Abstract
Chromatin-remodeling complexes can translocate nucleosomes along the DNA in an ATP-coupled reaction. This process is an important regulator of all DNA-dependent processes because it determines whether certain DNA sequences are found in regions between nucleosomes with increased accessibility for other factors or wrapped around the histone octamer complex. In a comparison of seven different chromatin-remodeling machines (ACF, ISWI, Snf2H, Chd1, Mi-2, Brg1, and NURF), it is demonstrated that these complexes can read out DNA sequence features to establish specific nucleosome-positioning patterns. For one of the remodelers, ACF, we identified a 40-bp DNA sequence element that directs nucleosome positioning. Furthermore, we show that nucleosome positioning by the remodelers ACF and Chd1 is determined by a reduced affinity to the end product of the translocation reaction. The results suggest that the linkage of differential remodeling activities with the intrinsic binding preferences of nucleosomes can result in establishing distinct chromatin structures that depend on the DNA sequence and define the DNA accessibility for other protein factors.
Keywords: ACF, nucleosome remodeling, nucleosome positioning
DNA packaging into nucleosomes has long been recognized as a mechanism to control the accessibility of protein–DNA interactions involved in processes like transcription, replication repair, and recombination (1). The specific location of nucleosomes on DNA may play both inhibitory and activating roles (2) and depends on the ATP-coupled activity of chromatin-remodeling complexes that reposition nucleosomes or evict them from the DNA (3). For example, nucleosomes can be located at silent yeast promoters to occlude binding of basal transcription factors (4, 5). In contrast, an alternative nucleosome position with transcription factor-binding sites in the flanking linker DNA was shown to stimulate transcription (6–8).
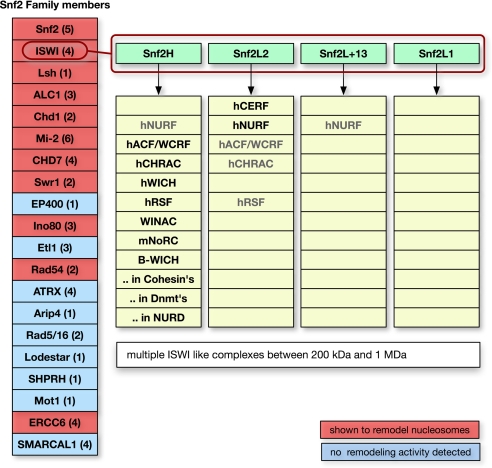
Because the cell harbors hundreds of different remodeling complexes, it appears likely that they possess distinct activities, rather than solely being unspecific nucleosome-moving entities. Indeed, recent results demonstrate that the majority of nucleosomes in yeast are found at well defined positions (9). These sites can be predicted, in part, from the analysis of DNA dinucleotide sequence motives (10, 11). However, it is currently unclear whether nucleosome positioning in the cell results only from sequence preferences of histone–DNA interactions or is directed by additional factors like the chromatin-remodeling complexes. As depicted in Fig. 1, these complexes comprise several groups of the Snf2-like ATPases and include the Snf2, ISWI, Mi-2, Chd1, Ino80, ERCC6, ALC1, CHD7, Swr1, RAD54, and Lsh subfamilies (12). Each subfamily consists of at least one to six similar ATPases, many of which have been shown to remodel nucleosomes, transfer histone octamers in trans, and generate superhelical torsion in DNA as reviewed previously (3, 13). The number of specific chromatin-remodeling activities in the cell is further increased by the assembly of the ATPases into large multiprotein complexes, where the same ATPase is shared within different remodeling complexes. For example, the human BAF and PBAF complexes differ by the subunits BAF250 and BAF180 present in the one, but not in the other, complex, and at least three different human NURD complexes containing the ATPase Mi-2 were described (14). In addition, the molecular motors of the complexes can be exchanged with other ATPases of the same subfamily. This finding was documented for the human BAF complex that contains either the hBRM or BRG1 ATPase (15) and for several ISWI complexes.
Fig. 1.
Mammalian chromatin-remodeling complexes are highly diverse. (Left) The Snf2 family members present in humans, with the number of individual proteins within a subfamily in parentheses. The 11 subfamilies marked in red were shown to possess ATP-dependent chromatin-remodeling activities. Each of these subfamilies comprises many different members. (Right) The multiple ISWI ATPases complexes known to date. There are four ISWI subgroups (green boxes), and the known chromatin-remodeling complexes containing these ATPases are listed. Complexes in light gray type were shown to exist with the alternative ISWI-type ATPases.
The diversity of the mammalian ISWI-like chromatin-remodeling complexes is described in Fig. 1. The mammalian genome encodes for at least four ISWI-like ATPases, Snf2H, Snf2L1, SNF2L2, and the catalytically inactive splice-variant Snf2L + 13 (16–18). To date, about a dozen specific mammalian complexes containing one of these ATPases were purified. It was shown that at least four of these complexes (hCHRAC, hRSF, hNURF, and hACF) can exist as isoformic complexes (i.e., they contain alternative ISWI-like ATPases) (16, 19, 20). This result suggests that the exchange of ATPases is a common theme and increases the number and complexity of chromatin-remodeling complexes in the cell. In addition, many of the subunits of the remodeling complexes exist as multiple-splice variants, such as CERC2 (19), BPTF (21), Tip5 (22), and Baz2B (22). This feature would further increase the diversity of ISWI-remodeling complexes. In summary, the current data indicate that a human cell is likely to form >40 different ISWI-like complexes. Extrapolating this finding to other Snf2 subfamilies, it is estimated that the nucleus harbors hundreds or even thousands of different chromatin-remodeling complexes. Furthermore, some of these complexes are highly abundant. Quantification studies in yeast suggest that one chromatin-remodeling complex is present for ≈10 nucleosomes (23, 24). This diversity and high total concentration of remodeling complexes appear to be unnecessary for simply maintaining an unspecific fluid and easily accessible conformation of chromatin. Instead it suggests that chromatin-remodeling complexes provide a higher order regulatory level by establishing specific chromatin structures in the cell. This hypothesis was tested here by comparing seven different remodeling machines. It is demonstrated that each of those machines possesses unique nucleosome-positioning characteristics. For the ACF-remodeling reaction, it is shown that nucleosome positioning is sequence-dependent, in that a short DNA element can determine ACF-dependent nucleosome positions. Finally, the mechanism of remodeler-dependent nucleosome positioning was analyzed. It is concluded that, for the ACF- and Chd1-remodeling machines, differences in the affinity of the remodeler toward differently positioned nucleosomes determine the outcome of the remodeling reactions.
Results
Chromatin-Remodeling ATPases Establish Unique Nucleosome Positions.
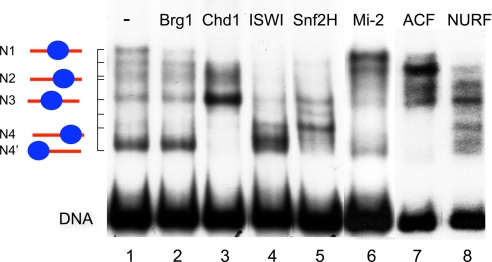
The DNA sequence-dependent specificity of the chromatin-remodeling reaction was examined in a comparison of seven different chromatin-remodeling machines (ACF, ISWI, Snf2H, Chd1, Mi-2, Brg1, and NURF) (18, 25–29). Two well characterized nucleosome substrates, the Drosophila hsp70 DNA fragment (28) and the murine rDNA promoter (30, 31), were used [Fig. 2 and supporting information (SI) Figs. 6 and 7A]. Nucleosome assembly on the hsp70 DNA fragment by salt dialysis gives rise to a distribution of nucleosomes positioned at five dominant positions. The different positions of the nucleosomes are designated as N1, N2, N3, N4, and N4′ and can be visualized by native PAGE because they show differences in their electrophoretic mobility (Fig. 2, lane 1) (28). This mixed nucleosome population was used as a substrate for the seven different chromatin-remodeling complexes listed previously. All remodeling complexes are capable of relocating nucleosomes on the substrate in an ATP-dependent reaction. The endpoint of this reaction obtained with a specific remodeling complex (lanes 2–8) was then compared with the distribution of nucleosomes on the input substrate (lane 1). The results of the remodeling reactions clearly demonstrate that every remodeling enzyme possesses a distinct nucleosome-positioning activity. This striking result cannot be explained by DNA sequence-dependent differences in the histone–DNA interactions. If the remodeling complexes would simply catalyze a transfer of the nucleosomes to their highest affinity histone-binding sites, the same end product should be obtained for the different remodeling complexes. This finding was clearly not the case. For example, NURF-dependent nucleosome remodeling was shown to position the nucleosomes efficiently from N1, N2, and N4 positions to the N3 position (lane 8) (28). In contrast, ACF, a similar remodeling complex harboring the identical ISWI ATPase, behaves differently and preferentially positions the nucleosomes at position N2 (lane 7). In addition, each isolated molecular motor subunit has a distinct positioning behavior. ISWI, the ATPase of ACF and NURF, positions nucleosomes at N4′ and N4 positions (lane 4). However, Snf2H preferentially places the nucleosomes on three sites between position N3 and a positioning site above N4 (lane 5). BRG1 does not change the nucleosome distribution significantly, but nucleosomes are displaced from the central position N1 (lane 2). Chd1 transfers the nucleosomes almost completely to position N3 (lane 3), whereas Mi-2 positions nucleosomes preferentially at the position N1 (lane 6). A similar complex-specific remodeling activity was observed in the analysis of the rDNA substrate (SI Fig. 7A). At the resolution of these experiments, the remodeling reactions led to a different distribution of nucleosomes at sites N1, N2, N3, N4, and N4′ (Fig. 2) or N1 and N2 (SI Fig. 7A), but no new nucleosome positions (except for the Snf2H reaction) were created (SI Fig. 7B).
Fig. 2.
Chromatin-remodeling complexes position nucleosomes in a DNA sequence-specific manner. The nucleosome substrate was reconstituted by salt dialysis on a radioactively labeled 350-bp fragment carrying the hsp70 promoter. A mixture of a single nucleosome at five different major positions, indicated as N1, N2, N3, N4, and N4′, was obtained (28). This mixed nucleosome population (lane 1) was used as the same substrate for all seven remodelers shown. The endpoint of the nucleosome translocation reaction obtained after incubation for 90 min at 26°C in the presence of ATP is shown for recombinant Brg1 (lane 2), Chd1 (lane 3), ISWI (lane 4), Snf2H (lane 5), Mi-2 (lane 6), ACF (lane 7), and NURF (lane 8).
It is concluded that remodeling machines do interpret the DNA sequence/structure information in different ways, establishing individual nucleosome-positioning patterns on a given DNA sequence. In particular, the nucleosome positioning depended on both the type of the ATPase motor protein as well as the composition of the multiprotein complex into which it is integrated (see also SI Fig. 7A). The nucleosome movements proceeded predominantly by positions characterized by an intrinsic nucleosome affinity preference (SI Fig. 7B) because the intermediate positions were mostly identical with the initial nucleosome positions generated by salt dialysis assembly. However, the relative occupancy of these sites can be strongly affected by specific chromatin-remodeling activities.
A Small DNA Element Directs ACF-Dependent Nucleosome Positioning.
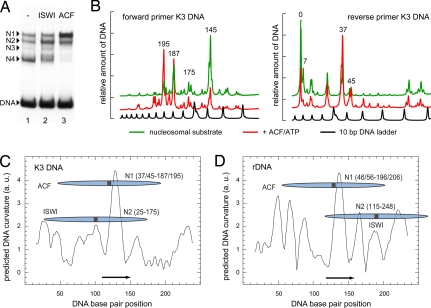
If the DNA sequence encodes information on the positioning of nucleosomes by remodeling complexes, it should be possible to identify specific DNA sequence elements that direct nucleosome positioning independent of the surrounding DNA sequence context. This prediction was tested with ACF in the experiments presented in Fig. 3. On the rDNA sequence, ACF moves the nucleosome from border positions to two rotationally spaced positions occupying nucleotides −190 to −40 and −180 to −30 relative to the rDNA gene transcription start site (30, 31). This finding corresponds to positions 46/56 to 196/206 (N1) on the 248-bp rDNA fragment studied in SI Fig. 7A. Previous studies established that the rDNA promoter of a variety of organisms exhibits a conserved sequence-dependent structure (32, 33). An analysis of this sequence for specific features revealed a strong correlation between ACF-dependent nucleosome positioning and the presence of an intrinsically curved DNA region (Fig. 3D). The dyad axis of the positioned nucleosome with the rDNA was close to the DNA curvature peak that was verified experimentally (SI Fig. 7C). Accordingly, a 40-bp fragment with the center of the curved region (positions 115–155) was then cloned into the DNA vector pT-K3 to examine it in a sequence environment with no apparent sequence similarities to the rDNA fragment. The resulting 253-bp-long DNA was used to assemble nucleosomes by salt dialysis. Nucleosomes were positioned at three major sites (N1, N2, and N4) and one minor site (N3) (Fig. 3A, lane 1). This distribution of differently positioned nucleosomes was used as substrate for the remodeling reaction with ISWI or ACF (Fig. 3A, lanes 2 and 3). In contrast to the 248-bp rDNA template (SI Fig. 7A), the ISWI-dependent nucleosome positioning did not place the nucleosomes to the extremes of the DNA, but preferentially to position N2 on the expense of nucleosomes positioned at the DNA border (N4). This result demonstrates that ISWI is not an unspecific, asymmetric molecular motor that moves any nucleosome to the ends of the DNA fragment, but can recognize certain DNA features (34). ACF, in contrast, translocated the nucleosomes predominantly to the central N1 site. The nucleosome positions involved in the remodeling reaction with the K3 DNA fragment were mapped at a high resolution (Fig. 3B). The major nucleosome positions were assigned to N1 being an unresolved mixture of two rotationally positioned nucleosomes at 37/45 to 195/187, N2 covering positions 25 to 175, and the nucleosomes positioned at the DNA ends (N4) consisting mainly of positions 0/7 to 151/157. The preferred sites found with the K3 DNA were superimposed with the DNA curvature prediction plot (Fig. 3C). From comparison with Fig. 3D, it is apparent that the 40-base pair sequence element from the rDNA promoter was sufficient to direct ACF-dependent nucleosome positioning also in the K3 fragment, with the dyad axis again being placed close to the region of highest DNA curvature. Due to the possibility that ACF simply moves the nucleosome to the center of the DNA because of its preference for sufficiently long (≈30-bp) DNA flanking the nucleosomes, two additional nucleosome substrates were examined (SI Fig. 8). The results clearly demonstrate that the 40-bp DNA element identified here is able to direct nucleosome positioning by ACF to a position closer to the border of the DNA.
Fig. 3.
A short DNA element can direct ACF-dependent nucleosome positioning. (A) Remodeling reaction with ACF or ISWI with a nucleosome substrate containing a 253-bp-long DNA fragment (K3 DNA) from the pT-K3 plasmid. After nucleosome assembly by salt dialysis on the K3 DNA, a mixed population of a single nucleosome with three main positions (N1, N2, and N4) and one minor position (N3, lane 1) was obtained. This substrate was used in a remodeling reaction with ISWI (lane 2) or ACF (lane 3). (B) High-resolution mapping of the remodeler-dependent nucleosome positions on the K3 DNA substrate. MNase protection and subsequent primer extension reactions were conducted. Scans for the primer extension reactions (Left, forward primer; Right, reverse primer) are shown for the nucleosomal input substrate (green, corresponding to A, lane 1) and the remodeling reaction for ACF (red, corresponding to A, lane 3). The black curve shows a 10-bp DNA marker. The same analysis was conducted with ISWI (data not shown). The peaks reflect nucleosomes positioned adjacent to this site. Considering that 147 bp of DNA are protected by the nucleosome, the major nucleosome positions were identified as 37/45 to 187/195 for N1, 25 to 175 for N2, and 0/7 to 151/157 for N4. (C) The ACF- and ISWI-dependent nucleosome positions determined on the 253-bp K3 DNA fragment were plotted together with the predicted DNA curvature. The black arrow refers to the 40-bp DNA sequence encompassing the region of maximal DNA curvature from the rDNA sequence that was cloned into the K3 DNA. (D) Same analysis as in C, but for the 248-bp rDNA promoter fragment with the previously determined nucleosome sites (30, 31).
Two Models Can Explain Remodeler-Dependent Nucleosome Positioning.
The comparison presented above revealed that the end product of the chromatin-remodeling reaction depends on both the type of chromatin remodeler and the DNA sequence (Figs. 2 and 3 and SI Figs. 7 and 8). To explain how a remodeling machine is able to direct the nucleosome to a specific position, the kinetic model presented in Fig. 4A was used. The approach considers the translocation of nucleosomes as an enzymatic reaction that follows a Michaelis–Menten-like model. This finding implies that “good” substrates for the enzyme (in this case, the chromatin-remodeling complex) are characterized by a high affinity of enzyme and its nucleosome substrate (low value of Michaelis–Menten constant KM) and a high catalytic conversion rate kcatof the enzyme–substrate complex to the product, which is the repositioned nucleosome. In this case, the kcat/KM ratio is high as expected for an efficient catalytic process. The opposite would be true for “bad” nucleosome-remodeling substrates (i.e., the kcat/KM ratio is low). This view leads to the proposal that the nucleosome-translocation reaction proceeds by moving nucleosomes from sites where they are good substrates to sites where they are bad substrates. Differences in the remodeling activity are due to DNA sequence-dependent differences for nucleosome locations that make them good and bad substrates for a given complex.
Fig. 4.
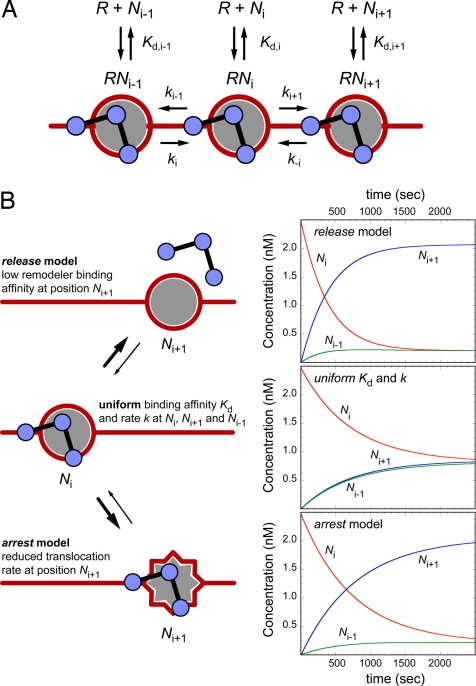
Mechanisms of nucleosome positioning by chromatin-remodeling complexes. (A) Scheme for the remodeling reaction. Three positions on the DNA (i − 1, i, and i + 1) are considered. The remodeler R can bind to a nucleosome N at each of these three positions with a dissociation constant Kd. Translocation to or from these positions occurs with rate constants k as described in the text. (B) Two models for nucleosome positioning are depicted. (Right) Corresponding time course of the concentrations of nucleosomes. The reaction starts with all nucleosomes at position i at an initial concentration of 2.5 × 10−9 M. If all binding constants and translocation rate constants are identical (uniform Kd and k), a homogenous distribution is obtained in equilibrium, where one-third of the nucleosomes is at positions i − 1, i, and i + 1, as expected. (Left) In the release model, the binding affinity to the nucleosome at position i + 1 is reduced by a 10-fold higher value of Kd,i+1, compared with positions i and i − 1. This result leads to a distribution in which ≈80% of the nucleosomes are at this site when an equilibrium is reached. For the arrest model, the rate constant k−1 that translocates the nucleosome from position i + 1 back to position i is 10 times reduced compared with the other translocation rates. Again ≈80% of the nucleosomes will be positioned at site i + 1 as the reaction reaches a steady state. The kinetic simulations were conducted with concentrations similar to those used in the in vitro reaction of ≈1 remodeler per 50 nucleosomes.
The reaction is described as follows. The nucleosome N can be at position i, i + 1, or i − 1, and the remodeler R can bind to nucleosomes at these positions. At the beginning of the reaction, all nucleosomes are at position i. The RNi complex can now translocate the nucleosome by a remodeler-specific number of base pairs (SWI/SNF ≈50 bp, ISW2 ≈10 bp) (35) to the other positions with the rate constants ki+1 and ki−1 or the RNi complex can dissociate with the equilibrium dissociation constant Kd,i into nucleosome Ni and free remodeler R with Kd,i = [R] × [Ni]/[RNi]. The same reaction can occur at positions i + 1 and i − 1. To position the nucleosome at a specific site on the DNA, we propose that certain DNA sequences make nucleosomes bad remodeling substrates with a low escape rate from these sites so that they constitute the preferred end points of the remodeling reaction. This finding is illustrated in Fig. 4B for a remodeling reaction that positions the nucleosome preferentially at position i + 1.
The escape rate kesc,i+1 from position i + 1 is given by the translocation rate constant k−i times the concentration of the RNi+1 complex, which in turn is determined by the corresponding equilibrium dissociation constant Kd,i+1. Accordingly, we can conclude that kesc,i+1 is proportional to k−i/Kd,i+1. To position the nucleosome at i + 1, either the translocation rate k−i away from this position or the binding affinity of the remodeler to the nucleosome at position i + 1 has to be reduced. The latter would be equivalent to increasing the value of Kd,i+1. This result is referred to here as a “release” mechanism. Its mode of operation is similar to transcription termination by specific DNA terminator sequences that form a hairpin structure in the RNA, which then disrupt the binding of RNA polymerase to the template so that the elongation reaction stops at this site (36, 37). For the second mechanism that involves a low translocation rate k−i away from the corresponding nucleosome position, the term “arrest” mechanism is used. It can again be described by analogy to the transcription reaction, where pausing/arrest of RNA polymerase can occur at certain DNA sequences because of a rearrangement of the enzyme's active site without the dissociation of the enzyme from the template (38). Accordingly, we can envision an arrest mechanism for the chromatin-remodeling reaction, in which a specific intermediate form interferes with the continuation of the translocation reaction. In contrast to the release model, the binding affinity of the remodeler is not reduced. It is noted that any increase of the binding affinity for the nucleosome at a given position will increase the likelihood of moving the nucleosome away from this position if the translocation rate remains unaffected. A particularly tight nucleosome remodeler reaction that induces a block of the translocation reaction (i.e., low rate k−i rate) could potentially position a nucleosome to a certain site. However, this anchoring mechanism would only work effectively under stoichiometric concentrations of remodeler and nucleosome, and its kinetics are not compatible with that of the in vitro remodeling reaction under enzymatic conditions studied here of ≈1 remodeling complex per 50 nucleosomes.
Nucleosome Positioning by Chd1 and ACF Follows the Release Model.
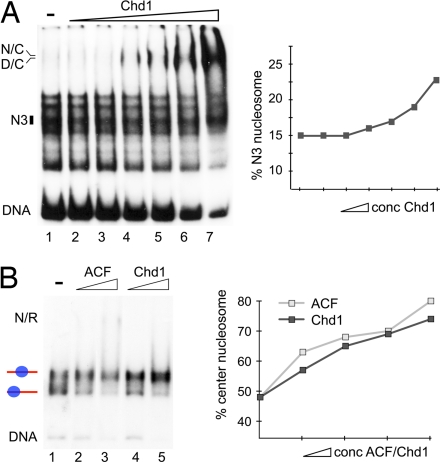
The release model predicts that the affinity of the remodeler to the nucleosome is reduced at the endpoint of the remodeling reaction. According to the arrest model, binding affinity at the terminal position is not lowered, but the translocation reaction is inhibited. These predictions were tested with Chd1 and ACF. The relative binding affinities of the remodelers to the initial mixture of nucleosome positions were analyzed in EMSAs in the absence of ATP so that only the remodeler nucleosome-binding event is examined. The addition of increasing amounts of Chd1 results in DNA and nucleosome binding and the appearance of DNA–Chd1 and nucleosome–Chd1 complexes (Fig. 5A). From a comparison of the relative intensities, it can be seen that nucleosomes at positions N1, N2, and N4 were preferential substrates to form Chd1–nucleosome complexes, whereas the signal for nucleosomes positioned at site N3 decreased to a much lower extent. As shown in Fig. 2, Chd1 positions nucleosomes at N3, the site with the lowest binding affinity for Chd1. This finding suggests that Chd1 translocates nucleosomes according to the release model. Similar results were obtained by ACF with this nucleosomal DNA (data not shown). The same behavior of the two remodelers was also observed for the rDNA nucleosome substrate. Purified nucleosomes positioned at either the center or border of the rDNA fragment were mixed in stoichiometric amounts and used for binding assays (Fig. 5B). At increasing concentrations, Chd1 and ACF bound preferentially to nucleosomes positioned at the border of the DNA fragment (lanes 2 to 5; the graph shows the fraction of the central nucleosome obtained with increasing ACF/Chd1 concentrations). Both remodelers displayed weaker binding affinities to the central nucleosome, which is the position to which they translocate the nucleosome in the remodeling reaction (SI Fig. 7A). Thus, for the two remodelers and two nucleosome substrates examined here, nucleosome positioning occurs by the release mechanism.
Fig. 5.
ACF and Chd1 position nucleosomes according to the release model. Nucleosome position-dependent differences in the affinity of the remodeling complexes to the nucleosomal substrate were analyzed by EMSAs. (A) (Left) A mixed nucleosomal species reconstituted on the hsp70 DNA (lane 1) was incubated with increasing concentrations of Chd1 (lanes 2–7) in the absence of ATP. The position of the appearing DNA–Chd1 (D/C) and the nucleosome–Chd1 (N/C) complexes are indicated. The position of the N3 nucleosome is shown by a black box. This position also is the preferred endpoint of the remodeling reaction (see Fig. 2). (Right) Percentage of nucleosomes at position N3 (radioactivity in the N3 band divided through the sum of the radioactivity of all nucleosome bands) is plotted versus the Chd1 concentration. An increase of the N3 fraction is apparent, suggesting that this site is the lowest affinity binding site for Chd1 with this substrate. (B) Chd1 and ACF binding to nucleosomes reconstituted at the rDNA promoter fragment. A purified mixture of nucleosomes positioned at the center and the border of the rDNA fragment (lane 1) was incubated with increasing concentrations of ACF (lanes 2 and 3) or Chd1 (lanes 4 and 5). The position of remodeler–nucleosome complexes (N/R) is indicated. The graph represents the fraction of nucleosomes at the center position with increasing concentrations of Chd1 or ACF. This lower affinity binding site also is the preferred endpoint of the reaction as shown in SI Fig. 7B.
Discussion
The present study demonstrates that the chromatin remodeler can establish specific local chromatin structures by reading out DNA features and targeting nucleosomes to specific positions. To exploit this differential activity of remodeling complexes in vivo, it appears necessary to spatially and temporally confine a given complex to certain chromatin regions. Indeed, an increasing number of reports describes such a targeting of chromatin remodelers to specific genomic loci that are characterized by their pattern of epigenetic markers as reviewed recently for Drosophila (39). Thus, targeting of chromatin-remodeling complexes in conjunction with their DNA sequence-directed activity would provide a mechanism for the gene-specific regulation of DNA-dependent processes by modulation of the DNA accessibility. For ACF and Chd1, this process follows a release mechanism, according to which the endpoint of the translocation reaction is determined by a reduced affinity of the remodeler to the nucleosome at this site.
The physiological relevance of specifically positioned nucleosomes for the organization of regulatory regions of eukaryotic genes has been demonstrated in numerous systems (40–44). However, although the ability of certain DNA sequences to position nucleosomes in vitro is well established, many of these sequences fail to precisely position nucleosomes in vivo (44). This result indicates that, in addition to DNA structure and flexibility, other mechanisms define nucleosome positioning in the cell. For example, it has been shown that DNA-binding factors like the α2–MCM1 complex actively position nucleosomes at repressed genes in yeast α-cells. This process requires the intact histone H4 tail (45, 46), a target of the ISWI-containing remodeling machines (47). Similarly, the Ssn6–Tup1 complex is a global corepressor responsible for nucleosome positioning at a number of genes and the recombination enhancer of the silent mating-type loci in budding yeast (48–52). For the RNR3 gene, the precise nucleosome positioning required the ISW2 chromatin-remodeling complex in addition to Ssn6–Tup1 (53). Furthermore, recent work demonstrates that the SNF2H-containing remodeling complex NoRC is involved in the repression of the rRNA genes that are characterized by two specific nucleosome positions discriminating between the active and inactive genes (54, 55). In this system, the recruitment of NoRC reorganizes the chromatin structure by moving the promoter-bound nucleosome ≈25 bp downstream to the position found at inactive genes (56).
In light of these findings and the results reported here, the high abundance and diversity of remodeling complexes suggests that chromatin-remodeling machines are specific chromatin organizers and not simply nucleosome mobilizers. Their activity seems to be directed by two additional information layers encoded in the DNA sequence. One would represent binding preferences of the histone octamer to certain DNA sequence elements. It has been shown for yeast that about half of the in vivo nucleosome positions can be predicted solely from the underlying DNA sequence (10, 11). These sites are likely to provide thermodynamically favorable histone–DNA interactions, and the data reported here suggest that they also are selected as preferred locations in the remodeling reaction (Fig. 3 and SI Fig. 7B). However, the relative occupancy of these sites can be strongly affected by the chromatin-remodeling complexes. The coupling of their specific activity with intrinsic nucleosome preferences for certain DNA sequences could contribute significantly to determining nucleosome locations in living cells. Accordingly, the targeting of a significant fraction of nucleosomes to their DNA sites in the cell cannot be predicted without including the characteristic activities of chromatin-remodeling complexes present at the respective genomic loci. This view is consistent with a recent analysis of nucleosome locations in yeast that points to the involvement of additional factors in the determination of nucleosome positions (57). As demonstrated here, one important parameter to be considered is the binding affinity of the remodeler and the nucleosome. Either because of a sequence-specificity of remodeler–DNA interactions or more indirectly by an altered nucleosome structure, a reduced remodeler–nucleosome interaction leads to the release of nucleosomes to these sites. Thus, the positioning of nucleosomes in the cell could involve a chromatin-remodeling code. Features encoded by the DNA sequence are recognized by chromatin-remodeling complexes to establish specific nucleosome-positioning patterns that define the accessibility of DNA and with it on or off states for DNA-dependent processes.
Materials and Methods
Nucleosome-Remodeling Experiments.
Recombinant ISWI, ACF, Brg1, Chd1, Snf2H, and Mi-2 were expressed in Sf9 cells and prepared as described previously (27, 34). The hsp70 DNA fragment was generated by PCR with [α-32P]dCTP for labeling (28). The 40-bp fragment encompassing the major DNA bending peak (CTGGGGAGGT GGCCCCAAAA ATGACCCCAT AACGAAAAGA) of this DNA was cloned into the pT7 blue3 Vector. From this vector, the 253-bp-long pT-K3 fragment with the insert was generated by PCR. Nucleosome-reconstitution and nucleosome-remodeling reactions were performed according to the protocol of Längst et al. (30). Briefly, nucleosomes and DNA were incubated at ratio of ≈1 remodeler complex per 50 nucleosomes for 90 min at 26°C in the presence of 1 mM ATP, and nucleosome positions were analyzed by native PAGE. EMSAs with reconstituted nucleosomes and remodeling ATPases were performed as previously described (27, 34). For mapping the nucleosome positions, 1.5 units MNase (Sigma–Aldrich, St. Louis, MO) were added for 40 sec to remodeling reactions. The protected nucleosomal core DNA was isolated and analyzed by a single round of PCR (denaturation, 5 min at 95°C; annealing, 2 min at 56°C; extension, 1 min at 72°C) by using at least three different 32P-labeled oligonucleotides hybridizing to different positions on the DNA fragment. Primer extension fragments were resolved on 8% sequencing gels and quantified with a phosphorimager by using the Aida software (Fuji, Tokyo, Japan). For further details, see SI Materials and Methods.
Theoretical Analysis of Nucleosome-Remodeling Reaction.
The DNA curvature from the beginning of the enhancer to the end of the promoter was analyzed with the NA-Bench program (M. Busch, R. Kochinke, K.R., and G. Wedemann, unpublished data). The program uses different algorithms for curvature prediction that are reviewed in ref. 58. For the analysis shown here, the parameter set from Bolshoy et al. (59) was used. Kinetic simulations were conducted with the COPASI software package, version 4.0 (60) by using the model depicted in Fig. 4, which describes the translocation reaction according to Eqs. 1–5. The remodeler R can bind to nucleosomes N at positions i, i − 1, and i + 1. Under the conditions of the in vitro experiments, the binding reaction is fast, compared with the nucleosome-translocation reaction, so that it can be described by an preequilibrium with the equilibrium constant Kd for the dissociation of the RN complex:
Initial conditions for the simulations were a concentration of 2.5 × 10−9 M nucleosomes at position i and a concentration of 5 × 10−11 M remodeler. Translocations of the nucleosomes were described by the Eqs. 4 and 5 with the indicated rate constants.
 |
 |
Default values for the dissociation constants were Kd,i = Kd,i+1 = Kd,i−1 = 10−9 M and for the rate constants ki+1 = ki−1 = k−i = ki = 1 sec−1 (Fig. 4B). For the simulations of the release model, Kd,i+1 was changed to 10−8 M, and the arrest mechanism simulation corresponded to k−i = 0.1 sec−1.
Supplementary Material
Acknowledgments
We thank Bob Kingston (Massachusetts General Hospital, Boston, MA) for the Snf2H and Brg1 constructs, Alexander Brehm (IMT, Marburg, Germany) for Mi-2 constructs, Alexandra Lusser (Biocenter, Innsbruck, Austria) for Chd1 constructs, Carl Wu (National Cancer Institute, Bethesda, MD) for the purified NURF complex, Michael Meisterernst (GSF, Munich, Germany) for the XUMEI construct, and Jacek Mazurkiewicz and Ramona Ettig for helpful discussions. This work was supported by Deutsche Forschungsgemeinschaft Grants LA1331/6-1 (to G.L.) and Ri 1283/8-1 (to K.R.), the European Molecular Biology Organization Young Investigator Program (G.L.), Fonds der Chemischen Industrie (G.L.), and the Junior Research Groups at German Universities program of the Volkswagen Foundation (K.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702430104/DC1.
References
- 1.Muchardt C, Yaniv M. J Mol Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 2.Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 3.Becker PB, Hörz W. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 4.Han M, Kim UJ, Kayne P, Grunstein M. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth SY, Shimizu M, Johnson L, Grunstein M, Simpson RT. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 6.Lu Q, Wallrath LL, Elgin SC. EMBO J. 1995;14:4738–4746. doi: 10.1002/j.1460-2075.1995.tb00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schild C, Claret FX, Wahli W, Wolffe AP. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson JR, Benyajati C. Nucleic Acids Res. 1993;21:957–967. doi: 10.1093/nar/21.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 10.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nat Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 12.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaus A, Owen-Hughes T. Curr Opin Genet Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Bowen NJ, Fujita N, Kajita M, Wade PA. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Mohrmann L, Verrijzer CP. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Barak O, Lazzaro MA, Cooch NS, Picketts DJ, Shiekhattar R. J Biol Chem. 2004;279:45130–45138. doi: 10.1074/jbc.M406212200. [DOI] [PubMed] [Google Scholar]
- 17.Okabe I, Bailey LC, Attree O, Srinivasan S, Perkel JM, Laurent BC, Carlson M, Nelson DL, Nussbaum RL. Nucleic Acids Res. 1992;20:4649–4655. doi: 10.1093/nar/20.17.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aihara T, Miyoshi Y, Koyama K, Suzuki M, Takahashi E, Monden M, Nakamura Y. Cytogenet Cell Genet. 1998;81:191–193. doi: 10.1159/000015027. [DOI] [PubMed] [Google Scholar]
- 19.Banting GS, Barak O, Ames TM, Burnham AC, Kardel MD, Cooch NS, Davidson CE, Godbout R, McDermid HE, Shiekhattar R. Hum Mol Genet. 2005;14:513–524. doi: 10.1093/hmg/ddi048. [DOI] [PubMed] [Google Scholar]
- 20.Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones MH, Hamana N, Nezu J, Shimane M. Genomics. 2000;63:40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- 23.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 24.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 26.Lusser A, Urwin DL, Kadonaga JT. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 27.Brehm A, Längst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. EMBO J. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 29.Phelan ML, Schnitzler GR, Kingston RE. Mol Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Längst G, Bonte EJ, Corona DF, Becker PB. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 31.Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Längst G. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 32.Längst G, Schatz T, Langowski J, Grummt I. Nucleic Acids Res. 1997;25:511–517. doi: 10.1093/nar/25.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marilley M, Pasero P. Nucleic Acids Res. 1996;24:2204–2211. doi: 10.1093/nar/24.12.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Längst G, Becker PB. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- 35.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 36.von Hippel PH, Yager TD. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 37.Greive SJ, von Hippel PH. Nat Rev Mol Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 38.Landick R. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 39.Bouazoune K, Brehm A. Chromosome Res. 2006;14:433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- 40.Simpson RT. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 41.Grunstein M. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 42.Simpson RT. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Wrange O, Eriksson P. Int J Biochem Cell Biol. 1997;29:731–742. doi: 10.1016/s1357-2725(97)00016-2. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth SY, Dean A, Simpson RT. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper JP, Roth SY, Simpson RT. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 49.Weiss K, Simpson RT. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kastaniotis AJ, Mennella TA, Konrad C, Torres AM, Zitomer RS. Mol Cell Biol. 2000;20:7088–7098. doi: 10.1128/mcb.20.19.7088-7098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fleming AB, Pennings S. EMBO J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Reese JC. J Biol Chem. 2001;276:33788–33797. doi: 10.1074/jbc.M104220200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Reese JC. J Biol Chem. 2004;279:39240–39250. doi: 10.1074/jbc.M407159200. [DOI] [PubMed] [Google Scholar]
- 54.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santoro R, Li J, Grummt I. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Längst G, Grummt I. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodsell DS, Dickerson RE. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolshoy A, McNamara P, Harrington RE, Trifonov EN. Proc Natl Acad Sci USA. 1991;88:2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.