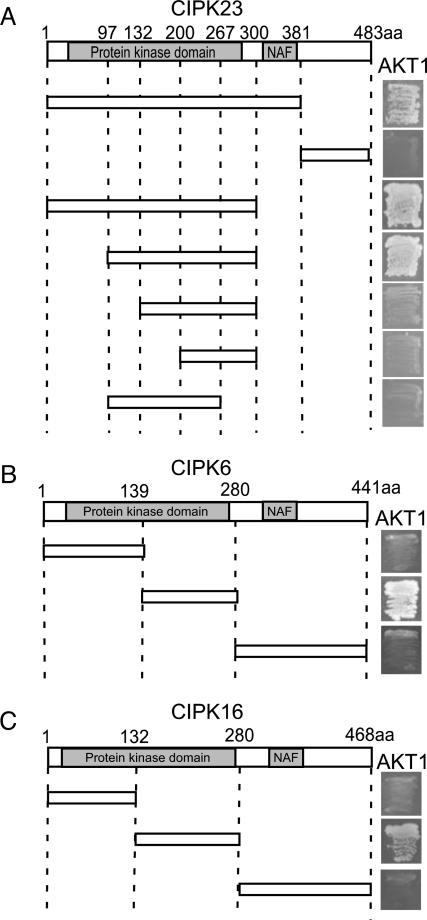
As AKT1 is one of the first physiological substrates identified for the CIPK family, it serves as a useful model system for further characterizing the kinase–substrate interaction. To this end, we dissected CIPK to identify the domain responsible for interacting with the channel protein. CIPKs consist of two major domains, the kinase and CBL-interacting domains. Using CIPK23, we found that the kinase domain interacted with the ankyrin repeat domain of the AKT1 substrate (Fig. 4A). On further analysis, we observed that the catalytic domain (the large lobe in the 3D structure) was involved in this interaction (Fig. 4A). Similarly, the kinase domain of CIPK6 and CIPK16 was responsible for their interaction with the ankyrin repeat domain of AKT1 (Fig. 4 B and C).
Fig. 4.
Comparative yeast two-hybrid interaction analysis of the catalytic domain of CIPKs with the ankyrin repeat domain of AKT1. Catalytic domain of CIPK23 (A), CIPK6 (B), or CIPK16 (C) is responsible for the interaction with the ankyrin repeat domain of AKT1.
The Myristoylation Site of the CBLs Contributes to the Activation of AKT1 by CIPKs.
Analysis of their structures suggested that CBL1 and CBL9 have a myristoylation site in the N terminus that could function to recruit these proteins and their interacting partners to the cell membranes. Both CBL1 and CBL9 have been shown to recruit CIPK23 to the plasma membrane of plant cells (20, 23). To examine the possibility that myristoylated CBLs recruit CIPKs to the plasma membrane to facilitate the interaction of CIPKs with AKT1, mutagenized forms of CBL1 and CBL9 (CBL1G2A and CBL9G2A) in which the glycine residue of the second amino acid from their N terminus was changed to alanine were expressed in the oocyte (Fig. 5A). Because this glycine is the site for myristoylation, the mutant CBLs should not be associated with cell membranes if this idea were valid. However, coexpression of CBL1G2A with CIPK23 and AKT1 still produced a significant level of inward current, ≈60% that of wild-type CBL1 (Fig. 5B). The CBL1G2A had a similar effect on currents produced by CIPK6- or the CIPK16–AKT1 complex (data not shown). The myristoylation mutant of CBL9 reduced the current produced by CIPK23–AKT1 to a similar degree (data not shown). A palmitoylation site at the cysteine residue adjacent to the myristoylated glycine was identified in the CBL1 (Fig. 5A). As palmitoylation has been shown to enhance the affinity of myristoylated proteins to the plasma membrane (24), we tested the potential role of palmitoylation by mutating the cysteine residue of CBL1G2A mutant to a glycine residue (Fig. 5A). This double mutant induced a level of AKT1 current similar to that of CBL1G2A coexpressed with CIPK23 and AKT1 (Fig. 5B). These results collectively indicated that, while contributing to the activity of CBL–CIPK complex, lipid modification of the CBLs was not absolutely required for activation of the AKT1 channel in the oocyte expression systems.
Fig. 5.
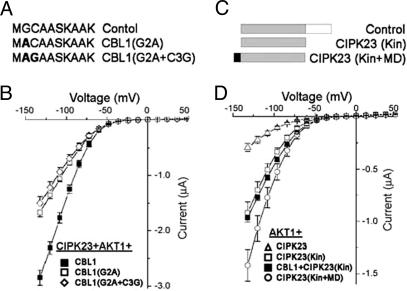
The MD of CBLs contributes to the activation of AKT1 by CBL–CIPK complexes. (A) Scheme of the amino acid sequences of CBL1 MD, from top to bottom, control CBL1, a mutant of Gly residue into Ala at the second amino acid of N-terminal CBL1 (G2A), double mutant of Gly-Ala and Cys-Gly in N-ternimal region of CBL1 (G2A+C3G). The mutant amino acids are highlighted in bold. (B) Current–voltage relationships deduced from recordings in oocytes expressing CIPK23+AKT1+CBL1, CIPK23+AKT1+CBL1 (G2A), or CIPK23+ AKT1+CBL1(G2A+C3G). (C) Scheme of, from top to bottom, full-length CIPK23 with kinase domain (shaded) and regulatory domain (open box), Control, CIPK23 kinase domain only, CIPK23(Kin), and CIPK23 kinase domain with CBL1 myristoylation site fused to the N terminus, CIPK23(Kin+MD). (D) Current–voltage relationships deduced from recordings in oocytes expressing AKT1+CIPK23, AKT1+CIPK23 (Kin), AKT1+CBL1+ CIPK23 (Kin), or AKT1+CIPK23 (Kin+MD).
To test the contribution of lipid modification and CIPK–AKT1 physical interaction in AKT1 activation, we coexpressed the truncated form of CIPK23 containing only the kinase domain with intact AKT1 (Fig. 5C). The kinase domain alone should be capable of physically interacting with AKT1 (Fig. 4) and more active than the wild type because it lacks the autoinhibitory domain in the C-terminal region. Although the full-length CIPK23 induced a low activity of AKT1, the CIPK23 kinase domain enhanced channel activity to a much greater degree (Fig. 5D). To analyze the role of myristoylation–directed membrane localization further, we constructed a plasmid to express the CIPK23 kinase domain fused with myristoylation domain (MD) derived from CBL1 (Fig. 5C) and expressed this fusion protein with the AKT1 channel in oocytes. The lipid modification of the kinase domain enhanced AKT1 activity compared with that induced with the unmodified CIPK23 kinase domain (Fig. 5D). However, the AKT1 current activated by the CBL1–CIPK23 complex was still greater than that produced with any of the other combinations (Fig. 5 B and D). We concluded that the modification of CBLs with lipids enhances CBL–CIPK activity by localizing the complex to the plasma membrane while, as shown above, the physical interaction between AKT1 and CIPKs is absolutely required for activation of the channel.
A PP2C Inactivates AKT1.
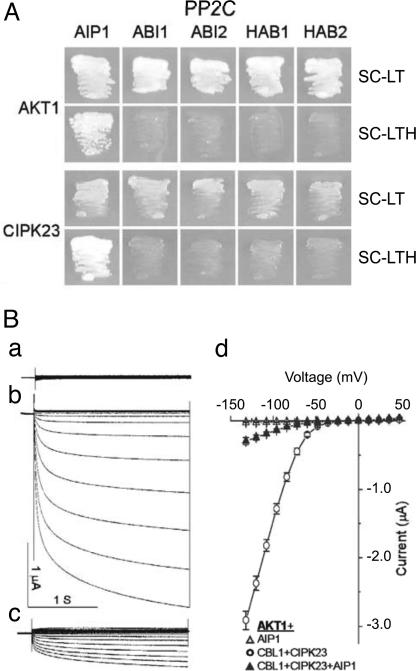
Identification of the CBL–CIPK network that regulates AKT1 activity provides a model system to study the regulation of plant ion channels by protein phosphorylation. When phosphorylation activates an ion channel in response to a signaling event, in this case calcium elevation in response to low K+, the event will be transient and countered by a signaling terminator leading to dephosphorylation, i.e., inactivation and return of the channel to the basal state. In search for a protein phosphatase that could serve as a signaling terminator, we applied the yeast two-hybrid assay to identify candidate protein phosphatases that interact directly with AKT1. Because earlier studies showed that AKT2, another AKT-type channel, interacts with PP2CA (25, 26), we focused on PP2CAs. Among the five PP2CA subfamily members tested (ABI1, ABI2, HAB1, HAB2, and a novel member, At1g07430), only the novel member interacted with AKT1 (Fig. 6A). We therefore named this PP2C as AIP1 (for AKT1-interacting PP2C 1). Because CIPKs are known to interact with PP2CAs such as ABI1 and ABI2 (27), we tested whether AIP1 and CIPK23 could interact with each other. The yeast two-hybrid results indicated that AIP1 (but not other PP2CAs) interacted with CIPK23 that also interacts with AKT1. The three components, CIPK23, AIP1, and AKT1, thus appeared to interact mutually and potentially form a molecular complex.
Fig. 6.
The PP2C AIP1 interacts with AKT1 and CIPK23 and inhibits AKT1 activity. (A) Yeast two-hybrid assay of AKT1 and CIPK23 interacting with various PP2Cs. Yeast strain PJ69-4A was transformed with various PP2Cs in the pGAD.GH vector and AKT1 and CIPK23 in the pGBT9 vector. The growth in selection medium indicates protein–protein interaction. SC-LT, SC minus leucine and tryptophan; SC-LTH, SC minus leucine, tryptophan, and histidine. (B) Modulation of CBL1–CIPK23–AKT1 currents by AIP1. (a) AKT1+AIP1. (b) AKT1+CBL1+CIPK23. (c) AKT1+CBL1+CIPK23+AIP1. (d) The current–voltage relationships recorded from oocytes expressing AKT1+AIP1, AKT1 +CBL1+CIPK23, or AKT1+CBL1+CIPK23+ AIP1. The current values were at 0.9 s of each voltage-clamp episode and presented as means ± SE (n > 7).
If AIP1 functions as an active AKT1 phosphatase in a manner opposing a corresponding kinase (CIPK23), AIP1 could potentially modify AKT1 channel activity. To test this possibility, we coexpressed AIP1 and each of the other PP2CAs tested above in the yeast two-hybrid system with AKT1 and CBL1–CIPK23 in the oocyte system. Fig. 6B shows that AIP1 significantly reduced AKT1 activity, suggesting that the phosphatase functions as a negative regulator of the channel by counteracting activity of the CBL–CIPK complexes. Other PP2CA members (ABI1, ABI2, HAB1, HAB2, and PP2CA) had no effect on AKT1 current (data not shown). These results suggested that the CIPK kinases and a PP2C (AIP1) physically interact with the AKT1 channel protein and modify its activity by phosphorylation and dephosphorylation.
Concluding Remarks
Recent studies have identified a number of key functions of the CBL–CIPK network in signaling environmental changes in plants. The network consists of at least 10 CBL and 25 CIPK members that act in combination with one another. Different pair combinations seem to participate in specific pathways with perhaps an overlap of function (21, 28–33). In the present study, we found that at least four CBLs and three CIPKs form 12 different CBL–CIPK complexes, each able to activate the voltage-gated K+ channel AKT1. Although the importance of the network in planta awaits confirmation by genetic analysis, the current results indicate a previously unsuspected degree of overlap and complexity in channel regulation by the CBL–CIPK complexes. Key to this regulation is the newly found physical interaction of the CIPKs with AKT1, a factor that seems to be important in determining the specificity of channel activation. To this end, the ankyrin repeat domain in the AKT1 protein appears to serve as a docking element for the kinase domain of the CIPK partner, thereby determining which specific CIPKs interact and attach to the AKT1 channel protein (Fig. 7). Modification of CBLs by lipids also contributes to the recruiting of specific CBL–CIPKs to the cell membrane. This aspect, however, seems less important than the interaction between CIPK and AKT1.
Fig. 7.
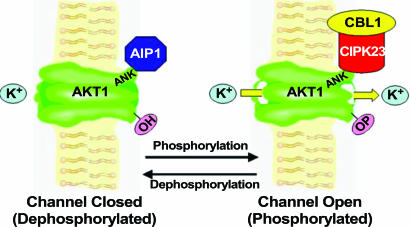
A working model for the regulation of AKT1 by CIPK(s) and PP2C(s). The AKT1 channel is activated when interacting with CIPK23–CBL1 (possibly through phosphorylation by CIPK23) (channel open). When AIP1 interacts with AKT1, the channel protein is believed to be dephosphorylated and inactivated (channel closed).
The finding that a specific PP2C, AIP1, inactivates AKT1 provides the other side of regulation, namely, channel inactivation by dephosphorylation (Fig. 7). Further biochemical analysis will illustrate whether AIP1 dephosphorylates the AKT1 channel protein at the same site(s) phosphorylated by the CIPKs. It is of particular interest that CIPK23 and AIP1 interact with each other and that both interact with AKT1. It is possible that the CIPK23 kinase and AIP1 phosphatase interact with AKT1 interchangeably to regulate the channel, or, alternatively, that the three components form an ensemble that regulates channel activity as a function of the relative activity of the kinase and phosphatase components. Future pursuit of these newly opened paths should lead to a greater understanding of the signaling pathways that regulate ion transport in plants.
Materials and Methods
Construction of Plasmids for Yeast Two-Hybrid and Oocyte Analysis.
The truncated AKT1 activation domain (AD) constructs were generated by PCR using different pairs of primers. Each of the PCR products was digested with EcoRI and SalI and cloned into pGAT7Rec vector. To create truncated CIPK23, CIPK6, and CIPK16, PCR was conducted with different pairs of primers. Each of the PCR products was digested with NdeI and SalI and cloned into a pGBKT7 vector. To express the proteins in oocytes for electrophysiological experiments, we cloned CBL2, CBL3, CBL5 and CIPK1, CIPK6, CIPK16, the kinase domains of CIPK23, and a full-length CIPK23 into the pGEMHE vector. Myristoylation and palmitoylation mutants of CBLs and the kinase domain with myristoylation site fusion were produced by using PCR-based mutagenesis and cloned into pGEMHE. The CBL1, CBL9, AKT1, and CIPK23 constructs for expression in Xenopus oocytes have been described (19).
Yeast Two-Hybrid Analysis.
The binding domain (BD) vector constructs and the AD vector constructs were used for yeast two-hybrid analysis in appropriate pairs described in the figure legends. To introduce AD and BD plasmids into yeast strain PJ69-4A, a lithium acetate-mediated transformation method was used. Transformants were selected on synthetic complete agar medium (SC)-leucin-tryptophan medium and then transferred to the interaction selection media (SC-leucine-tryptophan-histidine supplemented with 1.5 mM 3-amino-1,2,4-aminotriazole) to score growth as an indicator of protein–protein interaction (19).
Electrophysiological Procedures.
The concentration of the capped cRNA was determined by A260/A280 and adjusted to a final concentration of 0.5 μg/μl, except for that of AIP1 cRNA (1.5 μg/μl). The oocytes were bathed with a solution containing 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, and 10 mM Hepes (pH adjusted to 7.3 with 1 M KOH) and perfused with the same solution. The pipette solution contained 3 M KCl. Two-electrode voltage clamp recordings were performed to measure the activation and deactivation kinetics of currents, voltage steps of 48 to −132 mV (in 12-mV decrements, 2.25-s duration) were applied from a 0.25-s prepulse to −40 mV. Histograms were generated from the pooled current reached after 0.9 s of each voltage-clamp episode. Statistical analyses were performed by using either Student's paired test or unpaired t tests as appropriate, and differences were considered to be significant at P < 0.05. Data are presented as representative recordings or as mean ± SE of n observations, in which n is the number of samples.
Acknowledgments
We thank Dr. E. Y. Isacoff (University of California, Berkeley) for providing oocytes used in this study. S.C.L. was supported in part by Korea Research Foundation Grant KRF-2006-214-F00001 funded by the Korean Ministry of Education and Human Resource Development. This research was supported by grants from the National Science Foundation (to S.L.) and the U.S. Department of Agriculture (to B.B.B. and S.L.).
Abbreviations
- CBL
calcineurin B-like calcium sensor
- CIPK
CBL-interacting protein kinase
- PP2C
2C-type protein phosphatase
- AKT
Arabidopsis K+ transporter
- KAT
K+ transporters of Arabidopsis thaliana
- MD
myristoylation domain
- AIP1
AKT1-interacting PP2C 1
- SC
synthetic complete agar medium
- AD
activation domain
- BD
binding domain.
Footnotes
The authors declare no conflict of interest.
References
- 1.Leigh RA, Jones RGA. New Phytol. 1984;97:1–13. [Google Scholar]
- 2.Kochian LV. In: Biochemistry and Molecular Biology of Plants. Buchanan B, Gruissem W, Jones R, editors. Rockville, MD: Am Soc Plant Biol; 2000. pp. 1204–1249. [Google Scholar]
- 3.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 4.Very AA, Sentenac H. Annu Rev Plant Biol. 2003;54:575–603. doi: 10.1146/annurev.arplant.54.031902.134831. [DOI] [PubMed] [Google Scholar]
- 5.Schachtman DP. Biochim Biophys Acta. 2000;1465:127–139. doi: 10.1016/s0005-2736(00)00134-6. [DOI] [PubMed] [Google Scholar]
- 6.Very AA, Sentenac H. Trends Plants Sci. 2002;7:168–175. doi: 10.1016/s1360-1385(02)02262-8. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch RE, Lewis BD, Spalding EP, Sussman MR. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 8.Marten I, Hoth S, Deeken R, Ache P, Ketchum KA, Hoshi T, Hedrich R. Proc Natl Acad Sci USA. 1999;96:7581–7586. doi: 10.1073/pnas.96.13.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud JB. Plant Cell. 2000;12:837–851. doi: 10.1105/tpc.12.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouline K, Very AA, Gaymard F, Boucherez J, Pilot G, Devic M, Bouchez D, Thibaud JB, Sentenac H. Genes Dev. 2002;16:339–350. doi: 10.1101/gad.213902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R. Proc Natl Acad Sci USA. 2002;99:4079–4084. doi: 10.1073/pnas.052677799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippar K, Ivashikina N, Ache P, Christian M, Luthen H, Palme K, Hedrich R. Plant J. 2004;37:815–827. doi: 10.1111/j.1365-313x.2003.02006.x. [DOI] [PubMed] [Google Scholar]
- 13.Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud JB, Sentenac H. Cell. 1998;94:647–655. doi: 10.1016/s0092-8674(00)81606-2. [DOI] [PubMed] [Google Scholar]
- 14.Becker D, Hoth S, Ache P, Wenkel S, Roelfsema MR, Meyerhoff O, Hartung W, Hedrich R. FEBS Lett. 2003;554:119–126. doi: 10.1016/s0014-5793(03)01118-9. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann S, Sentenac H. Curr Opin Plant Biol. 1999;2:477–482. doi: 10.1016/s1369-5266(99)00020-5. [DOI] [PubMed] [Google Scholar]
- 16.Pilot G, Pratelli R, Gaymard F, Meyer Y, Sentenac H. J Mol Evol. 2003;56:418–434. doi: 10.1007/s00239-002-2413-2. [DOI] [PubMed] [Google Scholar]
- 17.Daram P, Urbach S, Gaymard F, Sentenac H, Cherel I. EMBO J. 1997;16:3455–3463. doi: 10.1093/emboj/16.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demidchik V, Davenport RJ, Tester M. Annu Rev Plant Biol. 2002;53:67–107. doi: 10.1146/annurev.arplant.53.091901.161540. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Kim BG, Cheong YH, Pandey GK, Luan S. Proc Natl Acad Sci USA. 2006;103:12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. Plant Cell. 2002;14(Suppl):S389–S400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Angelo C, Weinl S, Batistic O, Pandey GK, Cheong YH, Schultke S, Albrecht V, Ehlert B, Schulz B, Harter K, et al. Plant J. 2006;48:857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- 23.Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim B-G, Lee SC, Kudla J, Luan S. Plant J. 2007 doi: 10.1111/j.1365-313X.2007.03236.x. in press. [DOI] [PubMed] [Google Scholar]
- 24.Navarro-Lerida I, Alvarez-Barrientos A, Gavilanes F, Rodriguez-Crespo I. J Cell Sci. 2002;115:3119–3130. doi: 10.1242/jcs.209973. [DOI] [PubMed] [Google Scholar]
- 25.Vranova E, Tahtiharju S, Sriprang R, Willekens H, Heino P, Palva ET, Inze D, Van Camp W. J Exp Bot. 2001;52:181–182. [PubMed] [Google Scholar]
- 26.Cherel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB. Plant Cell. 2002;14:1133–1146. doi: 10.1105/tpc.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta M, Guo Y, Halfter U, Zhu JK. Proc Natl Acad Sci USA. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KN, Cheong YH, Gupta R, Luan S. Plant Physiol. 2000;124:1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batistic O, Kudla J. Planta. 2004;219:915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- 33.Gong D, Guo Y, Schumaker KS, Zhu JK. Plant Physiol. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]