Abstract
The JNK signaling pathway is implicated in the regulation of the AP1 transcription factor and cell proliferation. Here, we examine the role of JNK by using conditional and chemical genetic alleles of the ubiquitously expressed murine genes that encode the isoforms JNK1 and JNK2. Our analysis demonstrates that JNK is not essential for proliferation. However, JNK is required for expression of the cJun and JunD components of the AP1 transcription factor, and JNK-deficient cells exhibit early p53-dependent senescence. These data demonstrate that JNK can act as a negative regulator of the p53 tumor suppressor.
Keywords: AP1, cJun, cell cycle
It is established that the JNK signaling pathway is a critical regulator of AP1-dependent gene expression (1). JNK phosphorylation of a number of AP1 and AP1-related transcription factors (e.g., cJun, JunB, JunD, and ATF2) causes increased AP1 transcription activity. In addition, JNK can regulate AP1 activity by increasing the expression of cJun mRNA (by acting at AP1-like sites in the cJun promoter) and regulating the half-life of the cJun protein (1). This role of JNK to increase AP1-dependent gene expression suggests that JNK may play an important role in cellular responses to mitogens (2). Indeed, primary murine fibroblasts (MEF) isolated from compound mutant Jnk1−/− Jnk2−/− embryos proliferate slowly (3).
The observation that MEF that lack JNK expression grow slowly suggests that JNK may function to regulate cell-cycle progression. However, the target of JNK signaling that mediates this effect of JNK on proliferation has not been defined. In addition, it is unclear whether the requirement of JNK for proliferation reflects a role for JNK-mediated phosphorylation of a substrate that is critically required for cell-cycle progression or whether it is the prolonged absence of JNK that causes slow growth.
To test whether JNK is required for proliferation, we examined the effect of acute loss of JNK function on cellular proliferation. A drug with validated specificity for JNK is not yet available (4). We therefore used genetic approaches to examine the effect of acute loss of JNK function on proliferation.
Results and Discussion
Construction of a Conditional Jnk1 Allele.
Mice with a floxed allele of Jnk1 were constructed by using standard techniques (Fig. 1 A–C). These mice were crossed with JNK2-deficient mice to create Jnk1LoxP/LoxP Jnk2−/− mice. Primary MEF prepared from these animals expressed JNK1. Alternative splicing of Jnk1 mRNA results in the expression of 46-kDa isoforms of JNK1 with a small amount of 54-kDa JNK1 isoforms (5). Cre-mediated excision of the floxed Jnk1 alleles resulted in primary MEF that lack detectable JNK expression (Fig. 1D). We have previously reported that JNK-deficient primary MEF display reduced UV-stimulated apoptosis and increased TNF-stimulated apoptosis (3, 6). The compound knockout MEF derived from conditional JNK mice similarly exhibited reduced UV-stimulated apoptosis and increased TNF-stimulated apoptosis [supporting information (SI) Figs. 7 and 8]. These data indicate that primary MEF isolated from conditional JNK-deficient mice are suitable for the analysis of the acute effects of compound JNK deficiency.
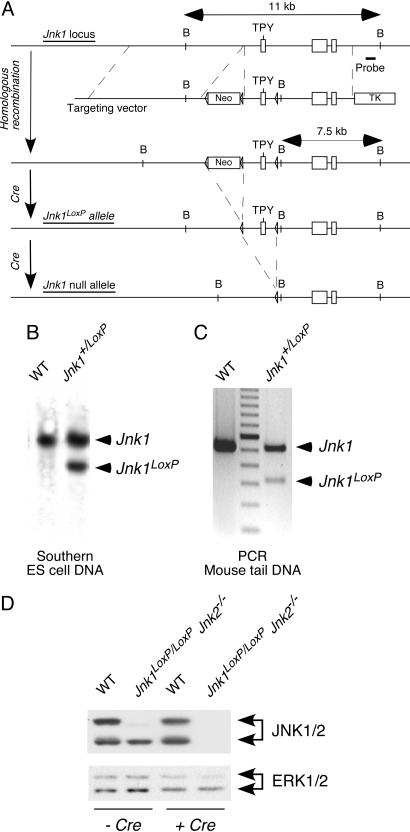
Fig. 1.
Construction of a conditional Jnk1 allele in mice. (A) The strategy used for targeting the Jnk1 gene to create a conditional Jnk1LoxP allele is illustrated. A probe used for Southern blot analysis and the expected BamH1 restriction fragments are shown. (B) Southern blot analysis of BamH1-restricted genomic DNA isolated from WT and Jnk1+/LoxP ES cells is shown. (C) Genomic DNA was isolated from the tails of WT and Jnk1+/LoxP mice and examined by PCR analysis. (D) Primary MEF were isolated from WT mice and Jnk1LoxP/LoxP Jnk2−/− mice. The MEF were transduced by using a retroviral Cre expression vector. The MEF were cultured (7 days) and extracts were examined by immunoblot analysis using antibodies to JNK1/2 and ERK1/2.
PCR analysis of genomic DNA demonstrated that the Cre-mediated deletion of floxed Jnk1 alleles was rapid. However, immunoblot analysis indicated that JNK1 expression slowly decreased after Jnk1 gene ablation (Fig. 2A). Indeed, low levels of JNK1 protein were detected at 3 days after Cre-mediated deletion. This slow loss of JNK1 protein expression is consistent with the half-life of the JNK1 protein (≈8 h) (1). Nevertheless, MEF without JNK (Jnk-null MEF) were obtained within 6 days after exposure to Cre-recombinase (Fig. 2A).
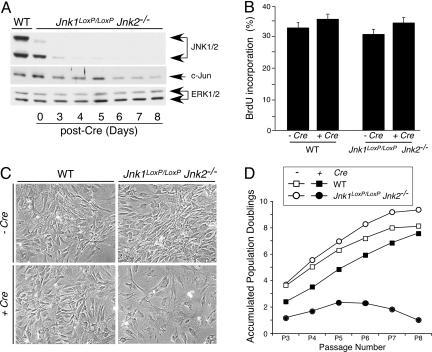
Fig. 2.
JNK deficiency causes a delayed reduction in cellular proliferation. (A) Time-course analysis of JNK expression after gene ablation. Jnk1LoxP/LoxP Jnk2−/− MEF were transduced by using a retroviral Cre expression vector. Cell extracts were prepared on different days after infection and examined by immunoblot analysis using antibodies to JNK, cJun, and ERK. Extracts prepared from WT MEF were also examined. (B) The effect of retroviral tranduction of Cre in WT and Jnk1LoxP/LoxP Jnk2−/− MEF was examined 10 days after infection by measurement of the incorporation of BrdU by flow cytometry. (C) The morphology of WT and Jnk1LoxP/LoxP Jnk2−/− MEF at passage 7 after retroviral transduction of Cre was examined by phase-contrast microscopy. (Magnification: ×100.) (D) The growth of WT and Jnk1LoxP/LoxP Jnk2−/− MEF was examined by using a 3T3 assay. The data are presented as the accumulated population doublings during eight passages in culture. The effect of retroviral transduction of Cre was investigated.
JNK Deficiency Causes a Delayed Decrease in Proliferation.
We have previously reported that MEF isolated from Jnk1−/− Jnk2−/− embryos grow slowly (3). In contrast, flow cytometry analysis of BrdU incorporation into DNA demonstrated that JNK deficiency caused no decrease in DNA synthesis at passage 3 (9 days) after Cre-mediated abation of JNK expression (Fig. 2B). This observation indicates that there is no essential requirement of JNK for proliferation. The slow growth of MEF isolated from Jnk1−/− Jnk2−/− embryos (3) contrasts with the rapid proliferation of Jnk-null MEF created by using Cre-recombinase. One possible explanation for the different phenotypes of these MEF is that slow growth is a delayed consequence of JNK deficiency. To test this hypothesis, we passaged MEF by using a 3T3 protocol (Fig. 2 C and D). Studies of Jnk-null MEF demonstrated a progressive decrease in growth with increasing passage number. Indeed, at passage 7 (21 days after Cre-mediated JNK ablation) the Jnk-null MEF exhibited a flattened morphology (Fig. 2C) and proliferated very slowly (Fig. 2D). Together, these data indicate that JNK is not essential for proliferation, but long-term JNK deficiency causes a severe decrease in proliferation.
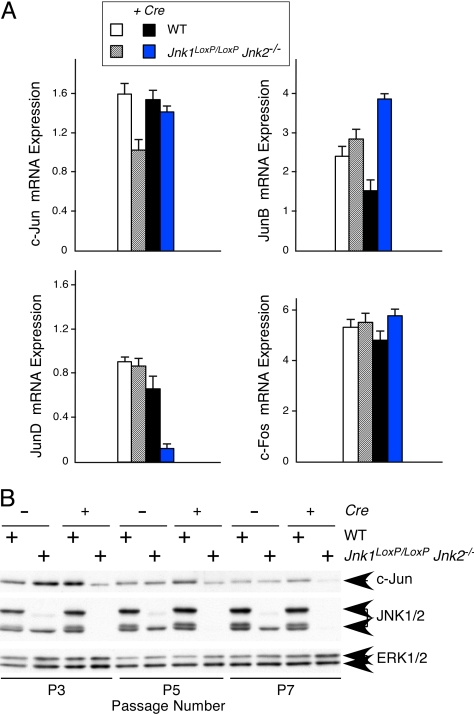
The mechanism that accounts for the delayed requirement of JNK for proliferation is unclear. One possible explanation is that JNK deficiency causes changes in gene expression. It is established that JNK is a critical regulator of the AP1 transcription factor (1) and that AP1 is important for proliferation (2). Decreased AP1 in Jnk-null MEF may therefore contribute to the observed reduction in proliferation. To test whether Jnk-null MEF expressed lower levels of AP1 proteins, we examined the expression of cJun, JunB, JunD, and cFos mRNA. Analysis of Jnk-null MEF demonstrated a selective loss of JunD mRNA (Fig. 3A). It is established that JNK can stabilize the cJun protein (7, 8). Thus, although Jnk-null MEF did not exhibit a decrease in cJun mRNA, reduced expression of cJun protein was anticipated. Indeed, immunoblot analysis demonstrated that the amount of cJun protein was markedly reduced in Jnk-null MEF. Together, these data indicate that Jnk-null MEF express decreased amounts of both cJun and JunD (Fig. 3). Interestingly, both cJun−/− MEF and JunD−/− MEF exhibit profoundly reduced proliferation (9, 10). Thus, reduced expression of cJun and JunD may contribute to the reduced proliferation of Jnk-null MEF.
Fig. 3.
JNK deficiency causes reduced expression of cJun and JunD. (A) WT and Jnk1LoxP/LoxP Jnk2−/− MEF were transduced by using a Cre retroviral expression vector. RNA was isolated from these cells at passage 7 postinfection and examined by quantitative RT-PCR (Taqman) analysis using probes for AP-1 transcription factors (cJun, JunB, JunD, and cFos) and GAPDH. The data are presented as the normalized ratio of [AP-1 mRNA]/[GAPDH mRNA]. (B) Protein extracts were prepared from MEF at 3T3 passages 3, 5, and 7 postinfection and examined by immunoblot analysis using antibodies to cJun, JNK, and ERK.
JNK Deficiency Causes Senescence.
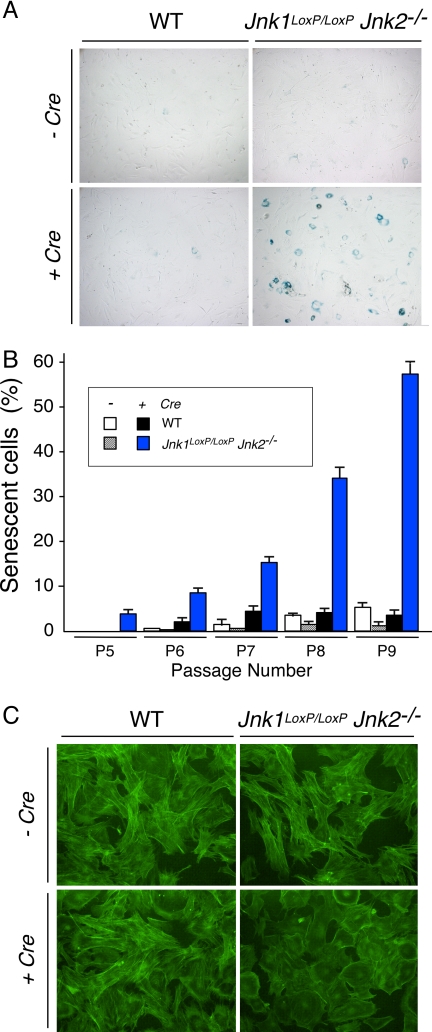
The delayed defect in the growth of Jnk-null MEF suggests that JNK deficiency may cause senescence. To test this hypothesis, we stained MEF for acidic β-gal, a biochemical marker of senescence (11). This analysis confirmed that the Jnk-null MEF exhibited a progressive increase in senescence when passaged with a 3T3 protocol (Fig. 4 A and B) with a flattened morphology that is characteristic of senescent cells (Fig. 4C). These data indicate that JNK deficiency causes senescence.
Fig. 4.
JNK deficiency causes senescence. (A) WT and Jnk1LoxP/LoxP Jnk2−/− MEF were transduced by using a Cre retroviral expression vector and stained for a senescence marker (acidic β-gal) using X-Gal. A culture stained at 3T3 passage 9 postinfection is illustrated. (B) The percentage of cells that stained with X-Gal was examined at passages 5, 6, 7, 8, and 9 postinfection was quantitated. The data shown represent the mean ± SD of three independent experiments. (C) MEF at 3T3 passage 9 postinfection were stained with phalloidin (green) and examined by fluorescence microscopy. (Magnification: ×100.)
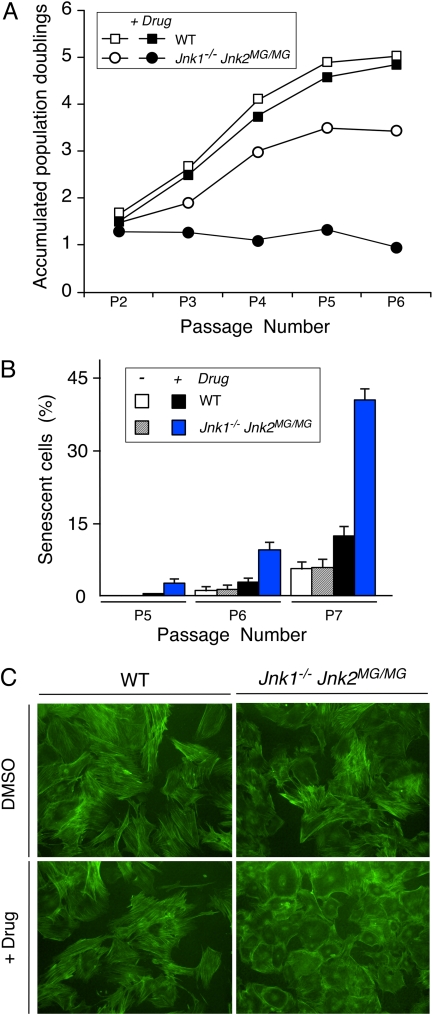
JNK is required to prevent premature senescence (Fig. 4). However, it is unclear whether this reflects a requirement for the kinase activity of JNK, because kinase-independent actions of JNK have been reported (12). To test whether the protein kinase activity of JNK is required to prevent early senescence, we used a chemical genetic approach to inhibit JNK activity with the drug 1-naphthylmethyl-4-amino-1-tert-butyl-3-(p-methylphenyl) pyrazolo[3,4-d]pyrimidine (1NM-PP1) (13). This drug does not inhibit the activity of WT JNK, but does inhibit the activity of a mutant form of JNK (Met-108 replaced with Gly) that has an expanded ATP pocket (14). Mice with this point mutation in the Jnk2 gene (Jnk2MG) have been described (13). We prepared MEF from WT embryos and Jnk1−/− Jnk2MG/MG embryos. Treatment of WT MEF with the drug 1NM-PP1 during a 3T3 protocol caused no marked changes in proliferation (Fig. 5A). In contrast, treatment of Jnk1−/− Jnk2MG/MG MEF with 1NM-PP1 caused severely reduced growth in a 3T3 protocol (Fig. 5A), increased expression of the senescence marker acidic β-gal (Fig. 5B), and a senescent morphology (Fig. 5C). Together, these data indicate that JNK activity is required to prevent early senescence.
Fig. 5.
Inhibition of the protein kinase activity of JNK causes senescence. (A) The growth of WT and Jnk1−/− Jnk2MG/MG MEF was examined by using a 3T3 assay. The data are presented as the accumulated population doublings during six passages in culture. The effect of addition of solvent (DMSO) or drug (1 μM 1NM-PP1) was investigated. (B) WT and Jnk1−/− Jnk2MG/MG MEF were stained for a senescence marker (acidic β-gal) by using X-Gal. The percentage of cells that stained with X-Gal was examined at 3T3 passages 5, 6, and 7 and quantitated. The data shown represent the mean ± SD of three independent experiments. (C) MEF at 3T3 passage 7 were stained with phalloidin (green) and examined by fluorescence microscopy. (Magnification: ×100.)
JNK Deficiency Increases p53 Expression and Activity.
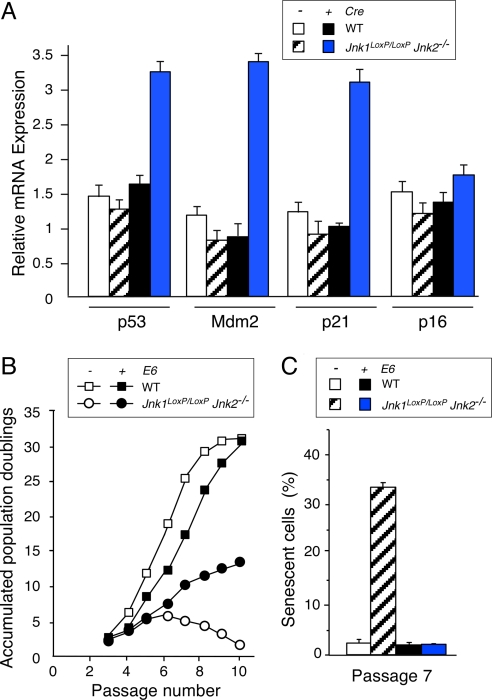
MEF prepared from Jnk1−/− Jnk2−/− embryos express increased amounts of the Mdm2 inhibitor p19ARF and increased amounts of p53 protein (3). The increased expression of p53 is most likely caused by both increased p53 protein stability, as a result of increased p19ARF expression (15), and an increase in p53 mRNA expression (Fig. 6A) caused by reduced cJun expression and derepression of the p53 promoter (16). To test whether the increased expression of p53 in Jnk-null MEF is functionally significant, we examined p53-dependent gene expression. These studies demonstrated that Jnk-null MEF exhibited increased expression of the p53 target genes Mdm2 and p21 (Fig. 6A). These data demonstrate that there is an increase in p53 expression and function in Jnk-null MEF.
Fig. 6.
JNK deficiency causes p53-dependent senescence. (A) WT and Jnk1LoxP/LoxP Jnk2−/− MEF were transduced by using a Cre retroviral expression vector. Total RNA was isolated from MEF at 3T3 passage 7 postinfection and the amount of p53, Mdm2, p21, p16, and GAPDH mRNA was measured by quantitative RT-PCR (Taqman assays). The data are presented as the normalized ratio of [p53, Mdm2, p21, or p16 mRNA]/[GAPDH mRNA]. (B) The growth of WT and Jnk1LoxP/LoxP Jnk2−/− MEF (transduced with a retroviral Cre vector) was examined by using a 3T3 assay. The effect of expression of the HPV16 E6 protein is presented. The data are presented as the accumulated population doublings during 10 passages in culture postinfection. (C) WT and Jnk1LoxP/LoxP Jnk2−/− MEF (transduced with a retroviral Cre vector) were stained for a senescence marker (acidic β-gal) by using X-Gal. The effect of expression of the HPV16 E6 protein is presented. The percentage of cells that stained with X-Gal was examined at 3T3 passage 7 postinfection and quantitated. The data shown represent the mean ± SD of three independent experiments.
JNK Deficiency Causes p53-Dependent Senescence.
The p53 tumor suppressor is implicated in senescence. The increased expression and function of p53 in Jnk-null MEF may therefore contribute to the early senescent phenotype of these cells. To test this hypothesis, we examined the effect of p53 down-regulation by using the human papilloma virus (HPV)-16 E6 protein. The expression of E6 in WT MEF caused no major change in the proliferation of WT MEF in a 3T3 protocol. In contrast, expression of E6 in Jnk-null MEF prevented the early senescent phenotype of these cells (Fig. 6 B and C). Thus, E6-mediated down-regulation of p53 prevents senescence caused by JNK deficiency. Together, these data indicate that JNK is not required for proliferation, but JNK does function to prevent p53-mediated senescence.
Role of JNK in Cell Proliferation and Senescence.
The conclusion that JNK is not required for proliferation markedly contrasts with conclusions drawn from previous studies (3). Acute loss of JNK function does not inhibit proliferation (Fig. 2B). However, JNK deficiency does cause p53-dependent senescence (Fig. 4). The slow proliferation of JNK-deficient MEF isolated from Jnk1−/− Jnk2−/− embryos that was reported previously (3) is most likely a consequence of chronic JNK deficiency and p53-dependent senescence rather than a direct role for JNK in the cell cycle. Interestingly, the effect of JNK deficiency to cause early p53-dependent senescence is similar to that caused by deficiency of the AP1 proteins cJun (10, 16) and JunD (9). The large decrease in cJun and JunD expression caused by JNK deficiency (Fig. 3) may therefore contribute to the engagement of the p53 senescence pathway in Jnk-null MEF. Indeed, ectopic expression of JunD in Jnk-null MEF partially suppressed p53-mediated senescence (SI Fig. 9).
The progressive decrease in AP1 protein expression after loss of JNK over several days (Figs. 2A and 3B) may explain the finding that loss of JNK does not immediately inhibit proliferation (Fig. 2B), but p53-induced senescence is observed later (Fig. 4). Collectively, these data indicate that JNK can act as a negative regulator of the p53 tumor suppressor. This conclusion is consistent with recent reports that have implicated a role for the JNK signaling pathway in some human tumors (17).
Materials and Methods
Animal Studies.
Mice with disruptions of the Jnk1 or Jnk2 genes or a knock-in mutation in the Jnk2 gene (Jnk2MG) have been described (13, 18, 19). Mice expressing a 4-hydroxytamoxifen-inducible Cre recombinase from the endogenous Gt(ROSA)26Sor promoter (20) were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice with a germ-line mutation in the Jnk1 gene with LoxP elements inserted into two different introns (Jnk1LoxP) were constructed by using homologous recombination in ES cells. A mouse strain 129/Svev genomic BAC clone containing the Jnk1 gene was used to create a targeting vector with a floxed NeoR cassette and a thymidine kinase cassette (Fig. 1A). ES cells were electroporated with this vector and selected with 200 μg/ml G418 and 2 μM gangcyclovir. Two correctly targeted ES cell clones were identified by Southern blot analysis and PCR analysis. These ES cell clones were injected into C57BL/6J blastocysts to create chimeric mice that transmitted the mutated Jnk1 allele through the germ line. The floxed NeoR cassette was excised by using Cre recombinase. The mice were backcrossed to the C57BL/6J strain (Jackson Laboratory) and housed in a facility accredited by the American Association for Laboratory Animal Care. These studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Genotype Analysis.
The genotype at the Jnk1 locus was examined by Southern blot analysis of BamHI-restricted genomic DNA by probing with a random-primed 32P-labeled probe (320 bp) that was isolated by PCR using a Jnk1 genomic clone as the template and the primers 5′-GGTACTGTGTCAGCCTGGTCAAG-3′ and 5′-GGGGGAACCAAACCAAAAAAC-3′. The WT and conditional alleles of Jnk1 were also detected by PCR amplification of genomic DNA using the primers 5′-GGATTTATGCCCTCTGCTTGTC-3′ and 5′-GAACCACTGTTCCAATTTCCATCC-3′; agarose gel electrophoresis indicates the presence of a 520-bp DNA fragment (WT Jnk1 allele) or a 330-bp DNA fragment (floxed Jnk1 allele). The deleted Jnk1 allele (ΔJnk1) was detected by PCR amplification of genomic DNA using the primers 5′-CCTCAGGAAGAAAGGGCTTATTTC-3′ and 5′-GAACCACTGTTCCAATTTCCATCC-3′; agarose gel electrophoresis indicates the presence of a 1,550-bp fragment (WT Jnk1 allele), a 1,095-bp DNA fragment (floxed Jnk1 allele), or a 395-bp DNA fragment (ΔJnk1 allele).
Primary Cell Culture.
MEF were isolated from E13.5 embryos and cultured in DMEM supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen, Carlsbad, CA) at 37°C in a humidified atmosphere with 5% CO2. Similar data were obtained in experiments using independently isolated MEF cultures. The cells were treated without or with the drug 1NM-PP1 (13). Retroviral transduction experiments were performed with a self-inactivating vector (HR-MMPCreGFP) that transiently expresses Cre recombinase (21), a vector (pLXSN16E6) that expresses HPV-16 E6 (22), and a vector (pMSCV-JunD-IRES-GFP) that expresses JunD (6). MEF proliferation was monitored with a 3T3 protocol (23). Proliferation assays were performed by staining with crystal violet (3), measurement of cell number with a hemacytometer, and measurement of BrdU incorporation by flow cytometry (24). Senescent cells were detected by staining for β-gal using X-Gal (Cell Signaling, Danvers, MA).
Fluorescence Microscopy.
MEF were fixed with 2% formaldehyde (15 min), permeabilized with 0.1% Triton X-100/PBS (5 min), stained with Alexa Fluor 488-conjugated phalloidin (Invitrogen) diluted 1:1,000 in PBS (15 min), and washed three times with PBS. The stained MEF were examined by fluorescence microscopy.
Biochemical Analysis.
Gene expression was examined by measurement of the amount of mRNA using quantitative RT–PCR analysis with Taqman probes (Applied Biosystems, Foster City, CA). Apoptotic cell death was examined by measurement of DNA fragmentation using the Cell Death Detection Elisaplus kit (Roche, Indianapolis, IN) following the manufacturer's recommendations (6, 25). Immunoblot analysis was performed by probing with antibodies to JNK1/2 (Pharmingen, Franklin Lakes, NJ), cJun (Santa Cruz Biotechnology, Santa Cruz, CA), JunD (Santa Cruz Biotechnology), and ERK1/2 (Cell Signaling).
Supplementary Material
Acknowledgments
We thank Vicky Benoit, Tamera Barrett, Julie Cavanagh-Kyros, Jien-Hua Liu, and Judy Reilly for expert technical assistance and Kathy Gemme for administrative assistance. These studies were supported by a grant from the National Institutes of Health. K.M.S., R.A.F., and R.J.D. are Investigators of the Howard Hughes Medical Institute.
Abbreviations
- MEF
murine fibroblasts
- 1NM-PP1
1-naphthylmethyl-4-amino-1-tert-butyl-3-(p-methylphenyl)pyrazolo[3,4-d]pyrimidine
- HPV
human papilloma virus.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707782104/DC1.
References
- 1.Davis RJ. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 2.Jochum W, Passegue E, Wagner EF. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 3.Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 4.Manning AM, Davis RJ. Nat Rev Drug Discov. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- 5.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb JA, Ventura JJ, Hess P, Flavell RA, Davis RJ. Mol Cell. 2003;11:1479–1489. doi: 10.1016/s1097-2765(03)00203-x. [DOI] [PubMed] [Google Scholar]
- 7.Musti AM, Treier M, Bohmann D. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs SY, Dolan L, Davis RJ, Ronai Z. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 9.Weitzman JB, Fiette L, Matsuo K, Yaniv M. Mol Cell. 2000;6:1109–1119. doi: 10.1016/s1097-2765(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 11.Dimri GP, Campisi J. Cold Spring Harb Symp Quant Biol. 1994;59:67–73. doi: 10.1101/sqb.1994.059.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs SY, Fried VA, Ronai Z. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 13.Jaeschke A, Karasarides M, Ventura J-J, Ehrhardt A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Mol Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 14.Ventura JJ, Hubner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Sherr CJ. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weston CR, Davis RJ. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Dong C, Yang DD, Tournier C, Whitmarsh AJ, Xu J, Davis RJ, Flavell RA. Nature. 2000;405:91–94. doi: 10.1038/35011091. [DOI] [PubMed] [Google Scholar]
- 19.Yang DD, Conze D, Whitmarsh AJ, Barrett T, Davis RJ, Rincon M, Flavell RA. Immunity. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 20.Badea TC, Wang Y, Nathans J. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silver DP, Livingston DM. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 22.Halbert CL, Demers GW, Galloway DA. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todaro GJ, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess P, Pihan G, Sawyers CL, Flavell RA, Davis RJ. Nat Genet. 2002;32:201–205. doi: 10.1038/ng946. [DOI] [PubMed] [Google Scholar]
- 25.Ventura JJ, Cogswell P, Flavell RA, Baldwin AS, Jr, Davis RJ. Genes Dev. 2004;18:2905–2915. doi: 10.1101/gad.1223004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.