Investigations into the biosynthesis of natural compounds such as antibiotics and other drugs have made major contributions to our understanding of complex biological reaction schemes and novel chemical mechanisms. The formation of many of these complicated natural compounds often occurs while tethered to peptidyl carrier protein (PCP) domains or in polyketide synthase (PKS) multidomain synthetic units where synthesis can be thought of as occurring somewhat like solid-phase methods used by organic chemists. Many unusual biological reactions have been characterized, including halogenations by flavin-dependent enzymes and by non-heme iron-containing enzymes, oxidative formation of carbon–carbon bonds by using P450 systems, and diverse stereo- and regiospecific reactions that are very difficult to carry out in the laboratory, especially at mild temperatures and pH values. One diverse class of natural compounds is the indolocarbazoles that have been found to have a broad spectrum of biological effects, leading to their potential use in antibacterial, antifungal, and antitumor treatments (1). In fact, several of these compounds are currently being clinically tested for their efficacy in treating cancer and other diseases (1). Tryptophan is used as the starting compound for many indolocarbazoles (2, 3). Recently, substantial progress has been made in understanding the detailed pathways for the biosynthesis of rebeccamycin and its close relative staurosporine (4–15). Rebeccamycin is an antitumor agent that binds to DNA–topoisomerase I complexes to prevent replication of DNA (16). It is therefore a promising candidate for antitumor therapies. The highly efficient biosynthetic pathway for rebeccamycin consists of a small number of soluble enzymes (7) that catalyze the reactions to form the basic aglycone scaffold from tryptophan, as well as the formation of N-glycosidic bonds to the indole nitrogens of the aglycone unit (Fig. 1A). The overall conversion of tryptophan to rebeccamycin consists of several unique biochemical transformations; it involves RebH and RebF, a two-component flavin-dependent enzyme system that halogenates tryptophan (10, 11), a flavoenzyme amino acid oxidase, RebO (12), a cytochrome P450 (RebD) that catalyzes the oxidative initial linking of two tryptophan skeletons (6, 13, 14), and a second cytochrome P450, RebP, that participates with a putative flavoprotein, RebC, to form the basic aglycone scaffold (Fig. 1A) (7, 15). However, until now, the particular reactions that RebP and RebC individually catalyze were a complete mystery. The work of Ryan et al. (17) in a recent issue of PNAS has provided new insights into this chemical conversion. Ryan et al. used x-ray crystallography to show that RebC has all of the characteristics of a single-component flavoprotein hydroxylase. In addition, they have found by some unusually astute crystallographic insights that the enzyme, while in its crystalline form, serendipitously “purifies” and binds a particular trace compound from a potential substrate mixture. This bound impurity suggests the nature of the true substrate and how RebP and RebC might cooperate to catalyze the formation of the aglycone of rebeccamycin.
Fig. 1.
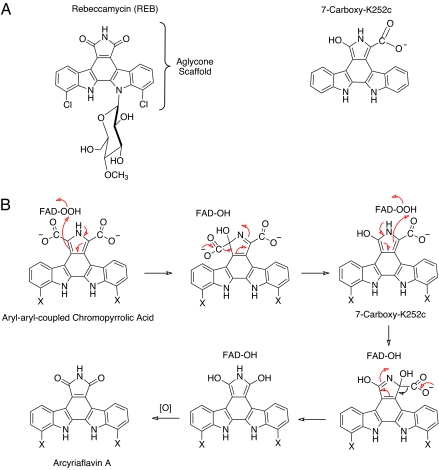
Structures of rebeccamycin and 7-carboxy-K252c, and postulated mechanism for converting aryl-aryl-coupled chromopyrrolic acid to arcyriaflavin A. (A) Structures of rebeccamycin and the molecule, 7-carboxy-K252c, bound to the active site of RebC. Rebeccamycin is halogenated as shown, but for simplicity the molecules used in the studies were not. Hydrogen was substituted. The P450 enzymes StaP and RebC readily accept the nonhalogenated derivatives as substrates (7). (B) Postulated reaction scheme for converting the aryl-aryl chromopyrrolic acid to the rebeccamycin precursor arcyriaflavin A. FAD-OOH represents a C(4a)-hydroperoxy-FAD, which is the oxygenating agent involved in flavin-dependent hydroxylations. In rebeccamycin, X is chloride. In the studies reported in ref. 17, X is hydrogen.
The crystallographic studies showed that RebC has a bound flavin adenine dinucleotide (FAD), a derivative of vitamin b2, and that this cofactor is in an OUT position exposed to solvent when no substrate is present and in a solvent-inaccessible IN position when a substrate-like molecule is bound. This property is a characteristic of the class A flavoprotein hydroxylases that typically hydroxylate activated aromatic compounds in biodegradative metabolism (18). The OUT position may facilitate the access of the large substrate to the active site and to the reductant NADPH, whereas the IN position protects reactive oxygen adduct intermediates of the FAD from solvent (19). Overall, the structure resembles that of phenol hydroxylase (20), m-hydroxybenzoate hydroxylase (21), and p-hydroxybenzoate hydroxylase (22), the latter of which is the prototypic, extensively studied flavin-dependent hydroxylase. Because the true substrate for RebC was a mystery, Ryan et al. (17) soaked the crystals of RebC with chromopyrrolic acid, the substrate for the combined steps that RebP and RebC catalyze. When they determined the structure of RebC after soaking the crystals, a molecule that was clearly different from the chromopyrrolic acid in which the crystal was soaked was found in the active site of the enzyme, and it was in a position suitable for the flavin to carry out its hydroxylation. The FAD was in the IN conformation. They showed that this bound molecule was most likely 7-carboxy-K252 (Fig. 1A), which could be a substrate for RebC. Upon further investigation, they determined that chromopyrrolic acid slowly decomposes while standing under aerobic conditions, forming a small amount of the bound K252 derivative; presumably, during the soaking and possibly while in the x-ray beam, the enzyme in the crystal acted as a miniature affinity-resin and selectively bound traces of the appropriate degradation compound from the mixture. Curiously, when K252c was bound, the flavin remained in the OUT position. Apparently, the carboxyl group(s) is required to trigger the movement of the FAD to the IN conformation. This research provides a novel example of x-ray structure determination leading to the characterization of an enzyme as a flavoprotein hydroxylase and, in addition, revealing some properties of a likely substrate. Thus, x-ray structural analysis was used for determining what is normally done first by bench-top enzymological procedures.
Conversion of tryptophan to rebeccamycin consists of several unique biochemical transformations.
Previous work had shown that when StaP (an analog of RebP used in the biosynthesis of staurosporin) was incubated with chromopyrrolic acid, three major products were formed, 7-hydroxy-K252c, arcyriaflavin A, and K252c in a 7:1:1 ratio (7). However, when StaP and RebC were both incubated with this substrate, the overall reactions were accelerated, and the primary product was arcyriaflavin A (Fig. 1B). In light of the work by Ryan et al. (17), these results suggest that StaP (and, by analogy, RebP) produces a reactive intermediate that is a substrate for RebC, and that RebC carries out specific reactions to produce arcyriaflavin A, the precursor to rebeccamycin. When RebC is not present, the reactive intermediate decomposes into several products, including those mentioned above. The high-affinity binding of 7-carboxy-K252c to RebC suggests that the true substrate is planar and contains one or more carboxylates. The first flavoprotein hydroxylase to be studied in detail, and the protein for which the term monooxygenase was first coined by Hayaishi and colleagues (23) more than 50 years ago, was salicylate (2-hydroxybenzoic acid) hydroxylase. This enzyme catalyzes the hydroxylation and decarboxylation of salicylate to yield catechol (1,2-dihydroxybenzene). The results reported by Ryan et al. are consistent with RebC carrying out a salicylate hydroxylase type of reaction as shown in Fig. 1B. Flavoprotein hydroxylases all form C4a-flavin hydroperoxides (FAD-OOH in Fig. 1B) that constitute the active form of oxygen for the hydroxylation of substrates. Fig. 1B shows how such a species could electrophilically attack the aryl-aryl-coupled chromopyrrolic acid that is the likely unstable product of RebP (or StaP). This would result in hydroxylation and decarboxylation to give 7-carboxy-K252c, which could also be a substrate for a second phase of hydroxylation and decarboxylation, resulting in the formation of arcyriaflavin A. Overall, this would consume two NADPH molecules required for reducing the FAD bound to RebC before it reacted with oxygen to form the FAD-OOH hydroxylating species. Flavoprotein hydroxylases that have as substrates aromatic compounds all require such substrates to be activated by  OH,
OH,  SH, or
SH, or  NH2 groups. However, the high reactivity of these indolocarbazoles (they react spontaneously with oxygen) is probably sufficient for RebC to carry out such classic hydroxylations. Although this story is in its infancy, details will surely be coming forth soon, stimulated by the crystallographic enzymology presented by Ryan et al. The understanding of these processes may lead to the development of a variety of pharmaceutical derivatives of these indolocarbazoles using modified precursors by means of methods such as those described by Salas and colleagues (15).
NH2 groups. However, the high reactivity of these indolocarbazoles (they react spontaneously with oxygen) is probably sufficient for RebC to carry out such classic hydroxylations. Although this story is in its infancy, details will surely be coming forth soon, stimulated by the crystallographic enzymology presented by Ryan et al. The understanding of these processes may lead to the development of a variety of pharmaceutical derivatives of these indolocarbazoles using modified precursors by means of methods such as those described by Salas and colleagues (15).
Acknowledgments
This work was supported by National Institutes of Health Grant GM64711 (to D.P.B.).
Footnotes
The author declares no conflict of interest.
See companion article on page 15311 in issue 39 of volume 104.
References
- 1.Sanchez C, Mendez C, Salas JA. Nat Prod Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 2.Meksuriyen D, Cordell GA. J Nat Prod. 1989;51:893–899. doi: 10.1021/np50059a013. [DOI] [PubMed] [Google Scholar]
- 3.Pearce CJ, Doyle TW, Forenza S, Lam KS, Schroeder DR. J Nat Prod. 1989;51:937–940. doi: 10.1021/np50059a020. [DOI] [PubMed] [Google Scholar]
- 4.Salas AP, Zhu L, Sanchez AC, Brana AF, Rohr J, Mendez C, Salas JA. Mol Microbiol. 2005;58:17–27. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onaka H, Asamizu S, Igarashi Y, Yoshida R, Furumai T. Biosci Biotechnol Biochem. 2005;69:1753–1759. doi: 10.1271/bbb.69.1753. [DOI] [PubMed] [Google Scholar]
- 6.Howard-Jones AR, Walsh CT. Biochemistry. 2005;44:15652–15663. doi: 10.1021/bi051706e. [DOI] [PubMed] [Google Scholar]
- 7.Howard-Jones AR, Walsh CT. J Am Chem Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 9.Onaka H, Taniguchi S-I, Igarashi Y, Furumai T. Biotechnol Biochem. 2003;67:127–138. doi: 10.1271/bbb.67.127. [DOI] [PubMed] [Google Scholar]
- 10.Yeh E, Garneau S, Walsh CT. Proc Natl Acad Sci USA. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh E, Cole LJ, Barr EW, Bollinger JM, Jr, Ballou DP, Walsh CT. Biochemistry. 2006;45:7904–7912. doi: 10.1021/bi060607d. [DOI] [PubMed] [Google Scholar]
- 12.Nishizawa T, Aldrich CC, Sherman DH. J Bacteriol. 2005;187:2084–2092. doi: 10.1128/JB.187.6.2084-2092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizawa T, Grüschow S, Jayamaha D-HE, Nishizawa-Harada C, Sherman DH. J Am Chem Soc. 2005;128:724–725. doi: 10.1021/ja056749x. [DOI] [PubMed] [Google Scholar]
- 14.Asamizu S, Kato Y, Igarashi Y, Furumai T, Onaka H. Tetrahedron Lett. 2006;47:473–475. [Google Scholar]
- 15.Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc Natl Acad Sci USA. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. J Med Chem. 2005;38:123–140. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 17.Ryan KS, Howard-Jones AR, Hamill MJ, Elliott SJ, Walsh CT, Drennan CL. Proc Natl Acad Sci USA. 2007;104:15311–15316. doi: 10.1073/pnas.0707190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joosten V, van Berkel JH. Curr Opin Chem Biol. 2007;11:195–202. doi: 10.1016/j.cbpa.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Entsch B, Cole LJ, Ballou DP. Arch Biochem Biophys. 2005;433:297–311. doi: 10.1016/j.abb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Enroth C. Acta Crystallogr D. 2003;59:1597–1602. doi: 10.1107/s0907444903014902. [DOI] [PubMed] [Google Scholar]
- 21.Hiromoto T, Fujiwara S, Hosokawa K, Yamaguchi H. J Mol Biol. 2006;364:878–896. doi: 10.1016/j.jmb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Gatti DL, Palfey BA, Lah MS, Entsch B, Massey V, Ballou DP, Ludwig ML. Science. 1994;266:110–114. doi: 10.1126/science.7939628. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri M, Yamamoto S, Hayaishi O. J Biol Chem. 1962;237:PC2413. [PubMed] [Google Scholar]