Abstract
Airway inflammation and epithelial remodeling are two key features of asthma. IL-13 and other cytokines produced during T helper type 2 cell-driven allergic inflammation contribute to airway epithelial goblet cell metaplasia and may alter epithelial–mesenchymal signaling, leading to increased subepithelial fibrosis or hyperplasia of smooth muscle. The beneficial effects of corticosteroids in asthma could relate to their ability to directly or indirectly decrease epithelial cell activation by inflammatory cells and cytokines. To identify markers of epithelial cell dysfunction and the effects of corticosteroids on epithelial cells in asthma, we studied airway epithelial cells collected from asthmatic subjects enrolled in a randomized controlled trial of inhaled corticosteroids, from healthy subjects and from smokers (disease control). By using gene expression microarrays, we found that chloride channel, calcium-activated, family member 1 (CLCA1), periostin, and serine peptidase inhibitor, clade B (ovalbumin), member 2 (serpinB2) were up-regulated in asthma but not in smokers. Corticosteroid treatment down-regulated expression of these three genes and markedly up-regulated expression of FK506-binding protein 51 (FKBP51). Whereas high baseline expression of CLCA1, periostin, and serpinB2 was associated with a good clinical response to corticosteroids, high expression of FKBP51 was associated with a poor response. By using airway epithelial cells in culture, we found that IL-13 increased expression of CLCA1, periostin, and serpinB2, an effect that was suppressed by corticosteroids. Corticosteroids also induced expression of FKBP51. Taken together, our findings show that airway epithelial cells in asthma have a distinct activation profile and identify direct and cell-autonomous effects of corticosteroid treatment on airway epithelial cells that relate to treatment responses and can now be the focus of specific mechanistic studies.
Keywords: gene expression microarray, serpinB2, CLCA1, FKBP51
Asthma is characterized by inflammation of the airways with eosinophils and mast cells, by airflow obstruction that is reversible by β-adrenergic agonists, and by bronchial hyperresponsiveness to stimuli such as cold air, methacholine, and histamine (1). Allergic airway inflammation in asthma is orchestrated by T helper type 2 cells, which are defined by their secretion of a panel of cytokines, including IL-4, IL-5, and IL-13. These cytokines promote the accumulation of eosinophils and mast cells in the asthmatic airway. In addition, resident lung cells, including airway epithelial cells and smooth muscle cells, appear to play important roles in effecting or perpetuating the airway response to T helper type 2 cytokines (2, 3).
Dysfunction of airway epithelial cells in asthma is thought to contribute to activation of innate immune responses, perpetuation of adaptive immune responses, increased mucus production, and production of mediators that may influence the underlying mesenchyme (4–7). These epithelial cell actions are likely triggered by T helper type 2 cytokines and the products of consequent infiltration by eosinophils, mast cells, and other inflammatory cells.
Asthma is typically responsive to treatment with inhaled or oral corticosteroids (8). At least some of the beneficial effects of corticosteroids may be due to inhibition of inflammatory cell function and thus decreased stimulation of epithelial cells. In addition, corticosteroids may have direct effects on the epithelial cells themselves.
To explore epithelial cell dysfunction in asthma and the therapeutic actions of corticosteroids, we collected epithelial cells from human airways by using bronchoscopy and examined expression profiles on genome-wide microarrays. In particular, we compared gene expression in epithelial cells from subjects with asthma before and after treatment with either an inhaled corticosteroid or placebo in a double-blind randomized controlled trial. We also analyzed gene expression in airway epithelial cells from two control groups: healthy nonsmokers and habitual smokers, who exhibit airway epithelial dysfunction not caused by asthma, to identify epithelial abnormalities that are specific to asthma and not simply a general response to airway inflammation. Finally, to assess the cell autonomy of any asthma-specific corticosteroid-sensitive responses detected, we examined expression in airway epithelial cells exposed in culture to IL-13 and to corticosteroids.
Results
Study Population.
During bronchoscopy, we collected airway epithelial brushings from 42 nonsmoking subjects with asthma, 28 nonsmoking healthy controls, and 16 current smokers without asthma but with mild to moderate airflow obstruction (disease controls) [supporting information (SI) Table 2]. Across all groups, the airway epithelial brushings yielded 1.3 ± 0.8 × 106 cells, of which 97 ± 3% were epithelial cells.
Genes Differentially Expressed in Asthmatic Airway Epithelial Cells.
Gene expression was first analyzed in subjects with asthma before treatment with inhaled corticosteroids (or placebo) by using healthy nonsmokers as a control group. By using the conservative Bonferroni correction for multiple comparisons, we found differential expression for 26 probe sets representing 22 genes in asthma compared with healthy controls (Table 1). Thirteen genes displayed increased expression in asthmatics compared with healthy subjects and nine genes displayed decreased expression. The differential expression of eight of these genes was reassessed and confirmed by quantitative real-time PCR (qPCR) (Fig. 1A).
Table 1.
Genes differentially expressed in epithelial brushings by microarray analysis in subjects with asthma compared with healthy control subjects
| Affymetrix ID | Gene symbol | Gene name | Entrez gene ID | Fold difference | P value |
|---|---|---|---|---|---|
| 210107_at | CLCA1 | Chloride channel, Ca2+-activated 1 | 1179 | 6.2 | 0.033 |
| 210809_s_at | POSTN | Periostin, osteoblast-specific factor | 10631 | 4.4 | 0.0015 |
| 204919_at | PRR4 | Proline rich 4 (lacrimal) | 11272 | 3.9 | 0.049 |
| 204614_at | SERPINB2 | Serine peptidase inhibitor B2 | 5055 | 3.5 | 0.047 |
| 205624_at | CPA3 | Carboxypeptidase A3 (mast cell) | 1359 | 3.4 | 0.0069 |
| 207134_x_at | TPSB2 | Tryptase β2 | 64499 | 2.2 | 0.0062 |
| 205683_x_at | TPSAB1 | Tryptase α/β1 | 7177 | 2.1 | 0.022 |
| 1559584_a_at | C16orf54 | Chromosome 16 orf 54 | 283897 | 1.4 | 0.015 |
| 201061_s_at | STOM | Stomatin | 2040 | 1.3 | 0.043 |
| 224782_at | ZMAT2 | Zinc finger, matrin type 2 | 153527 | 1.3 | 0.038 |
| 205002_at | DJ159A19.3 | Hypothetical protein DJ159A19.3 | 27245 | 1.2 | 0.026 |
| 217313_at | — | — | — | 1.2 | 0.0074 |
| 1555256_at | EVC2 | Ellis van Creveld syndrome 2 | 132884 | 1.1 | 0.032 |
| 237690_at | GPR115 | G protein-coupled receptor 115 | 221393 | −1.2 | 0.019 |
| 212820_at | DMXL2 | Dmx-like 2 | 23312 | −1.2 | 0.0003 |
| 241774_at | — | — | — | −1.3 | 0.016 |
| 225987_at | TNFAIP9 | TNFα-induced protein 9 | 79689 | −1.7 | 0.0078 |
| 213432_at | MUC5B | Mucin 5, subtype B | 4587 | −1.9 | 0.016 |
| 241436_at | SCNN1G | Na+ channel, nonvoltage-gated 1G | 6340 | −1.9 | 0.0027 |
| 219410_at | TMEM45A | Transmembrane protein 45A | 55076 | −2.0 | 0.0006 |
| 1556185_a_at | — | Clone IMAGE:5260162, mRNA | — | −2.0 | 0.0061 |
| 230378_at | SCGB3A1 | Secretoglobin 3A1 | 92304 | −2.1 | 0.0096 |
Fig. 1.
PCR validation of microarray findings. PCR validation of selected genes differentially expressed in epithelial brushings from asthmatic subjects compared with healthy control subjects (A) and genes responsive to corticosteroids in the clinical trial of inhaled fluticasone in asthmatics (B).§§ Fold induction by microarray was statistically significant (P < 0.05, Bonferroni corrected) for all genes shown. Fold induction by PCR (for validation) was statistically significant (P < 0.05) for all genes except the periostin response to fluticasone (POSTN, P = 0.064).
Among the most highly induced genes were chloride channel, calcium-activated, family member 1 (CLCA1) (6.2-fold), periostin (4.4-fold), and serine peptidase inhibitor, clade B (ovalbumin), member 2 (serpinB2) (also known as plasminogen activator inhibitor-2) (3.5-fold). Originally described as a calcium-activated chloride channel, CLCA1 likely functions indirectly in chloride transport (9) and has previously been reported to be up-regulated in asthma (10–13). Periostin is an integrin ligand and extracellular matrix protein with roles in cell adhesion, cell motility, and matrix remodeling. SerpinB2 is a member of the serpin class of proteases and functions to inhibit plasminogen activation and promote fibrin formation and deposition. Among the other most differentially expressed genes in the epithelial brushings were three mast cell markers: carboxypeptidase A3 (3.4-fold induced), tryptase β2 (2.2-fold induced), and tryptase α/β1 (2.1-fold induced).
Protein Confirmation.
Because CLCA1 is well recognized as an asthma-associated gene (10–13), we focused our confirmatory protein analyses on periostin and serpinB2. Periostin protein expression was confirmed by immunohistochemistry in bronchial biopsies from a randomly selected subset of asthmatic and healthy control subjects. Periostin immunolocalized to the basement membrane zone immediately below the epithelium and in the mesenchymal tissue compartment in all subjects, and quantification confirmed increased expression in asthma (SI Fig. 4). SerpinB2 protein expression was confirmed by ELISA (American Diagnostica, Stamford, CT). We found that serpinB2 levels in bronchoalveaolar lavage fluid were below the lower limit of detection (50 pg/ml) in all of the 13 healthy control subjects but were detectable in 13 of 27 asthmatics (median, 0.05; range, 0–3.3 ng/ml; P = 0.002).
Genes Differentially Expressed in Airway Epithelial Cells from Smokers.
We next analyzed gene expression in nonasthmatic habitual smokers compared with the healthy nonsmoking controls and found differential expression for 54 probe sets representing 40 genes (SI Table 3); 27 genes displayed increased expression and 13 were decreased. Twelve of the up-regulated genes are oxidoreductases, including AKR1B10 (13.6-fold induced), CYP1B1 (10.4-fold), ALDH3A1 (5.1-fold), GPX2 (4.9-fold), AKR1C2 (3.9-fold), and ME1 (3.4-fold). Transmembrane protein 45 (TMEM45A), a gene of unknown function, was the only gene differentially expressed in both asthma vs. healthy and smokers vs. healthy comparisons (repressed by 2.0-fold in asthma and 2.8-fold in smokers).
Genes Responsive to Corticosteroids.
Analysis of gene expression in asthmatic subjects treated either with inhaled fluticasone or placebo control revealed 33 probe sets representing 30 genes with at least a 2-fold change in expression in the fluticasone group compared with the placebo group (SI Table 4). The expression levels of six of these genes were reassessed by qPCR (Fig. 1B) and yielded statistically significant confirmatory changes for all genes except periostin, which showed a strong trend (P = 0.064) for reduced expression after inhaled corticosteroid treatment. The most highly induced known gene was FK506-binding protein 51 (FKBP51), which reportedly modulates glucocorticoid receptor (GR) activity (14). Baseline FKBP51 expression, as measured by qPCR, correlated inversely with lung function response to fluticasone treatment, as assessed in a standard test that measures the change in the volume of air that is exhaled in the first second of a forced exhalation (FEV1) (Fig. 2 A and B).
Fig. 2.
Baseline FKBP51 expression correlates inversely with lung function response to fluticasone. Baseline (pretreatment) FKBP51 mRNA expression levels (as measured by PCR) in subjects subsequently randomized to inhaled fluticasone correlated with treatment-related improvements in lung function at 4 weeks (r = −0.62, P = 0.007) (A) and at 8 weeks (r = −0.63, P = 0.009) (B).
To identify genes that are both associated with asthma and are corticosteroid-responsive, we compared the 22 genes differentially expressed in asthma (Table 1) with the 30 genes affected by corticosteroid treatment (SI Table 4). The following three genes met both criteria: CLCA1, periostin, and serpinB2. All three were up-regulated in asthma and down-regulated by corticosteroids. In addition, changes in the expression of these genes (as measured by qPCR) were associated with improvements in lung function in the fluticasone group. Specifically, the baseline expression of CLCA1, periostin, and serpinB2 correlated directly with improvements in lung function (FEV1) in the corticosteroid treated subjects at 4 weeks (r = 0.60, P = 0.011; r = 0.49, P = 0.048; and r = 0.53, P = 0.027, respectively). Furthermore, decreases in serpinB2 expression with fluticasone treatment (difference between log mRNA copy number at the second vs. the first bronchoscopy) correlated with improvements in FEV1 at both 4 and 8 weeks (r = −0.62, P = 0.01, and r = −0.65, P = 0.009, respectively).
In Vitro Effects of IL-13 and Corticosteroids on Lung Epithelial Cells.
Whereas CLCA1 is an airway epithelial cell product induced in asthma (10, 12), it is unknown whether airway epithelial cells are a source of serpinB2 and periostin, whether these genes are induced by T helper type 2 cytokines, or whether their expression is directly modulated by corticosteroids. We therefore tested the effects of IL-13 and corticosteroids on expression of these genes in two cell culture preparations: (i) BEAS-2B cells, an SV-40 transformed human airway epithelial cell line, in monolayer culture, and (ii) primary human airway epithelial cells grown in an air–liquid interface (ALI), which promotes cell differentiation and which allows for IL-13 responses similar to those seen in vivo in murine airway epithelial cells (15).
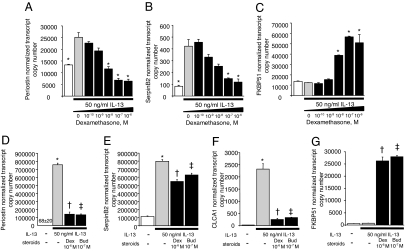
We found that IL-13 (50 ng/ml for 4 days) induced periostin and serpinB2 expression in BEAS-2B cells and that dexamethasone, added for the last 24 h, produced a dose-dependent inhibition of IL-13-induced gene expression (Fig. 3 A and B). Also parallel with our in vivo findings, FKBP51 expression was not induced by IL-13 in BEAS-2B, but was induced by dexamethasone in a dose-dependent manner (Fig. 3C). However, we failed to detect CLCA1 transcripts in BEAS-2B cells under any conditions. CLCA1 is thought to play a role in mucus production, and lack of expression of CLCA1 in BEAS-2B cells grown in monolayer may reflect the inability of these cells to acquire features of differentiated mucus-producing goblet cells.
Fig. 3.
In vitro effects of IL-13 and corticosteroids on lung epithelial cells. Dexamethasone inhibits induction of periostin (A) and serpinB2 (B) by IL-13 in BEAS-2B cells grown in monolayer (*, P < 0.05 compared with IL-13-exposed cells without dexamethasone). FKBP51 is not induced by IL-13 but is induced by dexamethasone in a dose-dependent manner (C) (*, P < 0.05 compared with cells not exposed to dexamethasone). Dexamethasone (Dex) and budesonide (Bud) inhibit induction of periostin (D) and serpinB2 (E) and CLCA1 (F) by IL-13 and induce expression of FKBP51 (G) in primary airway epithelial cells grown at an ALI (*, P < 0.05 compared with all other groups; †, P < 0.05 compared with all groups except Bud; ‡, P < 0.05 compared with all groups except Dex).
In primary human airway epithelial cells grown in an ALI, we found that expression of CLCA1 in addition to periostin and serpinB2 was induced by IL-13 (50 ng/ml for 4 days) (Fig. 3 D–F), and that either dexamethasone or budesonide (an inhaled corticosteroid preparation commonly used in asthma therapy) added for the last 24 h inhibited these IL-13 effects (Fig. 3 D–F). Dexamethasone and budesonide also induced expression of FKBP51 in these primary cells (Fig. 3G). We conclude that periostin, serpinB2, and CLCA1 expression in airway epithelial cells can be directly regulated by IL-13, and that repression of periostin, serpinB2, and CLCA1 and induction of FKBP51 are direct and cell-autonomous effects of corticosteroids on those cells.
Discussion
In cells obtained from airway epithelial brushings, we identified 22 genes differentially expressed in asthma compared with healthy controls; with one exception, these genes were distinct from 40 genes that we found to be differentially expressed in habitual smokers. Three of the 22 asthma-associated genes were proteases that are known products of mast cells and basophils (tryptase β2, tryptase α/β1, and carboxypeptidase A3), indicating that these cells are present in the epithelial compartment in asthma and that their secreted proteases, cytokines, and lipid mediators likely contribute to the epithelial cell activation (16, 17) and dysfunction that characterizes asthma.
Three of the asthma-associated genes (CLCA1, periostin, and serpinB2) changed in expression by at least 2-fold with corticosteroid treatment, and all were repressed. Based on the actions of the products of these genes, it is tempting to speculate that their regulation may produce at least part of the therapeutic effects of corticosteroids in the asthmatic airway. Thus, CLCA1 is implicated in mechanisms of mucus hypersecretion, bronchial hyperreactivity, and eosinophil accumulation in asthma (10). In addition, periostin secreted by lung fibroblasts has been proposed to promote subepithelial fibrosis in asthma (18), and SerpinB2 is induced by allergen challenge in airway epithelial cells from asthmatics (19).
It is conceivable that periostin and serpinB2 contribute to airway remodeling. Periostin is an extracellular matrix protein and integrin ligand originally identified in osteoblasts but known to be secreted by malignant epithelial cells, periodontal ligaments, and pericardial and heart valve tissue (20–22). Interestingly, periostin appears to stimulate adhesion and migration of malignant epithelial cells through interaction with multiple integrins, including αvβ3, αvβ5, and αvβ6 (20, 23, 24), and to enhance collagen fibrillogenesis and the biomechanical properties of connective tissues (21). Periostin expression is induced by IL-13 in lung fibroblasts in vitro and is elevated in the airways of ovalbumin-challenged mice and in human subjects with asthma, localizing to the subepithelial compartment (18). We found that IL-13 markedly up-regulated periostin gene expression in cultured airway epithelial cells as well, and the immunolocalization of periostin to the subepithelial region in airway biopsies suggests that it is secreted basolaterally into the underlying matrix. From this location, periostin could interact with matrix proteins to promote cell motility or epithelial–mesenchymal signaling interactions that produce airway remodeling.
SerpinB2 [also known as plasminogen activator inhibitor-2 (PAI-2)] is a serine protease inhibitor, which exists largely as a 47-kDa intracellular form and may also be secreted as a 60-kDa glycosylated protein (25). Like plasminogen activator inhibitor-1 (PAI-1), SerpinB2 inhibits tissue-type and urokinase-type plasminogen activators (tPA and uPA), prevents activation of plasmin, and promotes fibrin formation. Fibrinogen and thrombin accumulation on the airway surface can contribute to the pathogenesis of airway hyperresponsiveness (26), and PAI-1 has been linked to asthma in both genetic studies (27–30) and in mouse models (31). However, whether serpinB2 functions primarily in extracellular fibrin deposition is uncertain, particularly because it is predominantly intracellular. Provisionally, we suggest that serpinB2 represents a novel effector of the multiple airway remodeling actions provoked by IL-13 (32).
FKBP51 was among the genes most highly induced by corticosteroids. FKBP51 is an immunophilin chaperone protein, a subunit of the multiprotein GR “aporeceptor” complex that resides in the cytoplasm before hormone binding. On association with an agonist such as dexamethasone, GR dissociates from the chaperone complex, translocates to the nucleus, and modulates transcription. FKBP51 overexpression appears to inhibit GR signaling by impairing nuclear translocation (33) and reducing hormone binding (34). Moreover, FKBP51 is potently induced by corticosteroids (35, 36), suggesting that FKBP51 may function in a negative-feedback loop to limit GR signaling within a permitted range. Indeed, FKBP51 overexpression may contribute to glucocorticoid resistance syndrome in primates (37, 38), and FKBP51 mRNA expression levels in peripheral blood mononuclear cells may provide an assay for individual sensitivity to corticosteroids (39, 40). In principle, FKBP51 expression might affect clinical responsiveness to corticosteroids in asthma.
New ways to characterize asthma phenotypes are needed, so that treatment can be better tailored to individuals with disease. In this study, we have used gene expression microarray analysis of airway epithelial cells from patients and normal subjects to initiate a molecular analysis that can progress to mechanistic studies. We describe periostin, serpinB2, and CLCA1 as biomarkers of classic IL-13-driven, corticosteroid-responsive asthma and as potential mediators of disease. These proteins can plausibly be inserted in pathways of pathology in the epithelium and submucosa. In addition, we identify FKBP51 as a biomarker of corticosteroid responsiveness and a potential mediator of corticosteroid-resistant disease. Finally, we demonstrate that bronchial epithelial cells are direct targets both for IL-13-mediated gene induction in asthma, and for corticosteroid repression of those induced genes in treatment of this disease. It should now be possible to use this information to develop focused strategies to determine disease mechanisms and potential selective therapeutics.
Materials and Methods
Human Subjects and Protocols.
We collected epithelial brushings by bronchoscopy from 42 subjects with asthma, 16 current smokers without asthma, and 28 healthy controls. Healthy control subjects and smokers were enrolled in one of three cross-sectional studies, which comprised two visits each, the first for characterization and the second for bronchoscopy 1 week later. Subjects with asthma were enrolled in a 10-week, randomized, double-blind, placebo-controlled trial specifically designed to determine the effects of inhaled fluticasone (500 μg, twice daily) on airway gene expression and to relate gene expression changes to improvements in lung function (SI Fig. 5). Bronchoscopy was performed at baseline and repeated 1 week after starting study drug. Thirty-five subjects had adequate baseline bronchoscopy, and 32 had RNA available from epithelial brushings at both bronchoscopies. Lung function was measured at intervals over the 8-week treatment period. Epithelial RNA from a subset of the subjects studied by microarray here have been used in other PCR-based gene expression studies performed by our group (41). Methods for bronchoscopy, epithelial brushing, bronchoalveaolar lavage, and sample handling were identical across all studies. All clinical studies were approved by the University of California at San Francisco Committee on Human Research, written informed consent was obtained from all subjects, and all studies were performed in accordance with the principles expressed in the Declaration of Helsinki.
In Vitro Studies.
BEAS-2B cells were maintained on DME-H16 10% FBS medium and changed to DME-H16 medium containing 10% activated charcoal-stripped FBS the day before they reach confluence. All further experiments are carried out in stripped serum medium. Cells were treated with IL-13 (50 ng/ml) for 4 days, with the addition of dexamethasone at a dose range in the last 24 h before cell lysis and RNA extraction using the Qiagen RNeasy mini-kit (Qiagen, Valencia, CA). All conditions were performed in triplicate. Human bronchial epithelial cells were obtained at autopsy (courtesy of Walter Finkbeiner, University of California, San Francisco) by enzymatic digestion as described previously in ref. 42 and plated in a 1:1 mixture of DMEM/Ham's F-12 plus 5% FBS, penicillin (100 units/ml), streptomycin (1 mg/ml), fungizone (2.5 μg/ml), gentamicin (50 μg/ml) (PSFG), and onto polycarbonate inserts (0.4-μm pore size; Costar, Corning, MA) coated with 20 μg/ml human placental collagen at 9.1 × 105 cells per filter. For 2 successive days, filters and wells were rinsed twice with PBS with PSFG, and 1 ml of ALI medium (43) was added to each well. Then, the medium was changed every other day. On days 13 and 14, medium was switched to ALI (−) hydrocortisone/(+) 50 ng/ml IL-13 excepting four control filters, which were switched to ALI (−) hydrocortisone. On day 15, either 1 μM dexamethasone (Sigma–Aldrich, St. Louis, MO) reconstituted in DMSO or 100 nM budesonide (AstraZeneca, Wilmington, DE) diluted in PBS were added as appropriate. All conditions were performed in quadruplicate. On day 16, cells were lysed, and RNA was extracted as described above.
Gene Expression Microarray Analysis.
For microarray analysis, 25 ng of total epithelial RNA was amplified by using NuGen Ovation RNA amplification system. Then, 2.75 μg of single-stranded cDNA was hybridized to Affymetrix (Santa Clara, CA) U133 plus 2.0 arrays (>54,000 probe sets coding for 38,500 genes). Array images were analyzed by using Affymetrix GeneChip Expression Analysis Software. Bioconductor (44) was used for quality control (affyPLM algorithm), preprocessing (RMA algorithm), cluster analysis, and linear modeling (44–46). Raw array data are available from the Gene Expression Omnibus public database (www.ncbi.nlm.nih.gov/geo, accession number GSE4302).
Real-Time PCR (qPCR).
For analysis of epithelial brushings and primary human airway epithelial cells, two-step qPCR was performed as described previously in ref. 47. cDNA synthesis was carried out by using 20 ng of total RNA and BD Clontech (Mountain View, CA) Powerscript Reverse Transcriptase with random hexamers for priming. Then, multiplex preamplification was performed by using one-fifth of the resultant cDNA, Advantage 2 Polymerase (BD Clontech), and 5 pmol of each outflanking primer. Multiplex hot-start amplification was done for 5, 10, 15, and 20 cycles to ensure that the reaction remained in the exponential phase of PCR and the substrates were not limiting (47). Real-time PCR gene quantification was then performed on the amplified cDNA by using TaqMan probes (Applied Biosystems, Foster City, CA) and Universal Master Mix (Platinum Quantitative PCR SuperMix-UDG with ROX; Invitrogen, Carlsbad, CA). Transcript quantification was run on an ABI Prism 7900 Sequence Detection System (Applied Biosystems). Cycle threshold values obtained for each gene were then converted into relative transcript copy numbers based on logarithmic transformation and linear regression of prior data, as described previously in ref. 47. Transcript copy numbers were normalized by using a two-step approach. First, the amount of amplification product used in TaqMan profiling was normalized on the basis of housekeeping gene expression. Then a panel of four housekeeping genes was measured during TaqMan profiling (GAPDH, ubiquitin, EEF1A1, and PPIA), and the geometric mean value of the two housekeeping genes most stably expressed across the samples was used for normalization (48). For BEAS-2B cells, qPCR was performed without multiplex preamplification. First-strand cDNA synthesis was carried out by using 10 μg of starting RNA, and then one-fourth of the resultant cDNA was used for qPCR gene quantification by using the methods described above (including the same four housekeeping genes for normalization).
Immunohistochemistry.
Sections were deparaffinized, rehydrated, incubated in 3% hydrogen peroxide/absolute methanol for 10 min, and then blocked with 1% goat serum for 30 min at 23°C. Sections were then blotted and incubated in primary antibody [“anti-OSF2” (periostin); BioVendor, Candler, NC] diluted in 1% goat serum/0.3% Tween 20/PBS at 1:16,000 for 1 h at 23°C. Sections were then incubated for 1 h at 23°C in biotinylated goat anti-rabbit secondary antibody (Vector Laboratories, Burlingame, CA). Next, sections were incubated in ABC reagent (Vector Laboratories) for 1 h at 23°C, followed by DAB Plus reagent (Zymed, San Francisco, CA) for 10 min, and counterstaining with Gill's no. 3 hematoxylin.
Design-Based Stereology.
Six endobronchial biopsies were obtained from second- through fifth-order carinae, formalin-fixed, and paraffin-embedded, and then analyzed by using immunohistochemistry (described above) and design-based stereology (49). First, the volume of immunohistochemical staining, the volume of submucosal tissue surveyed, and the surface area of the epithelial basal lamina surveyed were determined by point and intersection counting using an integrated microscope (Olympus, Albertslund, Denmark), video camera (JVC Digital Color; JVC, Tatstrup, Denmark), automated microscope stage, and computer [Dell Optiplex GS270 PC running Computer-Assisted Stereology Toolbox (C.A.S.T.) software; Olympus, Albertslund, Denmark]. A line segment grid was superimposed on systematically randomly selected microscopic fields. Points overlying submucosal tissue that is immunostained and unstained were counted separately along with intersections of test lines with basal lamina. The measurements were recorded by a blinded investigator using a ×20 objective lens. The volume density of immunostaining was calculated by quantification of the volume of stained submucosa referenced to the surface area of the basal lamina surveyed (cubic micrometer per square micrometer).
ELISA.
ELISA for serpinB2 protein levels in bronchoalveolar lavage fluid was performed by using a commercially available kit (Imubind PAI-2 ELISA; American Diagnostica, Stamford, CT) according to the manufacturer's directions.
Statistical Analysis.
For between-group comparisons, t test, rank-sum test, or ANOVA followed by pairwise analyses with Sidak correction were used as appropriate. Correlation of mRNA copy number by PCR (log transformed for normality) and lung function was performed by using Pearson's correlation. STATA, version 9, software was used (StataCorp, College Station, TX), and P < 0.05 was taken as statistically significant. In microarray analyses, differential gene expression was assessed by using linear models (controlling for age and gender). For between-group comparisons, a Bonferroni-corrected value of P < 0.05 was used to identify differentially expressed genes. For the placebo-controlled trial, changes in gene expression from baseline to postintervention were calculated and the mean changes in the fluticasone and placebo groups were compared. A 2-fold change (either higher or lower in the fluticasone group compared with placebo) was used to identify differentially expressed genes. Microarray analyses were performed by using Bioconductor open source software (44) in the R statistical environment. For cell culture experiments, between-group comparisons of gene expression (using copy number, log-transformed for normality) were performed by using ANOVA followed by pairwise comparisons by using t test with the Sidak correction for multiple comparisons.
Supplementary Material
Acknowledgments
Inhaled fluticasone and matching placebo were kindly provided as a gift from GlaxoSmithKline (Uxbridge, U.K.). We are grateful for the contributions of members of the University of California at San Francisco Airway Clinical Research Center, including Hofer Wong, Peggy Cadbury, Roderick Carter, Stephen Lazarus, Steven Hays, and Anh Innes, and to Walter Finkbeiner and Alicia Goodwin for assistance with primary epithelial cell culture. Research support was from National Institutes of Health Grants CA020535 (to K.R.Y.); HL5683, HL080414, and HL66564 (to J.V.F.); HL72301 (to D.J.E.); HL72301 (to C.S.B.); and RR17002 (to P.G.W.); from The Sandler Asthma Basic Research Center (K.R.Y., J.V.F., D.J.E., and Y.H.Y.); and from the General Clinical Research Centers of Moffitt Hospital (Grant RR-00079) and San Francisco General Hospital (Grant RR-00083).
Abbreviations
- qPCR
quantitative real-time PCR
- FKBP51
FK506-binding protein 51
- GR
glucocorticoid receptor
- ALI
air–liquid interface.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE4302).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707413104/DC1.
The clinical trial registration information is as follows: ClinicalTrials.gov identifier NCT00187499; study ID nos. H6788-20160-04; P50HL56385; last updated December 8, 2005; record first received September 13, 2005.
References
- 1.Boushey HA, Corry DB, Fahy JV, Burchard EG, Woodruff PG. In: Murray and Nadel's Textbook of Respiratory Medicine. Mason RJ, Broaddus CV, Murray JF, Nadel JA, editors. Vol 1. Philadelphia: Elsevier Saunders; 2005. pp. 1168–1216. [Google Scholar]
- 2.Cookson W. Nat Rev Immunol. 2004;4:978–988. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 3.Cockcroft DW, Davis BE. J Allergy Clin Immunol. 2006;118:551–559. doi: 10.1016/j.jaci.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Holgate ST, Holloway J, Wilson S, Bucchieri F, Puddicombe S, Davies DE. Proc Am Thorac Soc. 2004;1:93–98. doi: 10.1513/pats.2306034. [DOI] [PubMed] [Google Scholar]
- 5.Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, Erle DJ. Nat Med. 2002;8:885–889. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 6.Fahy JV. Am J Respir Crit Care Med. 2001;164:S46–S51. doi: 10.1164/ajrccm.164.supplement_2.2106066. [DOI] [PubMed] [Google Scholar]
- 7.Holtzman MJ, Look DC, Sampath D, Castro M, Koga T, Walter MJ. Proc Assoc Am Physicians. 1998;110:1–11. [PubMed] [Google Scholar]
- 8.Barnes PJ. N Engl J Med. 1995;332:868–875. doi: 10.1056/NEJM199503303321307. [DOI] [PubMed] [Google Scholar]
- 9.Erle DJ, Zhen G. Am J Respir Crit Care Med. 2006;173:1181–1182. doi: 10.1164/rccm.2603006. [DOI] [PubMed] [Google Scholar]
- 10.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, Fujino M. Proc Natl Acad Sci USA. 2001;98:5175–5180. doi: 10.1073/pnas.081510898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Dong Q, Louahed J, Dragwa C, Savio D, Huang M, Weiss C, Tomer Y, McLane MP, Nicolaides NC, Levitt RC. Am J Respir Cell Mol Biol. 2001;25:486–491. doi: 10.1165/ajrcmb.25.4.4578. [DOI] [PubMed] [Google Scholar]
- 12.Toda M, Tulic MK, Levitt RC, Hamid Q. J Allergy Clin Immunol. 2002;109:246–250. doi: 10.1067/mai.2002.121555. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino M, Morita S, Iwashita H, Sagiya Y, Nagi T, Nakanishi A, Ashida Y, Nishimura O, Fujisawa Y, Fujino M. Am J Respir Crit Care Med. 2002;165:1132–1136. doi: 10.1164/ajrccm.165.8.2107068. [DOI] [PubMed] [Google Scholar]
- 14.Davies TH, Ning Y-M, Sanchez ER. J Biol Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- 15.Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, Paquet AC, Erle DJ. Am J Respir Cell Mol Biol. 2007;36:244–253. doi: 10.1165/rcmb.2006-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce JA. Prostaglandins Leukotrienes Essent Fatty Acids. 2003;69:195–205. doi: 10.1016/s0952-3278(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 17.Marone G, Triggiani M, Genovese A, Paulis AD. Adv Immunol. 2005;88:97–160. doi: 10.1016/S0065-2776(05)88004-6. [DOI] [PubMed] [Google Scholar]
- 18.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, McKenzie AN, Nagai H, Hotokebuchi T, Izuhara K. J Allergy Clin Immunol. 2006;118:98–104. doi: 10.1016/j.jaci.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 19.Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA. Am J Respir Crit Care Med. 2005;171:579–586. doi: 10.1164/rccm.200404-532OC. [DOI] [PubMed] [Google Scholar]
- 20.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 21.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, et al. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, II, Conway SJ, et al. Circ Res. 2007;101:313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 24.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medcalf RL, Stasinopoulos SJ. FEBS J. 2005;272:4858–4867. doi: 10.1111/j.1742-4658.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 26.Wagers SS, Norton RJ, Rinaldi LM, Bates JH, Sobel BE, Irvin CG. J Clin Invest. 2004;114:104–111. doi: 10.1172/JCI19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckova D, Izakovicova Holla L, Vacha J. Allergy. 2002;57:446–448. doi: 10.1034/j.1398-9995.2002.03582.x. [DOI] [PubMed] [Google Scholar]
- 28.Cho SH, Hall IP, Wheatley A, Dewar J, Abraha D, Del Mundo J, Lee H, Oh CK. J Allergy Clin Immunol. 2001;108:212–214. doi: 10.1067/mai.2001.117260. [DOI] [PubMed] [Google Scholar]
- 29.Hizawa N, Maeda Y, Konno S, Fukui Y, Takahashi D, Nishimura M. Clin Exp Allergy. 2006;36:872–876. doi: 10.1111/j.1365-2222.2006.02413.x. [DOI] [PubMed] [Google Scholar]
- 30.Pampuch A, Kowal K, Bodzenta-Lukaszyk A, Di Castelnuovo A, Chyczewski L, Donati MB, Iacoviello L. Allergy. 2006;61:234–238. doi: 10.1111/j.1398-9995.2005.00948.x. [DOI] [PubMed] [Google Scholar]
- 31.Oh CK, Ariue B, Alban RF, Shaw B, Cho SH. Biochem Biophys Res Commun. 2002;294:1155–1160. doi: 10.1016/S0006-291X(02)00577-6. [DOI] [PubMed] [Google Scholar]
- 32.Cohn L, Elias JA, Chupp GL. Annu Rev Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- 33.Wochnik GM, Ruegg J, Abel GA, Schmidt U, Holsboer F, Rein T. J Biol Chem. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 34.Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 35.Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR. Proc Natl Acad Sci USA. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. J Clin Endocrinol Metab. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds PD, Ruan Y, Smith DF, Scammell JG. J Clin Endocrinol Metab. 1999;84:663–669. doi: 10.1210/jcem.84.2.5429. [DOI] [PubMed] [Google Scholar]
- 38.Denny WB, Prapapanich V, Smith DF, Scammell JG. Endocrinology. 2005;146:3194–3201. doi: 10.1210/en.2005-0027. [DOI] [PubMed] [Google Scholar]
- 39.Vermeer H, Hendriks-Stegeman BI, van Suylekom D, Rijkers GT, van Buul-Offers SC, Jansen M. Mol Cell Endocrinol. 2004;218:49–55. doi: 10.1016/j.mce.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Vermeer H, Hendriks-Stegeman BI, Verrijn Stuart AA, van Buul-Offers SC, Jansen M. Eur J Endocrinol. 2004;150:41–47. doi: 10.1530/eje.0.1500041. [DOI] [PubMed] [Google Scholar]
- 41.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ. J Allergy Clin Immunol. 2005;116:305–311. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Yamaya M, Finkbeiner WE, Chun SY, Widdicombe JH. Am J Physiol. 1992;262:L713–L724. doi: 10.1152/ajplung.1992.262.6.L713. [DOI] [PubMed] [Google Scholar]
- 43.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 44.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Astrand M, Speed TP. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolganov GM, Woodruff PG, Novikov AA, Zhang Y, Ferrando RE, Szubin R, Fahy JV. Genome Res. 2001;11:1473–1483. doi: 10.1101/gr.191301. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolender RP, Hyde DM, Dehoff RT. Am J Physiol. 1993;265:L521–L548. doi: 10.1152/ajplung.1993.265.6.L521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.