Abstract
During mitosis in higher eukaryotic cells, transcription is silenced and transcription complexes are absent from promoters in the condensed chromosomes; however, epigenetic information concerning the pattern of expressed and silent genes must be preserved. Recently, it has been reported that CTCF, a major protein in vertebrate insulator elements, remains associated with mitotic chromatin. If the structure of insulators is preserved during mitosis, then it is possible that insulators can function as components or elements of the mechanism involved in the transfer of epigenetic information through the mitotic phase and can help guide the reconstitution of domain structure and nuclear organization after the completion of this phase. We have studied the chromatin structure of the insulator upstream of the c-MYC gene in mitotic HeLa cells. The region of the insulator corresponds to the DNase I hypersensitive site I, but Southern blot analysis revealed that hypersensitivity was lost during mitosis. High resolution in vivo footprinting analysis using dimethyl sulfate, UV light, psoralen, and DNase I also demonstrated the disappearance of the sequence-specific direct binding of CTCF and the absence of detectable structures during mitosis. Thus, it appears that the nucleoprotein complex involving this insulator element must be reassembled de novo with each new cell generation.
Keywords: epigenetics, in vivo footprinting
In higher eukaryotic cells, essentially all transcription is repressed during mitosis (1). This mitotic repression of transcription is accompanied by the displacement of most of the components of the transcriptional machinery from their templates. In vivo footprinting analysis has revealed that the footprints of sequence-specific transcription factors are absent from the repressed promoters on mitotic chromosomes (2–5). Upon exit from mitosis, previously active genes resume transcription, whereas previously inactive genes remain silent. This “cell memory” indicates that some kind of epigenetic information is maintained through mitosis and must be used to guide the cell in reestablishing transcription complexes only on previously active genes after the completion of mitosis. It has been established that DNA methylation and histone modifications play important roles in epigenetic control (6, 7). On the other hand, some recent models for mitotic cell memory assume that a subset of protein factors remains associated with previously active genes during mitosis and that these factors serve as epigenetic marks (8–14).The significance and mechanism of such protein associations are yet to be elucidated.
The domains containing individual genes in eukaryotic genomes often are demarcated by insulator elements (15). Two ways in which insulators protect genes from their surroundings have been described: (i) enhancer-blocking activity prevents the influence of outside enhancers and (ii) barrier activity prevents the advance of silencing heterochromatin. The enhancer-blocking activity of vertebrate insulators seems to be mediated by the protein CTCF, i.e., CCCTC binding factor (16). Recently, by fluorescence microscopy, biochemical fractionation, and ChIP, it has been shown that CTCF remains associated with mitotic chromatin (17). If the CTCF-associated local chromatin structure of insulator elements is preserved during mitosis, then it appears possible that insulators may serve as components of cellular mechanisms for the transfer of epigenetic information through the mitotic period and for the reestablishment of the domain structure of genes after this period.
An insulator element has been found ≈2 kb upstream of the major transcription start sites of the human c-MYC gene (18, 19). The enhancer-blocking activity of this element has been demonstrated by colony assays. The c-MYC insulator is located between a domain containing hyperacetylated histones and a domain containing hypoacetylated histones (19), and it corresponds to the DNase I hypersensitive site I, which is constitutive and pronounced (20–22). The region of the c-MYC insulator/DNase I hypersensitive site I contains a CTCF binding motif (c-MYC CTCF binding site N), a thyroid hormone response element, and a CCAAT box (18, 19, 23). We performed high-resolution in vivo footprinting analysis of this region to determine whether the protein–DNA architecture of the insulator was preserved during mitosis and to better understand the transfer of epigenetic information through mitosis. We observed the absence of transcription factor binding and the disappearance of detectable structures from this region in mitotic cells.
Results
Low-Resolution Southern Blot Analysis.
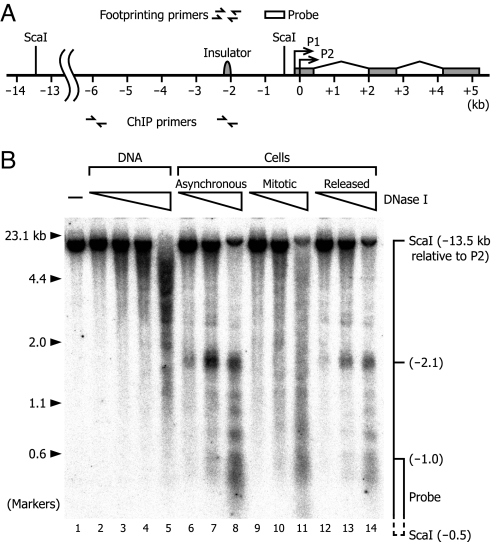
Before mapping protein footprints at high resolution, we performed low-resolution analysis to examine hypersensitivity in the region of the c-MYC insulator element/DNase I hypersensitive site I in three populations of HeLa cells: “asynchronous cells” (primarily interphase), “mitotic cells” (arrested with nocodazole), and “released cells” (incubated for 5 h after release from mitotic arrest). The cells were permeabilized and treated with DNase I. As a control, purified genomic DNA was also digested in vitro with DNase I. As shown in Fig. 1, the positions of DNase I cleavage in a 13-kb genomic ScaI fragment were determined by Southern blot analysis with indirect end-labeling (24). The analysis confirmed the presence of a prominent site of hypersensitivity, which appeared to coincide with the insulator element and the DNase I hypersensitive site I, in the area 2.2–2.0 kb upstream of the transcription start site of the P2 promoter in asynchronous cells (Fig. 1, lanes 6–8). In mitotic cells, however, this hypersensitive site disappeared, suggesting that at least some aspects of the chromatin structure of the insulator region were lost or altered (Fig. 1, lanes 9–11). In the cells released from mitotic arrest, the site became evident again, suggesting that restoration of the chromatin structure had occurred (Fig. 1, lanes 12–14). These results did not seem to be in full accord with previous studies, however. There have been few studies on the precise location of DNase I-hypersensitive sites in specific genes during mitosis, but there have been reports on preservation of the pattern of hypersensitive sites in the human heat shock protein 70 gene (3) and on partial alteration in the chicken ovalbumin and glyceraldehyde 3-phosphate dehydrogenase genes (25). Hypersensitivity in insulator elements in mitotic cells does not appear to have been studied.
Fig. 1.
Southern blot analysis of DNase I-hypersensitive sites upstream of the human c-MYC gene. (A) Map of the region of the c-MYC gene. Bent arrows indicate the transcription start sites of the P1 and P2 promoters. The positions of the three exons, the insulator element, the ScaI restriction sites, and the radioactive probe relative to the P2 start site are indicated. Small arrows indicate the positions of the primers used for footprinting and ChIP. (B) HeLa cells were arrested with nocodazole (mitotic cells) and allowed to proceed through the cell cycle after the arrest (released cells) or were not treated (asynchronous cells). The cells (lanes 6–14) and the purified genomic DNA (lanes 2–5) were treated with DNase I. Triangles indicate increasing concentrations of DNase I. The sample in lane 1 was not digested with DNase I. In this Southern blot hybridization experiment, a 13-kb genomic ScaI fragment was end-labeled indirectly with the probe, as represented by the diagram on the right. The positions of the molecular weight markers (λ/HindIII and φX174/HaeIII digests) are indicated on the left.
High-Resolution in Vivo Footprinting Analysis.
To examine whether the fine structure of the c-MYC insulator was preserved or altered during mitosis, we performed high-resolution in vivo footprinting analysis (26, 27), focusing on the binding of CTCF. Four footprinting agents were used: dimethyl sulfate (DMS), UV light, 4,5′,8-trimethylpsoralen, and DNase I. Asynchronous, mitotic, and released cells were treated with one of the footprinting agents, and genomic DNA was then extracted and subjected to ligation-mediated PCR (LM-PCR) or terminal transferase-dependent PCR (TD-PCR). Purified genomic DNA was treated in vitro with the same agents for a control. Footprints were recognized by comparing the pattern of in vivo formation of DNA damage, visualized as a sequencing ladder, with the pattern of in vitro formation. Only sites and areas reproducibly showing at least 2-fold differences in band intensity between the in vivo and in vitro lanes were considered to be footprints and are marked as such in Figs. 2 and 3, and also in supporting information (SI) Figs. 5 and 6. All of the in vivo footprints detected in this study were in asynchronous and released cells; no footprints were observed in mitotic cells. The details of the analysis are described below.
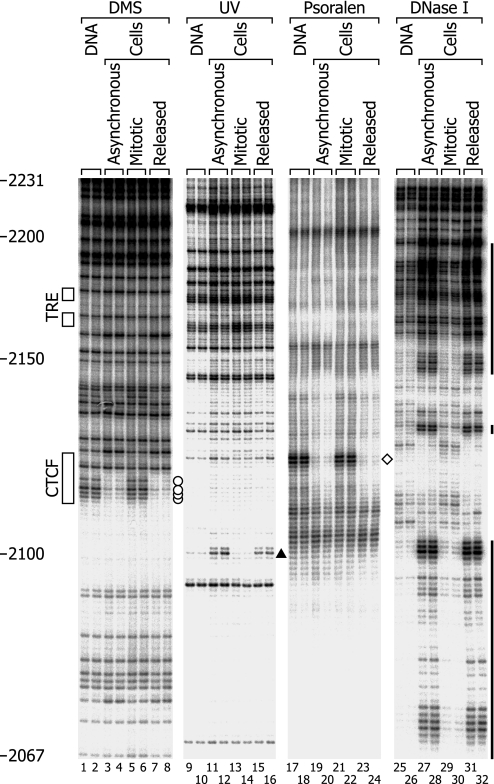
Fig. 2.
In vivo footprinting analysis of the upper strand of the region of the c-MYC insulator element. Primers MYCF1 and MYCF2 were used. The distribution of methylguanines induced by DMS, (6-4) photoproducts induced by UV light, and single-strand breaks induced by DNase I was determined by LM-PCR. The locations of interstrand cross-links induced by 4,5′,8-trimethyl psoralen were determined by TD-PCR. The four blots are from separate gels and are aligned. The numbers on the left indicate the nucleotide positions of the LM-PCR signals relative to the transcription start site of the P2 promoter. The products of TD-PCR are usually two or three nucleotides longer than those of LM-PCR. A band at a nucleotide position in a DNase I lane indicates the hydrolysis of the phosphodiester bond immediately 3′ to the nucleotide. Boxes on the left represent the consensus recognition sequences for transcription factors. Lanes labeled “DNA” denote samples of DNA treated in vitro, and lanes labeled “Cells” denote samples of cells treated in vivo. Open circles between lanes 8 and 9 indicate the sites of decreased formation of methylguanines in vivo (in asynchronous and released cells). The solid triangle between lanes 16 and 17 indicates the site of increased formation of (6-4) photoproducts in vivo. The open diamond between lanes 24 and 25 indicates the site of decreased formation of cross-links in vivo. Vertical bars on the right indicate the areas of increased formation of strand breaks in vivo. TRE, thyroid hormone response element.
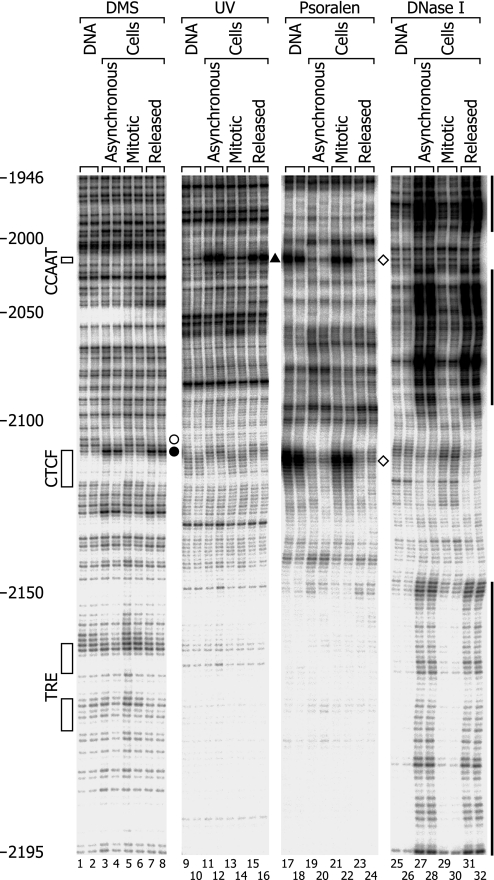
Fig. 3.
In vivo footprinting analysis of the lower strand of the region of the c-MYC insulator element. Primers MYCL1 and MYCL2 were used. The four blots are from two gels and are aligned. The solid circle between lanes 8 and 9 indicates the site of increased formation of methylguanines in vivo. Other notations are similar to those in Fig. 2.
Fig. 2 shows the results of analysis of the upper strand of the c-MYC insulator region. DMS, which alkylates guanines, is the most commonly used agent for the detection of the sequence-specific binding of transcription factors (26). We found clear protection of guanines from alkylation in the area from −2116 to −2112, within the CTCF binding motif in asynchronous cells (Fig. 2, lanes 3 and 4, marked with circles). This footprint disappeared in mitotic cells (Fig. 2, lanes 5 and 6) but reappeared in released cells (lanes 7 and 8).
Short-wavelength UV light efficiently induces cyclobutane pyrimidine dimers and (6-4) photoproducts at the sites of adjacent pyrimidines. The conformational changes of DNA caused by the binding of transcription factors can modulate the induction of these lesions (28). Cleavage of UV-irradiated DNA at the sites of (6-4) photoproducts by piperidine treatment enables the detection of this type of lesions by LM-PCR. We noticed that in vivo induction of (6-4) photoproducts was enhanced at the 5′-TT dipyrimidine at position −2101 to −2100 in asynchronous and released cells but not in mitotic cells (Fig. 2, lanes 9–16, marked with a triangle).
The psoralen molecule, which has a planar tricyclic structure with two photoreactive sites, intercalates in DNA (29). Upon absorption of long-wavelength UV light, psoralen forms DNA interstrand cross-links at the sites of 5′-TA, 5′-AT, 5′-TG, and 5′-GT dinucleotides. The formation of cross-links is inhibited by transcription factors as well as by histone octamers. We observed that in vivo psoralen cross-linking was severely inhibited in the 5′-GTA sequence (containing GT and TA) from −2119 to −2117 in asynchronous and released cells but not in mitotic cells (Fig. 2, lanes 17–24, marked with a diamond).
LM-PCR analysis permits mapping of the fine structure of DNase I-hypersensitive sites. Most hypersensitive sites possess internal “cold spots” against hypersensitive backgrounds; hypersensitivity is considered to be caused by the absence of histones, whereas the relative nuclease resistance in cold spots is considered to be caused by the presence of bound trans-acting factors (30). Therefore, in the case of DNase I analysis, not the areas of hypersensitivity in vivo but those of relative resistance were considered to be footprints. In Fig. 2, the DNase I hypersensitive site in asynchronous and released cells extended from approximately −2200 to beyond the bottom of the panel (lanes 27, 28, 31, and 32). On this hypersensitive background, there was a large cold spot from −2144 to −2103, although it was interrupted by hypersensitivity at nucleotides −2129 and −2128. We presume that this large cold spot corresponds to the area occupied by CTCF because it has been reported that CTCF produces large footprints (16, 18, 31). The DMS, UV light, and psoralen footprints described above were within or were on the periphery of this large area of relative DNase I resistance. In contrast to the situation in asynchronous and released cells, the pattern of DNase I cleavage in mitotic cells (Fig. 2, lanes 29 and 30) was very similar to that of in vitro cleavage of purified DNA (Fig. 2, lanes 25 and 26); the hypersensitive site, together with the internal cold spot, disappeared during mitosis, confirming the results obtained from the low-resolution analysis.
The concentration of DNase I used for in vivo treatment of cells (3 units/ml, 23°C, 3 min) was much higher than that for in vitro treatment of DNA (0.1 unit/ml, 23°C, 5 min). It seems likely that this difference reflects the protection of DNA by the chromatin structure and by other structures in living cells. The presence of the hypersensitive site and the cold spot suggests that the protection in asynchronous and released cells is sequence-specific and nonuniform. The binding of transcription factors and the positioning (or exclusion) of nucleosomes appear to contribute to the nonuniformity. On the other hand, the similarity between the pattern of DNase I cleavage in mitotic cells and that of in vitro cleavage suggests that the protection is provided uniformly in mitotic cells. The loss of the sequence-specific binding of transcription factors during mitosis can certainly be a cause of the uniformity. We speculate that the loss of nucleosome positioning, which we previously observed in mitotic cells (see ref. 5), may also be a contributory factor; randomly distributed nucleosomes may uniformly protect DNA from DNase I cleavage in mitotic cells.
There is a thyroid hormone response element at nucleotides −2176 to −2161, near the upstream border of the hypersensitive site, but no footprints or cold spots were observed in this region. Thus, under the experimental conditions used here, this element did not detectably bind the thyroid hormone receptor, which is consistent with the reported unresponsiveness of HeLa cells to thyroid hormone (32).
Fig. 3 shows the results of analysis of the lower strand. The DNase I hypersensitive site in asynchronous and released cells extends over the entire length of the panel (Fig. 3, lanes 27, 28, 31, and 32). In this hypersensitive background, there were two cold spots: one was around the CTCF binding motif and corresponded approximately to the cold spot observed on the opposite strand, and the other was around a CCAAT box at nucleotides −2015 to −2019, which was outside the region analyzed on the opposite strand. Within the 5′-CCAAT sequence, we found a 5′-CC showing hyperreactivity for the in vivo formation of (6-4) photoproducts (Fig. 3, lanes 11, 12, 15, and 16) and a 5′-AT in which psoralen cross-linking was suppressed in vivo (lanes 19, 20, 23, and 24). These footprints are typical of CCAAT boxes (28, 29). Within or near the CTCF binding motif, there were guanines with increased (position −2111) and decreased (position −2107) DMS reactivity in vivo (Fig. 3, lanes 3, 4, 7, and 8). Inhibition of in vivo psoralen cross-linking was also seen at nucleotides −2117 to −2119 (Fig. 3, lanes 19, 20, 23, and 24), just opposite the psoralen footprint on the upper strand.
The results of the footprinting experiments on both strands are summarized in SI Fig. 5. Although the borders of the DNase I-hypersensitive site I were not mapped except for the upstream border on the upper strand, the analysis of the lower strand revealed that the hypersensitive region included at least 250 bases. This extensive region of hypersensitivity is probably nucleosome-free (30), and it has been proposed that the absence of nucleosomes from insulators is important for their function (33, 34). Within the hypersensitivite site, there were two cold spots or areas of relative resistance around the CCAAT box and around the CTCF binding motif but not around the thyroid hormone response element. The cold spots were accompanied by DMS, UV light, and psoralen footprints internally or on their periphery. The hypersensitive site and the footprints were observed in asynchronous and released cells but not in mitotic cells. These results indicate the loss of the sequence-specific direct binding of trans-acting factors to the CCAAT box and to the CTCF binding motif and the dissolution of the protein–DNA architecture of the c-MYC insulator element/DNase I-hypersensitive site I during mitosis. The restoration of protein binding and the reestablishment of the protein–DNA architecture within 5 h after the release from mitotic arrest were also observed.
As shown in SI Fig. 6, we confirmed the loss of the sequence-specific binding of CTCF during mitosis in another insulator in the DM1 locus (dystrophia myotonica protein kinase gene; see refs. 35 and 36) by in vivo DMS footprinting.
ChIP Analysis.
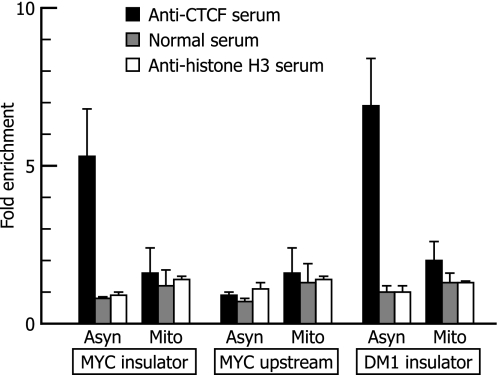
The results described above appeared somewhat inconsistent with previous work by Burke et al. (17), who had shown the retention of CTCF on mitotic chromosomes, so we examined the association of CTCF with the c-MYC gene by using ChIP, one of the methods used by Burke et al. (17). Chromatin from asynchronous and mitotic cells was precipitated with anti-CTCF serum. The relative abundance of specific DNA sequences in the immunoprecipitated chromatin compared with the nonimmunoprecipitated (input) chromatin was estimated by real-time quantitative PCR. In the c-MYC gene, the insulator region (−2.1 kb) and an upstream region (−5.9 kb) were analyzed. The upstream region was used as a negative control because ChIP analysis by Gombert et al. (19) demonstrated that CTCF is constitutively associated with the insulator region but not with the upstream region. The insulator region of the DM1 locus also was analyzed. Our ChIP analysis (Fig. 4) showed the enrichment of the DNA sequences of the insulator regions of the c-MYC gene and the DM1 locus in the immunoprecipitated fractions of the chromatin from asynchronous cells, suggesting the association of CTCF with these regions. However, the majority of the CTCF association with these regions appeared to be lost during mitosis. Thus, the results of our ChIP assays and those of our footprinting experiments are consistent with each other, but they do not seem to be completely in accord with the previous work. ChIP analysis by Burke et al. (17) has shown that there are substantial levels of CTCF association in the c-MYC and DM1 insulator regions in mitotic cells, although the levels are lower than those in asynchronous cells. In our ChIP analysis, the levels of CTCF association with these regions in mitotic cells appeared to be even lower, near the control levels. In these two studies, the same target sites in the same cells were analyzed by similar procedures, and the reason for the somewhat different results is not obvious. We consider that the negative results with mitotic cells in our ChIP analysis are not artifactual for several reasons: (i) agarose gel electrophoresis showed that the DNA in mitotic chromatin had been properly fragmented by sonication (SI Fig. 7); (ii) Western blot analysis demonstrated that full-length CTCF was present in the sonicated chromatin preparation from mitotic cells (SI Fig. 8); and (iii) conversion of the values expressed as fold enrichment in Fig. 4 into those expressed as percentage of input (plotted in SI Fig. 9) reveals that the insulator sequences in mitotic chromatin can be efficiently precipitated with an appropriate serum (anti-histone H3).
Fig. 4.
ChIP analysis of the association of CTCF with the c-MYC gene and the DM1 locus in asynchronous and mitotic cells. Values are expressed as “fold enrichment” over the input control. Chromatin from asynchronous (Asyn) and mitotic (Mito) HeLa cells was precipitated with anti-CTCF serum, normal serum, or anti-histone H3 serum. DNA was recovered from immunoprecipitated and nonimmunoprecipitated (input) chromatin and analyzed by real-time PCR. Primer pairs used for PCR were specific to the insulator or upstream region of the c-MYC gene (see Fig. 1A) or the insulator region of the DM1 locus. The fold enrichment of the target sequence in precipitated DNA compared with input DNA was calculated by comparison of the threshold cycle value of the sample of precipitated DNA with the standard curve generated from the threshold cycle values for input DNA. The means and the SD of data from three independent experiments with separate chromatin preparations are shown.
Discussion
The displacement of transcription complexes from promoters during mitosis, which has been observed in several laboratories (2–4), would seem to be an obstacle for the preservation of stable patterns of expressed and silent genes from one cell generation to the next. The loss of protein–DNA contacts in the c-MYC and DM1 insulators during mitosis reported here also seems to present an obstacle for the preservation of patterns of active and inactive domains from one cell generation to the next. In the c-FOS and U6 promoters, as we reported in ref. 5, not only transcription factor binding but also nucleosome positioning disappears during mitosis. It has also been reported in ref. 37 that the footprints of the origin recognition complex are absent from the replication origin in the lamin B2 gene during mitosis. An attractive scenario that could reconcile the transient disruption of preexisting complexes during mitosis and the stability of epigenetic cell memory has been suggested: only a subset of protein factors remains bound to mitotic chromosomes, and these serve as marks that then help reestablish complexes after mitosis (8–14). Although it has been proposed in ref. 17 that CTCF molecules can function as such epigenetic marks, the footprinting analysis reported here demonstrated the loss of sequence-specific binding of CTCF to the c-MYC and DM1 insulator regions during mitosis. Similarly, our UV light footprinting analysis in ref. 5 demonstrated the loss of sequence-specific binding of TBP (TATA binding protein) to the c-MYC P2 promoter and the c-FOS promoter during mitosis, although TBP has been proposed to mark active genes through this phase of the cell cycle (11, 12). However, these observations do not necessarily mean that there was a complete loss of the association of CTCF or TBP with regional chromatin. It was reported in refs. 38–40 that CTCF interacts with some nuclear or chromatin proteins. TBP is in a large complex (TFIID), and some members of the complex have the ability to interact with nucleosomes or to form a nucleosome-like structure (41–43). Assuming that the indirect associations of CTCF and TBP with regional chromatin through such protein–protein interactions are preserved during mitosis, even though direct binding of these proteins to DNA is lost, the possibility exists that the indirect associations are able to serve as epigenetic marks. If this is actually the situation, the association of CTCF with mitotic chromatin could be weak because of the absence of direct sequence-specific binding, and this could lead to variable experimental results. It was reported in refs. 17 and 44 that mitotic chromosomes can be stained with anti-CTCF antibodies after ethanol/acetic acid fixation but not after paraformaldehyde or methanol fixation. It has been pointed out in ref. 17 that the levels of the association of CTCF with individual genes in mitotic cells determined by ChIP are never the same as in asynchronous cells. In our ChIP analysis, the levels of the association of CTCF with the c-MYC and DM1 insulators in mitotic cells were very low and near the control levels. It is possible that these results reflect a weak or unstable CTCF–chromatin association, although the discrepancies might be explained by technical variations.
An alternative scenario might be suggested preferably by the absence of protein footprints in mitotic cells in this and previous studies (2–5, 37): the transmission of epigenetic information through mitosis depends mainly on classic memory devices such as DNA methylation and histone modifications (6, 7, 45–48). If this is the case, then after mitosis of each generation, cells could reassemble nucleoprotein complexes on promoters, insulators, and other chromosomal elements and reestablish their domain structure and nuclear organization completely de novo by deciphering the genetic and epigenetic information present on DNA and histones. Without the persistent binding of nucleating proteins, the reassembly of regulatory complexes and the resumption of nuclear functions could conceivably be a slow process. Indeed, it has been demonstrated that the recovery of transcriptional activity after mitosis takes several hours (49).
The roles of DNA methylation and histone modifications in modulating the structure and function of chromosomal elements, however, have not been fully elucidated. For example, although it has been reported that DNA methylation regulates both the CTCF binding at and the activity of some insulator elements, including the DM1 and H19 insulators (15, 16, 31, 36), the influence of DNA methylation on other insulators, including the c-MYC insulator, is not clear. The CTCF binding motif in the c-MYC insulator contains no 5′-CG dinucleotide; the 5′-CG dinucleotides in the areas adjacent to this CTCF binding motif were unmethylated in both asynchronous and mitotic cells (data not shown). For estimating the contribution to mitotic cell memory of various factors, including protein binding, DNA methylation, and histone modifications, analysis of a broader range of insulators as well as other various chromosomal elements will be helpful.
Materials and Methods
Cell Culture.
HeLa S3 cells were cultivated as described in ref. 5. Asynchronously growing cells were harvested by trypsinization. We collected mitotic cells by shaking them off the surfaces of the flasks after treatment with 50 ng/ml nocodazole for 16 h. For preparation of cells released from mitotic arrest, the mitotic cells collected as described above were allowed to proceed through the cell cycle and to adhere to the surfaces in nocodazole-free medium for 5 h. The cells then were harvested by trypsinization. The mitotic indexes of the asynchronous, mitotic, and released cell preparations were 4%, 98%, and 3%, respectively (5).
Treatment with DNA-Modifying Agents.
Cells and purified genomic DNA were treated with DMS, UV light, 4,5′,8-trimethylpsoralen, or DNase I essentially as described in refs. 5, 26, and 29. See SI Materials and Methods for details.
Footprinting Analysis by LM-PCR and TD-PCR.
LM-PCR and TD-PCR were carried out as described in refs. 5 and 29 by use of the Expand long-template PCR system (Roche Diagnostics, Indianapolis, IN) during the PCR step. Psoralen-treated DNA was digested with the restriction enzymes PvuII and BglI (New England Biolabs, Ipswich, MA) before alkaline reversal and TD-PCR. The primers MYCF1 (5′-GCAAACGCGGGGAGCAA; nucleotides −1998 to −2014 relative to the transcription start site of the P2 promoter) and MYCF2 (5′-GCGGGGAGCAACCAATCGCTATG; −2004 to −2026) were used for the analysis of the upper strand; MYCL1 (5′-GCAGCCAACCTGAAAGAATAACAAG; −2280 to −2256) and MYCL2 (5′-AAGGAGGTGGCTGGAAACTTGTTTTAAGGA; −2258 to −2229) were used for the analysis of the lower strand. In the primer extension step, MYCF1 and MYCL1 were used; for both PCR and probe labeling, MYCF2 and MYCL2 were used. A DNA fragment from −1934 to −2479 was used as a template in the probe-labeling reactions.
Southern Blot Analysis of DNase I-Hypersensitive Sites.
Southern blot analysis was performed essentially as described by Sambrook and Russel (50). DNase I-digested genomic DNA (10 μg per sample) was treated with the restriction enzyme ScaI (60 units per sample; New England Biolabs). The fragments were separated by electrophoresis through a 1.3% agarose gel in Tris/acetate/EDTA buffer, immobilized on a charged nylon membrane by alkaline capillary transfer, and hybridized to a single-stranded radioactive probe, which had been prepared by the method used in footprinting analysis (29). The primer MYCK1 (5′-ACCGCATCCTTGTCCTGTGA; −466 to −485) and a DNA fragment from −466 to −988 were used in the probe-labeling reaction. A genomic ScaI fragment from −454 to −13473 was end-labeled indirectly with this probe. Hybridization was performed in phosphate/SDS buffer at 65°C, and the final wash was in 4 mM sodium phosphate (pH 7.2)/0.2 mM EDTA/0.2% SDS at 65°C.
ChIP.
ChIP assays were performed with the ChIP assay kit (catalog no. 17−295; Upstate Biotechnology, Lake Placid, NY) essentially according to the manufacturer's instructions. Modifications to their protocol are described in SI Materials and Methods. Chromatin was precipitated with anti-CTCF rabbit serum (catalog no. 07-729; Upstate Biotechnology), normal rabbit serum (Wako Pure Chemical, Osaka, Japan), or anti- histone H3 (C terminus) rabbit serum (catalog no. 07-690; Upstate Biotechnology). The abundance of specific sequences in precipitated chromatin was estimated as described in SI Materials and Methods with a 7500 real-time PCR system (Applied Biosystems, Foster City, CA), with the SYBR premix Ex Taq reagent (Takara Bio, Otsu, Japan), and with the following primers: MYCF3 (5′-ACGCGGGGAGCAACCAATC; −2002 to −2020) and MYCL3 (5′-GCTCCTGCCCCCACCTGAC; −2181 to −2163) for amplification of the insulator region of the c-MYC gene; the H.1 primers of Gombert et al. (19) for amplification of the upstream −5.9-kb region of the c-MYC gene; and DMPKA3 (5′-ACCGCCTGCCAGTTCACAA) and DMPKD1 (5′-GCAGCATTCCCGGCTACAAG) for amplification of the insulator region of the DM1 locus.
Supplementary Material
Acknowledgments
We thank Dr. Wendy M. Gombert and Dr. Anton Krumm (both of University of Washington School of Medicine, Seattle, WA) for providing primer sequences, Dr. Leon N. Kapp for comments on the manuscript, and Ms. Yukiko Ikeda for help with real-time PCR. This work was supported by the Japan Society for the Promotion of Science.
Abbreviations
- DMS
dimethyl sulfate
- LM-PCR
ligation-mediated PCR
- TD-PCR
terminal transferase-dependent PCR
- TBP
TATA binding protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702363104/DC1.
References
- 1.Gottesfeld JM, Forbes DJ. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 2.Hershkovitz M, Riggs AD. Proc Natl Acad Sci USA. 1995;92:2379–2383. doi: 10.1073/pnas.92.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 4.Boyd DC, Pombo A, Murphy S. Gene. 2003;315:103–112. doi: 10.1016/s0378-1119(03)00717-0. [DOI] [PubMed] [Google Scholar]
- 5.Komura J, Ono T. J Biol Chem. 2005;280:14530–14535. doi: 10.1074/jbc.M500637200. [DOI] [PubMed] [Google Scholar]
- 6.Riggs AD. Cytogenet Genome Res. 2002;99:17–24. doi: 10.1159/000071569. [DOI] [PubMed] [Google Scholar]
- 7.Richards EJ, Elgin SCR. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 8.Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 9.Michelotti EF, Sanford S, Levens D. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 10.John S, Workman JL. BioEssays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 11.Christova R, Oelgeschläger T. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Hinkley CS, Henry RW, Huang S. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge O-K, Sarge KD. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 15.West AG, Gaszner M, Felsenfeld G. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 16.Ohlsson R, Renkawitz R, Lobanenkov V. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 17.Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, et al. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R. EMBO J. 2003;22:1579–1587. doi: 10.1093/emboj/cdg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gombert WM, Farris SD, Rubio ED, Morey-Rosler KM, Schubach WH, Krumm A. Mol Cell Biol. 2003;23:9338–9348. doi: 10.1128/MCB.23.24.9338-9348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siebenlist U, Hennighausen L, Battey J, Leder P. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 21.Dyson PJ, Littlewood TD, Forster A, Rabbitts TH. EMBO J. 1985;4:2885–2891. doi: 10.1002/j.1460-2075.1985.tb04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer CA, Groudine M. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 23.Arcinas M, Boxer LM. Oncogene. 1994;9:2699–2706. [PubMed] [Google Scholar]
- 24.Wu C. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 25.Kuo MT, Iyer B, Schwarz RJ. Nucleic Acids Res. 1982;10:4565–4579. doi: 10.1093/nar/10.15.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeifer GP, Chen HH, Komura J, Riggs AD. Methods Enzymol. 1999;304:548–571. doi: 10.1016/s0076-6879(99)04032-x. [DOI] [PubMed] [Google Scholar]
- 27.Zaret KS. Methods Enzymol. 1999;304:612–626. doi: 10.1016/s0076-6879(99)04036-7. [DOI] [PubMed] [Google Scholar]
- 28.Tornaletti S, Pfeifer GP. J Mol Biol. 1995;249:714–728. doi: 10.1006/jmbi.1995.0331. [DOI] [PubMed] [Google Scholar]
- 29.Komura J, Ikehata H, Hosoi Y, Riggs AD, Ono T. Biochemistry. 2001;40:4096–4105. doi: 10.1021/bi002539f. [DOI] [PubMed] [Google Scholar]
- 30.Gross DS, Garrard WT. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 31.Szabó PE, Tang S-HE, Rentsendorj A, Pfeifer GP, Mann JR. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 32.Fondell JD, Ge H, Roeder RG. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi X, Yu Q, Sandmeier JJ, Zou Y. Mol Cell Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Kim A, Song S, Dean A. J Biol Chem. 2006;281:30573–30580. doi: 10.1074/jbc.M606803200. [DOI] [PubMed] [Google Scholar]
- 35.Steinbach P, Gläser D, Vogel W, Wolf M, Schwemmle S. Am J Hum Genet. 1998;62:278–285. doi: 10.1086/301711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 37.Abdurashidova G, Riva S, Biamonti G, Giacca M, Falaschi A. EMBO J. 1998;17:2961–2969. doi: 10.1093/emboj/17.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutz M, Burke LJ, Barreto G, Goeman F, Greb H, Arnold R, Schultheiβ H, Brehm A, Kouzarides T, Lobanenkov V, Renkawitz R. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernukhin IV, Shamsuddin S, Robinson AF, Carne AF, Paul A, El-Kady AI, Lobanenkov VV, Klenova EM. J Biol Chem. 2000;275:29915–29921. doi: 10.1074/jbc.M001538200. [DOI] [PubMed] [Google Scholar]
- 40.Ishihara K, Oshimura M, Nakao M. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Burley SK, Roeder RG. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann A, Chiang C-M, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, Roeder RG. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson RH, Ladurner AG, King DS, Tjian R. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R, Burke LJ, Rasko JEJ, Lobanenkov V, Renkawitz R. Exp Cell Res. 2004;294:86–93. doi: 10.1016/j.yexcr.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang X-J, Verdin E, Bazett-Jones DP. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 46.Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 47.Peters AHFM, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 48.Kouskouti A, Talianidis I. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein J, Grummt I. Proc Natl Acad Sci USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.