Abstract
Ribosomal RNA (rRNA) gene transcription accounts for most of the RNA in prokaryotic and eukaryotic cells. In eukaryotes, there are hundreds (to thousands) of rRNA genes tandemly repeated head-to-tail within nucleolus organizer regions (NORs) that span millions of basepairs. These nucleolar rRNA genes are transcribed by RNA Polymerase I (Pol I) and their expression is regulated according to the physiological need for ribosomes. Regulation occurs at several levels, one of which is an epigenetic on/off switch that controls the number of active rRNA genes. Additional mechanisms then fine-tune transcription initiation and elongation rates to dictate the total amount of rRNA produced per gene. In this review, we focus on the DNA and histone modifications that comprise the epigenetic on/off switch. In both plants and animals, this system is important for controlling the dosage of active rRNA genes. The dosage control system is also responsible for the chromatin-mediated silencing of one parental set of rRNA genes in genetic hybrids, a large-scale epigenetic phenomenon known as nucleolar dominance.
Keywords: epigenetic phenomena, RNA polymerase I, DNA methylation, cytosine methylation, histone methylation, histone deacetylation, histone deacetylase, transcription, ribosomal RNA, nucleolus, nucleolus organizer region
Large scale cytological effects of rRNA gene regulation: nucleolus and secondary constriction formation
The most prominent feature of a eukaryotic nucleus during interphase is the nucleolus (Figure 1), a region that contains relatively little chromosomal DNA but is rich in RNAs, proteins and ribonucleoprotein particles, including the large and small ribosome subunits as they undergo assembly [1-3]. The location of a nucleolus is not determined randomly; instead its formation is determined by the position of a discrete chromosomal locus known as a nucleolus organizer region, or NOR [4]. This fact was discovered based on an intriguing property: upon condensation of chromosomes at metaphase, NORs fail to condense to the same extent as surrounding chromosomal sequences and thus give rise to “secondary constrictions”, with “primary constrictions” being defined as the centromeres (see Figure 1). Heitz initially noted that nucleoli form at, or very near, the sites of secondary constrictions [5] and McClintock soon thereafter obtained direct evidence [6]. She identified a maize line in which a chromosome break at or near the secondary constriction on chromosome 9, followed by translocation of one of the broken ends to another chromosome, caused the formation of two nucleoli during interphase and two secondary constrictions at metaphase. These observations pointed to a specific chromosomal locus that had been split into two still-functional entities, thereby defining a locus that McClintock named the “nucleolar organizer” [6].
Figure 1. Relationships between the nucleolus, secondary constriction, NOR and ribosomal RNA genes.
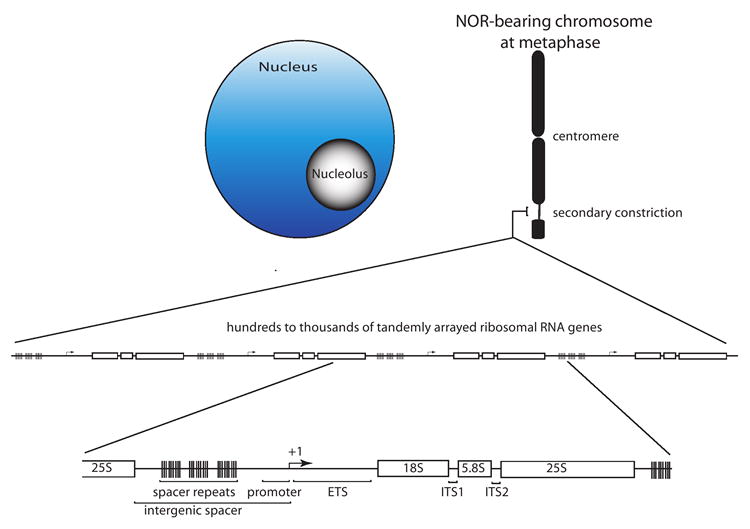
Typically, only a subset of the rRNA genes within a nucleolus organizer region (NOR) is active; subsequent transcription by RNA polymerase I and pre-rRNA processing causes the nucleolus to form. At metaphase, the rRNA genes that had been active during the preceding interphase give rise to a secondary constriction due to the persistent binding of transcription factors. The secondary constriction is decondensed to the point that it is generally not visible using standard light microscopy techniques. Within an NOR the tandemly arrayed rRNA genes are separated from one another by an intergenic spacer that typically contains repeated elements as well as the rRNA gene promoter, at which the transcription start site is denoted as +1. The transcribed portion of the gene includes an external transcribed sequence (ETS) and two internal transcribed sequences, ITS1 and ITS2 that separate the 18S, 5.8S and 25S structural rRNA sequences. Multiple RNA processing events are needed to generate the structural RNAs from the primary transcript.
In the same year that McClintock deduced the relationship between nucleolus formation and the NOR, Navashin noted an unusual behavior of secondary constrictions in hybrids that had been formed between different species of the plant genus Crepis [7]. In some F1 hybrids, secondary constrictions occurred on chromosomes inherited from both parental species, as one might expect if the constrictions were an invariant physical characteristic of the chromosomes. However, in many of the hybrids the secondary constrictions were absent from chromosomes inherited from one species, regardless of whether that species had served as the egg or pollen donor. Navashin called the phenomenon “differential amphiplasty” but the era of molecular biology ushered in a new name by which the phenomenon is better known: nucleolar dominance [8] (for recent reviews, see [9-11]). Navashin observed that the secondary constrictions reappeared in progeny whose karyotypes resembled the initial “pure species”, indicating that the chromosomes bearing the secondary constrictions had not been lost or damaged and that the absence of secondary constriction formation was reversible [7]. In hindsight, this was the first indication of the epigenetic nature of nucleolar dominance.
According to the most common use of the term today, epigenetic phenomena can be defined as heritable, but potentially reversible, alternative states of gene activity that are not explained by changes in gene sequence. Navashin’s observations in 1934 preceded the discovery that DNA is the genetic material or a molecular understanding of genes, so the significance of Navashin’s observations with respect to gene expression and epigenetics awaited the demonstration that NORs are loci at which repetitive, nearly identical, rRNA genes are clustered in long tandem arrays, typically spanning millions of basepairs [12, 13]. Each rRNA gene within an NOR encodes a precursor transcript that can be processed to generate one molecule each of 18S, 5.8S and 25-28S (the size is species-dependent) rRNA (Figure 1) [14, 15]. The understanding of NORs as loci composed of many hundreds of rRNA genes explained McClintock’s obervation that an NOR can be split and yet retain nucleolus organizer function, which was an early indication that the information at the locus was redundant. Moreover, the molecular understanding of the NOR suggested that Navashin’s observations of differential secondary constriction formation might be a visible manifestation of differential rRNA gene transcription at NORs derived from the two progenitors [16], a prediction that was subsequently verified by Honjo and Reeder in studies of Xenopus (frog) hybrids [8].
What explains secondary constrictions? The answer apparently lies in the persistence of RNA polymerase I transcription factors that associate with active rRNA genes and give them a unique chromatin structure that resists condensation to the same degree as adjacent chromosomal regions [4]. The best evidence supporting this hypothesis has come very recently by the demonstration that arrays of binding sites for the vertebrate RNA polymerase I transcription factor UBF (Upstream Binding Factor) cause the formation of ectopic secondary constrictions when integrated into novel (i.e. non-NOR) locations of mammalian chromosomes. The ectopic UBF binding sites are transcriptionally inert, indicating that it is protein binding, rather than RNA polymerase I transcription, that is responsible for secondary constriction formation [17]. UBF is an extremely abundant nuclear protein in vertebrates that apparently binds throughout intergenic spacer and transcribed regions of rRNA genes [18, 19] and appears to play an important architectural role [20]. UBF persists at NORs throughout metaphase [21-23], which is when secondary constrictions are apparent, and is absent from those mammalian NORs that are not active and that do not form secondary constrictions. UBF binds cooperatively to naked DNA [24] and wraps the DNA [25] in a right-handed direction, thereby constraining positive supercoils [26, 27], unlike histones which wrap DNA in a left-handed direction and constrain negative supercoils. However, UBF can also bind to nucleosomal DNA and displace linker histone H1, which plays a role in chromatin compaction [28]. Either of these binding behaviors may be sufficient for UBF to influence secondary constriction formation at NORs. Because UBF is found only in vertebrates, it is not clear what is responsible for secondary constriction formation at NORs in other organisms, including plants. However, UBF contains multiple HMG (High Mobility Group) DNA binding domains, so other HMG proteins may play UBF-like roles in non-vertebrate eukaryotes, as appears to be the case in budding yeast [29, 30]. The recent demonstration that the octameric form of the bacterial histone-like protein HU wraps DNA in a right-handed direction and constrains positive supercoils [31], reminiscent of UBF’s DNA binding behavior, also suggests the possibility that non-HMG proteins could serve structural roles similar to UBF.
Concerted changes in DNA methylation and histone modification comprise an epigenetic on/off switch controlling nucleolar dominance
In the past decade, an understanding of nucleolar dominance as a large-scale silencing phenomenon has provided additional insights into a chromatin-mediated on/off switch controlling rRNA genes and NORs. A number of early investigations in plants identified correlations between rRNA gene activity, cytosine methylation, and/or the accessibility of chromosomal rRNA genes to DNase digestion. For instance, in hexaploid wheat, which has multiple NORs that are differentially active, rRNA genes located within the most active NORs are hypomethylated and more DNase accessible than rRNA genes at inactive or less active NORs [32-34]. Likewise, in Triticale (wheat - rye hybrids), the inactive (underdominant) rRNA genes of rye are more heavily methylated within their intergenic spacers than are the dominant wheat rRNA genes [35, 36]. However, in Xenopus hybrids displaying nucleolar dominance, Macleod and Bird showed a correlation between DNase accessibility and rRNA gene transcriptional dominance, but no evidence for differential cytosine methylation was apparent [37]. Collectively, these and other studies indicated a difference in the chromatin compaction of rRNA genes subjected to nucleolar dominance, but the role of DNA methylation in the process was unclear. Direct evidence that cytosine methylation plays a role in rRNA gene expression and nucleolar dominance came from simple experiments in which treatment with 5-aza-2’-deoxycytosine (aza-dC), a chemical inhibitor of cytosine methylation, caused the cytological reactivation of rye NORs in Triticale [38] and the transcriptional derepression of silent rRNA genes in Brassica [39] or Arabidopsis [40] allotetraploid hybrids.
The fact that nucleolar dominance occurs in hybrids of Drosophila [41], which until recently were thought to lack genomic cytosine methylation entirely [42], suggested that DNA methylation alone was unlikely to explain nucleolar dominance in all organisms, prompting an investigation of histone modifications that might also play a role. Resulting studies revealed that rRNA genes subjected to nucleolar dominance in Brassica or Arabidopsis allotetraploids can be derepressed by treatment with the histone deacetylase inhibitors sodium butyrate or trichostatin A (TSA), similar to the derepression observed upon treatment with aza-dC [39, 40]. Significantly, no appreciable additivity or synergism between aza-dC and TSA occurs when plants are grown in the presence of both chemicals, indicating a partnership between DNA methylation and histone deacetylation within the same repression pathway.
The technique of chromatin immunoprecipitation (abbreviated as ChIP; see Figure 2) has made possible several key insights that reveal the partnership between DNA methylation and various histone modifications in Arabidopsis suecica, the allotetraploid hybrid of A. thaliana and A. arenosa. In A. suecica, the A. thaliana-derived rRNA genes are selectively repressed (underdominant) whereas the A. arenosa-derived genes are active (dominant) [43]. Using ChIP, Lawrence et al. [40] found that nucleosomes associated with the promoter regions of underdominant A. thaliana genes are enriched in histone H3 dimethylated on lysine 9 (H3K9me2), a canonical mark of transcriptionally repressed and highly condensed chromatin (heterochromatin) [44]. In contrast, A. arenosa rRNA gene promoter regions associate with both H3K4me3, a reliable mark of active decondensed chromatin (euchromatin) and H3K9me2, suggesting that among the dominant class of rRNA genes, some genes are present in a euchromatic environment and others are organized in heterochromatin [40]. The same situation holds true in non-hybrid A. thaliana, which has a fraction of its rRNA genes associated with H3K4me3 and a fraction associated with H3K9me2 [40]. These observations are consistent with substantial evidence that only a subset of the rRNA genes present in a eukaryotic genome are typically active (discussed in greater detail in a subsequent section).
Figure 2. The ChIP-chop assay.
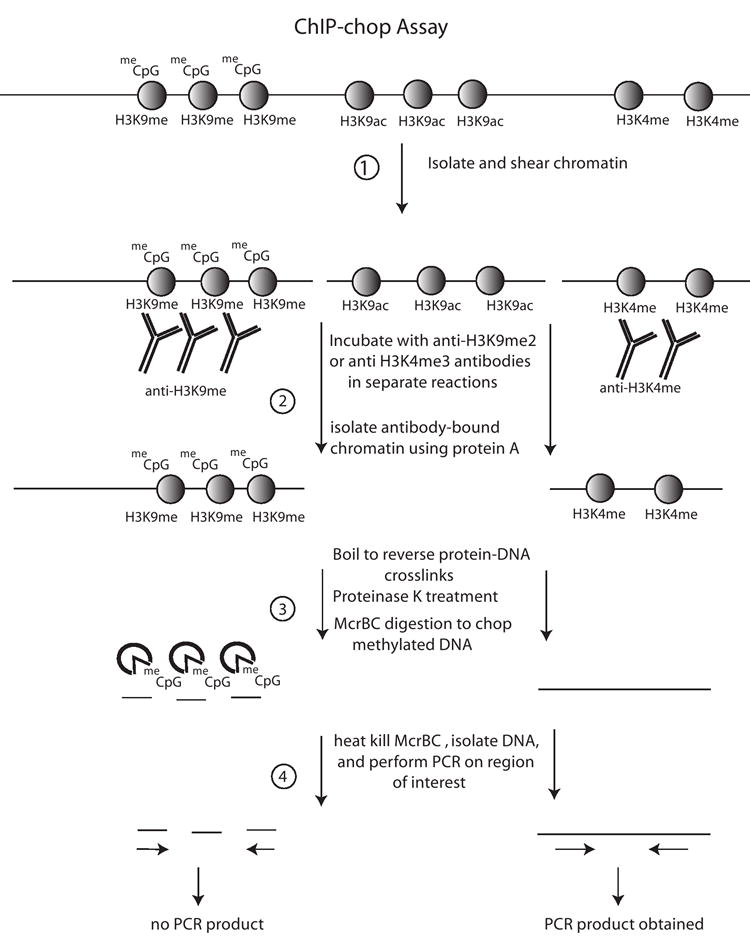
(1) Proteins are crosslinked to DNA using formaldehyde and chromatin is then isolated and sheared to generate oligonucleosomes. (2) The chromatin is then incubated with an antibody of choice, in this example antibodies specific for Histone H3 dimethylated on lysine 9 (H3K9me2) or H3K4me3. The antibody-bound chromatin is then immobilized on protein A agarose beads, followed by centrifugation and washing. (3) Crosslinked proteins are removed though proteinase K treatment and boiling, and the methylation-specific nuclease McrBC is incubated with the DNA. (4) Subsequent isolation of the DNA and PCR amplification of the region of interest allows one to determine if McrBC sensitivity, and therefore DNA methylation, correlates with the specific histone modifications queried.
A variation of the ChIP technique, which we dubbed ChIP-chop [40], allows an estimation of the density of cytosine methylation on any DNA associated with an immunoprecipitated protein (Figure 2). In this technique, ChIP’ed DNA is digested with McrBC, an enzyme that specifically cleaves DNA containing at least two purine-methylC dinucleotides which can be located at variable distances relative to one another [45]. The DNA is then amplified by PCR using primers that span the region of interest. If the DNA template is hypermethylated, it is digested (chopped) by McrBC and therefore PCR amplification of the interval fails. By contrast, hypomethylated templates are resistant to McrBC and PCR is successful in this case. ChIP-chop-PCR analysis of A. suecica chromatin revealed that those rRNA gene promoters associated with H3K9me2 are heavily methylated on cytosines whereas rRNA gene promoters associated with H3K4me3 are hypomethylated (see Figure 3) [40]. rRNA genes immunoprecipitated with an antibody specific for RNA polymerase I are also hypomethylated [40]. Collectively, these ChIP and ChIP-chop data indicate that active rRNA genes are cytosine- hypomethylated and associate with H3K4me3 as well as RNA polymerase I. By contrast, inactive genes are associated with H3K9me2 and their promoter cytosines tend to be hypermethylated. These observations have been extended to include additional modifications that help define the on and off states of rRNA genes, as summarized in Figure 3. For instance, in addition to H3K4me3, active rRNA genes are hyperacetylated on lysines 5, 8, 12 and 16 of histone H4 and on lysines 9 and 14 of histone H3 whereas inactive genes are depleted for these modifications [46].
Figure 3. A model for the epigenetic on/off switch regulating nucleolar dominance in Arabidopsis.
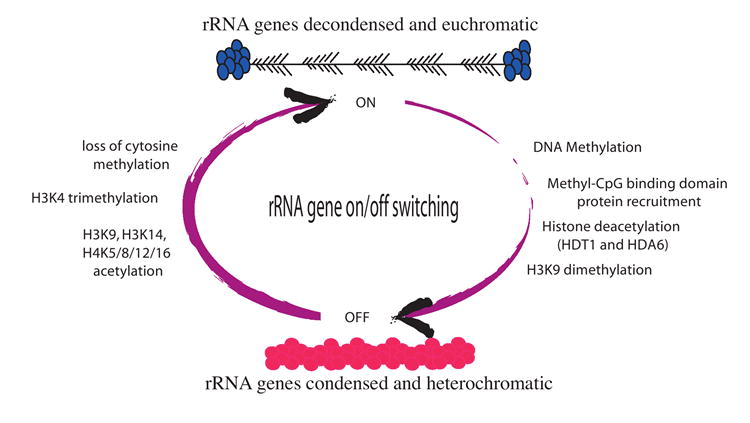
Changes in rRNA gene cytosine methylation, histone deacetylation and histone methylation occur in a concerted fashion. The “off” switch involves cytosine methylation, histone deacetylation, H3K9 dimethylation and condensation of the rRNA genes into heterochromatin. In contrast, the “on” switch involves the loss of cytosine methylation, histone H3 and H4 hyperacetylation, H3K4 trimethylation and decondensation of rRNA genes into euchromatin. In the cartoon of the “on” state, a series of tandem rRNA genes are depicted with progressively longer RNA transcripts emanating from the horizontal DNA axis such that each transcription unit has a “christmas-tree”-like appearance as observed in electron micrographs of transcribing rRNA genes.
Importantly, DNA methylation and histone modification states are interdependent and mutually reinforcing at rRNA genes. This was initially shown by experimentally turning on silenced rRNA genes using either aza-dC or TSA and showing that treatment with either chemical has identical consequences with respect to the suite of DNA and histone modifications that ensue [40]. Upon switching from “off” to “on”, promoters undergo a heterochromatin to euchromatin transition, losing their cytosine hypermethylation and H3K9me2 association and gaining association with H3K4me3 and other marks of active chromatin. Because inhibiting DNA methylation or histone deacetylation yields the same outcome, DNA methylation and repressive histone modifications apparently specify one another as part of a self-reinforcing cycle [40, 46].
Components of the rRNA gene off switch in Arabidopsis
Having established that epigenetic modifications are critical aspects of nucleolar dominance, identifying chromatin modifying proteins responsible for the phenomenon has become a priority. Derepression of underdominant rRNA genes upon treatment with the histone deacetylase inhibitor TSA indicated a role for at least one histone deacetylase in nucleolar dominance [39]. The Arabidopsis genome contains 16 putative histone deacetylases, including 10 members of the RPD3/HDA1 family, four HDT family members, and two genes related to the yeast NAD dependent histone deacetylase Sir2p. A systematic knockdown screen of predicted Arabidopsis histone deacetylase genes using transgene-induced RNA interference (RNAi) in A. suecica identified a role for two of the genes, HDT1 [40] and HDA6 [46] (see Figure 3). Knocking down either of these genes results in the loss of nucleolar dominance, with derepression of the underdominant ribosomal RNA genes accompanied by a loss of cytosine methylation in the vicinity of the rRNA gene promoter and a switch from H3K9me2 to H3K4me3 association, mimicking the effects of treatment with TSA (or aza-dC). In A. thaliana hda6 mutants, as in A. suecica HDA6-RNAi lines, NORs become partially decondensed and changes in rRNA gene methylation and histone modifications are observed, indicating that HDA6 plays similar roles in non-hybrid A. thaliana and in the allotetraploid hybrid, A. suecica [46, 47].
RNAi-based screens in A. suecica have been conducted to identify components of the DNA methylation machinery that might account for the derepression of silenced rRNA genes by aza-dC, similar to the screen described above to identify histone deacetylases that could account for the effects of TSA. Recent studies indicate that de-novo DNA methyltransferase activity and two different methylcytosine binding domain proteins are required to maintain nucleolar dominance (Preuss and Pikaard, unpublished; see Figure 3). Biochemical studies are now needed to understand how the DNA methylation and histone deacetylation machineries exert their effects on rRNA gene expression.
rRNA gene dosage control mechanisms and the mammalian NoRC complex
Findings in Arabidopsis concerning the molecular basis for nucleolar dominance fit well with studies of rRNA gene regulation in other systems, particularly in mammals. Ribosomal RNA gene transcription is tightly regulated in order to provide the proper amount of rRNA for ribosome assembly, according to the cellular need for protein synthesis [48-50]. Available evidence indicates that eukaryotes have more rRNA genes than are required for ribosome biogenesis under most physiological conditions such that they regulate the effective dosage of the rRNA genes they possess [51]. One level of control appears to be on/off switching to control the number of rRNA genes transcribed. This conclusion is supported by multiple lines of evidence including the molecular analyses of nucleolar dominance, as described above and summarized in Figure 3, the transcription-dependent accessibility of only a subset of rRNA genes to the DNA crosslinking agent psoralen [52-54] and direct electron microscopic observations of rRNA genes engaged in transcription by RNA polymerase I [55, 56]. All of these experimental approaches indicate that only a subset of the rRNA genes is active (for more extensive reviews on this subject, see [51] and [10]). Once the on/off decision has been made, eukaryotic cells then appear to regulate the frequency at which RNA polymerase I initiates on each active gene in order to fine-tune the amount of rRNA produced. Studies in yeast and mammals indicate that fine-tuning is accomplished primarily through signal transduction mechanisms that lead to the post-translational modification of essential RNA polymerase I transcription factors [57-62].
A key component of the rRNA gene on/off switch in mammals is a specialized chromatin remodelling activity known as NoRC (Nucleolar Remodelling Complex; see Figure 4) [63]. The discovery of NoRC in the Grummt lab came about in an interesting way. In mammals the transcriptional termination factor TTF-I binds to repeated RNA polymerase I transcription termination sequences at the end of the rRNA gene transcription unit [64] but also binds a site located just upstream of the minimal rRNA gene promoter where it is thought to prevent read-through of transcripts initiated from upstream. However, this latter TTF binding site also has characteristics of a position-dependent promoter element and is essential, along with added TTF-I, for transcription of minigenes assembled into nucleosomal templates [65, 66]. In the vicinity of the promoter, TTF-I influences nucleosome positioning in a way that allows transcription initiation to proceed. A yeast two hybrid screen for TTF-I-interacting partners identified TIP5 (TTF-I-interaction protein #5), which, in turn interacts with SNF2h, an ATP-dependent chromatin remodeling protein. Collectively, TIP5 and SNF2h comprise NoRC [63].
Figure 4. NoRC-mediated ribosomal RNA gene silencing in mouse.
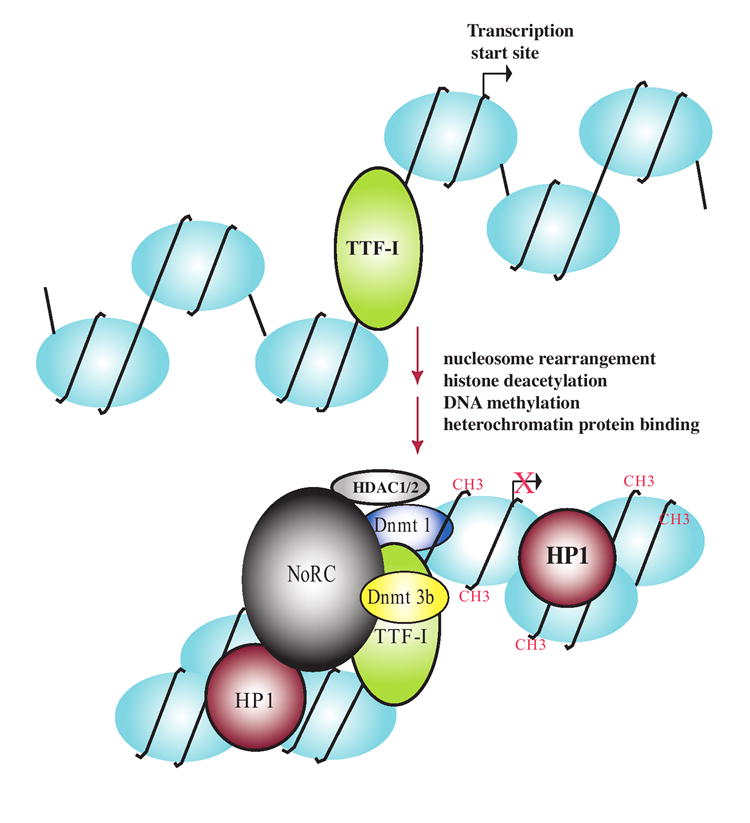
TTF-I binding upstream of the gene promoter recruits NoRC, an ATP-dependent chromatin remodeling complex that physically associates with DNA methyltransferases Dnmt 1 and Dnmt3b and histone deacetylases HDAC1 and HDAC2. Resulting DNA methylation (denoted by CH3) and histone deacetylation facilitates H3K9 methylation, Heterochromatin Protein 1 (HP1) binding and condensation of the nucleosomal DNA into silenced heterochromatin.
A prediction was that overexpressing NoRC components would increase transcription of co-transfected rRNA minigenes in cultured cells. Surprisingly, TIP5 over-expression inhibits transcription, and does so in an aza-dC and TSA-reversible manner, implicating DNA methylation and histone deacetylation in NoRC-mediated repression [67]. Subsequent biochemical studies have demonstrated that NoRC physically interacts with the Sin3 co-repressor complex that includes histone deacetylase HDAC1 and HDAC2 and also interacts with the DNA methyltransferases DNMT1 and DNMT3 [67, 68]. Collectively, the data suggest a mechanism whereby sequence-specific DNA binding by TTF-I can recruit NoRC to bring about histone deacetylation and de novo DNA methylation in order to help establish a repressive chromatin state, as summarized in Figure 4.
NoRC mediated repression of mammalian rRNA genes and nucleolar dominance have a number of interesting parallels. Although there is no obvious ortholog of TIP5 in Arabidopsis, and a NoRC-like complex has not yet been identified in plants, both nucleolar dominance and mammalian rRNA gene repression involve RPD3 class histone deacetylases (HDA6 in plants, HDAC1 and HDAC2 in mammals). Histone deacetylation is also mechanistically coupled to cytosine methylation in both cases and silencing can be inhibited by blocking either modification using aza-dC or TSA. Collectively, these observations suggest that similar mechanisms regulate rRNA genes in plants and mammals [9, 51]. Moreover, the chromatin modifications responsible for nucleolar dominance in Arabidopsis hybrids are also the same modifications that specify the active and inactive subsets of rRNA genes in non-hybrid A. thaliana, suggesting that nucleolar dominance is a manifestation of the rRNA gene dosage control system that operates in all eukaryotes [40].
Are NORs regulated as single loci, or is it every rRNA gene for itself?
Collectively, the available evidence points to rRNA gene dosage control as the reason that nucleolar dominance occurs. Nonetheless, it remains unclear how or why one parental set of rRNA genes should be selectively turned off as opposed to coordinately down-regulating the rRNA genes inherited from both progenitors. The coding regions of the rRNA genes are nearly identical in species that can interbreed, so it is highly unlikely that functional differences would exist in ribosomes derived from rRNA genes of either parent. Identifying the “choice” mechanism for selective rRNA/NOR inactivation is likely to reveal important insights into the overall regulation of rRNA genes.
How might choice come about? Because nucleolar dominance is independent of maternal or paternal effects, gametic imprinting [69-71] does not appear to be the explanation for initially choosing one progenitor’s NORs for inactivation. The choice of which NORs to inactivate is also not random, unlike X-chromosome inactivation in somatic cells of female mammals, one consequence of which is the randomly splotched coat coloration in calico cats due to the inactivation of different coat color genes on the two X chromosomes [72, 73]. The number of rRNA genes at an NOR is also not a clear predictor of nucleolar dominance because NORs with fewer rRNA genes are sometimes dominant; likewise, dominant NORs can often be outnumbered substantially upon changes in ploidy and yet remain dominant [7, 74-76].
For the reasons above, it has been attractive to look to sequence differences in the rRNA genes themselves in order to try to find explanations, with differences in intergenic spacer repeat type, or repeat number having been proposed to control nucleolar dominance in Xenopus, wheat, and Solanum [34, 77-80]. The only experimental evidence supporting this hypothesis has come from oocyte injection experiments in Xenopus using plasmid-encoded rRNA minigenes [78, 81]. There are differences in the numbers and types of repeated sequences in the intergenic spacers of X. laevis and X. borealis. Promoters with adjacent X. laevis spacer sequences cloned upstream are dominant over coinjected minigenes that have X. borealis spacer sequences linked to the promoter, mimicking the direction of nucleolar dominance in X. laevis - X. borealis hybrids [78]. The spacers include duplicated sequences that share sequence similarity with the gene promoter and have the properties of enhancers [82-84]. Therefore differences in binding affinity of one or more limiting transcription factors to the enhancers could explain the dominance of X. laevis rRNA genes. Initially, it was inferred that enhancers alone are sufficient to explain the nucleolar dominance-like phenomenon in injected oocytes [78], though subsequent tests have revealed the need for a spacer promoter upstream of the enhancer repeats in order for X. laevis spacers to confer dominance in injected oocytes [81], suggesting a need for transcription of the spacer repeats in order to stimulate the downstream gene promoter. The mechanism by which spacer transcription might stimulate pre-rRNA transcription initiated from the adjacent gene promoter is unclear, particularly in light of recent data from the Grummt lab suggesting that spacer transcription participates in NoRC-dependent rRNA gene silencing [85].
Despite the elegance of the oocyte injection experiments in Xenopus, these experiments are not without their caveats. Foremost of these is the fact that a ten million fold excess of rRNA minigenes is typically injected relative to the number of endogenous rRNA genes, which may artificially create transcription factor deficiencies that endogenous rRNA genes never experience. Analogous tests in plants in which minigenes were transfected into protoplasts in numbers similar to the number of endogenous rRNA genes failed to reveal any differences in the competitive strength of dominant and underdominant genes, nor did competitive in vitro transcription assays [43, 86]. Moreover, the demonstration that underdominant rRNA genes are abundantly expressed in Arabidopsis and Brassica upon blocking DNA methylation or histone deacetylation, and dominant genes are also up-regulated, suggests that transcription factors are present in sufficient amounts to support very high levels of transcription provided that the genes are in an accessible chromatin state [39, 40, 46]. The demonstration that the direction of nucleolar dominance can be reversed in response to changes in ploidy in Arabidopsis thaliana x A. arenosa hybrids bearing 1:3, 2:2 or 3:1 genome dosages of the two progenitors [43] also does not fit with the idea that one parental type of rRNA gene has an inherently higher binding affinty for a limiting transcription factor, in which case that gene type would always recruit transcription factors and be transcribed to some degree. There is no correlation between intergenic spacer length or repeat number in Brassica allotetraploids that display nucleolar dominance [76]. Likewise, nucleolar dominance occurs in marine copepods whose rRNA gene spacers lack repeated sequences entirely [87]. Collectively, these lines of evidence suggest that something other than transcription factor binding affinities account for the complete, or nearly complete, silencing of entire NORs in nucleolar dominance.
It is plausible that finding sequence differences among the rRNA genes of two different progenitors has lured us, in many cases, into focusing on those differences within each rRNA gene without appreciating the possibility that these sequence polymorphisms may only be markers linked to distant regulatory loci that might be millions of basepairs away, possibly at the edges of the NORs, or beyond. Recombination is notoriously suppressed on chromosome arms in the vicinity of NORs in plants as in other eukaryotes [88], making the possibility of a NOR locus control region linked to the observed rRNA gene sequence polymorphisms worth considering. In other words, rather than being controlled at the level of individual rRNA genes, an alternative possibility is that nucleolar dominance results from controls exerted on NORs as the regulatory units.
Evidence for the possibility that NORs are the units of regulation in nucleolar dominance has come from genetic studies of Arabidopsis thaliana inter-ecotype hybrids. In hybrids of the Cape Verde island (Cvi) and Landsberg erecta (Ler) ecotypes, Ler rRNA genes tend to be repressed and Cvi rRNA genes tend to be dominant [89]. In a Cvi/Ler recombinant inbred population carried to the F8 generation, in which essentially all loci are homozygous for alleles derived from one ecotype or the other, Ler rRNA genes were found to be silenced when Cvi contributed the NOR on chromosome 4 (NOR4) and the NOR on chromosome 2 (NOR2) came from Ler [89]. The opposite combination did not trigger any appreciable rRNA gene silencing, even though the rRNA genes within an ecotype are essentially identical at both NORs. Furthermore, the presence of the Cvi NOR4/Ler NOR2 combination did not always trigger efficient silencing, pointing to the involvement of other modifier loci. QTL mapping identified one locus, on chromosome three, which somehow boosts expression at NOR4, thereby enhancing nucleolar dominance when Cvi contributes NOR4 and diminishing nucleolar dominance when NOR4 comes from Ler and NOR2 comes from Cvi [89].
The Arabidopsis inter-ecotype data do not fit easily into models in which nucleolar dominance is regulated at the level of individual rRNA genes. The sequences of rRNA genes are >95% identical across A. thaliana ecotypes [90] in our experience and the rRNA genes within an ecotype (or species) are nearly identical due to the phenomenon of concerted evolution [91, 92]. Therefore it seems unlikely that sufficient differences exist between the rRNA genes on NOR2 and NOR4 in either Cvi or Ler plants to explain why only the Cvi NOR4/Ler NOR2 combination triggers efficient nucleolar dominance. Instead, the data point to larger scale regulation of the NORs, dictated by the involvement of unlinked loci.
Challenges ahead
The epigenetic control of rRNA gene expression appears to play an important role in the normal physiology of all eukaryotes and the selective use of this dosage control system to preferentially inactive rRNA genes from one progenitor provides a molecular explanation for nucleolar dominance in hybrids. In plants and in mammals, essentially identical cytosine methylation and histone modifications are involved in dictating the rRNA gene on and off states, indicating an ancient origin for the epigenetic control system. As additional components of the dosage control system are identified, it will be interesting and important to determine how the various chromatin modifying activities work together in order to couple changes in DNA methylation and histone modification.
The choice mechanism(s) by which whole NORs or subsets of rRNA genes are selected for inactivation remains an intriguing mystery. Understanding these mechanisms would no doubt shed new light on how nucleolar dominance is initially established and would also tell us more about how rRNA gene dosage levels are set and maintained and/or readjusted through multiple cell cycles in non-hybrids. The possibility that regulation occurs at the level of whole NORs rather than one rRNA gene at a time deserves additional thought and experimental testing.
There is a long, rich history of cytological and molecular research into the functions of NORs and rRNA genes, but there is still much that we don’t know and much more to be learned about the chromosomal control of these dynamic loci.
Acknowledgments
Work on nucleolar dominance in the C.S.P. lab is funded by National Institutes of Health grant R01-GM60380. S.P. was also supported by an NIH National Research Service Award (1 F32 GM69206). Any opinions, findings, and conclusions or recommendations expressed in this article are those of the author(s) and do not necessarily reflect the views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Verdun D, Roussel P. Regulators of nucleolar functions. Prog Cell Cycle Res. 2003;5:301–308. [PubMed] [Google Scholar]
- 3.Leung AK, Lamond AI. The dynamics of the nucleolus. Crit Rev Eukaryot Gene Expr. 2003;13:39–54. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.40. [DOI] [PubMed] [Google Scholar]
- 4.Prieto JL, McStay B. Nucleolar biogenesis: the first small steps. Biochem Soc Trans. 2005;33:1441–1443. doi: 10.1042/BST0331441. [DOI] [PubMed] [Google Scholar]
- 5.Heitz E. Nukleolen und chromosomen in der Gattung Vicia. Planta. 1931;15:495–505. [Google Scholar]
- 6.McClintock B. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Zeit Zellforsch Mik Anat. 1934;21:294–328. [Google Scholar]
- 7.Navashin M. Chromosomal alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia. 1934;5:169–203. [Google Scholar]
- 8.Honjo T, Reeder RH. Preferential transcription of Xenopus laevis ribosomal RNA in interspecies hybrids between Xenopus laevis and Xenopus mulleri. J Mol Biol. 1973;80:217–228. doi: 10.1016/0022-2836(73)90168-x. [DOI] [PubMed] [Google Scholar]
- 9.McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 10.Neves N, Viegas W, Pikaard CS. Nucleolar dominance and rRNA gene dosage control: a paradigm for transcriptional regulation via an epigenetic on/off switch. In: Meyer P, editor. Plant Epigenetics. Blackwell Publishing; Sheffield: 2005. pp. 201–222. [Google Scholar]
- 11.Pikaard CS. Nucleolar dominance: uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol Biol. 2000;43:163–177. doi: 10.1023/a:1006471009225. [DOI] [PubMed] [Google Scholar]
- 12.Ritossa FM, Spiegelman S. Localization of DNA complementary to ribosomal RNA in the nucleolus organizer region of Drosophila melanogaster. Proc Natl Acad Sci USA. 1965;53:737–745. doi: 10.1073/pnas.53.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace H, Birnstiel ML. Ribosomal cistrons and the nucleolar organizer. Biochem Biophys Acta. 1966;114:296–310. doi: 10.1016/0005-2787(66)90311-x. [DOI] [PubMed] [Google Scholar]
- 14.Reeder RH. Ribosomes from eukaryotes: genetics. In: Nomura M, editor. Ribosomes. Cold Spring Harbor; NY: 1974. pp. 489–519. [Google Scholar]
- 15.Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- 16.Wallace H, Langridge WHR. Differential amphiplasty and the control of ribosomal RNA synthesis. Heredity. 1971;27:1–13. [Google Scholar]
- 17.Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pikaard CS, McStay B, Schultz MC, Bell SP, Reeder RH. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes Dev. 1989;3:1779–1788. doi: 10.1101/gad.3.11.1779. [DOI] [PubMed] [Google Scholar]
- 19.O’Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeder RH, Pikaard CS, McStay B. UBF, an architectural element for RNA polymerase I promoters. In: Eckstein F, Lilley DMJ, editors. Nucl Acids and Mol Biol. Springer-Verlag; Berlin: 1995. pp. 251–263. [Google Scholar]
- 21.Wright JE, Mais C, Prieto JL, McStay B. A role for upstream binding factor in organizing ribosomal gene chromatin. Biochem Soc Symp. 2006:77–84. doi: 10.1042/bss0730077. [DOI] [PubMed] [Google Scholar]
- 22.Roussel P, Andre C, Masson C, Geraud G, Hernandez-Verdun D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci. 1993;104(Pt 2):327–337. doi: 10.1242/jcs.104.2.327. [DOI] [PubMed] [Google Scholar]
- 23.Gebrane-Younes J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110(Pt 19):2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- 24.Putnam CD, Pikaard CS. Cooperative binding of the Xenopus RNA polymerase I transcription factor xUBF to repetitive ribosomal gene enhancers. Mol Cell Biol. 1992;12:4970–4980. doi: 10.1128/mcb.12.11.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bazett-Jones D, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 26.Putnam CD, Copenhaver GP, Denton ML, Pikaard CS. The RNA polymerase I transactivator UBF requires its dimerization domain and HMG-box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol. 1994;14:6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neil KJ, Ridsdale RA, Rutherford B, Taylor L, Larson DE, Glibetic M, Rothblum L, Harauz G. Structure of recombinant UBF by electron image analysis and homology modelling. Nucleic Acids Res. 1996;24:1472–1480. doi: 10.1093/nar/24.8.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kermekchiev M, Workman JL, Pikaard CS. Nucleosome binding by the pol I transcription factor UBF displaces linker histone H1. Mol Cell Biol. 1997;17:5833–5842. doi: 10.1128/mcb.17.10.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadal O, Labarre S, Boschiero C, Thuriaux P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. Embo J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. J Biol Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 32.Thompson WF, Flavell RB. DNase I sensitivity of ribosomal RNA genes in chromatin and nucleolar dominance in wheat. J Mol Biol. 1988;204:535–548. doi: 10.1016/0022-2836(88)90353-1. [DOI] [PubMed] [Google Scholar]
- 33.Flavell RB, O’Dell M, Thompson WF. Regulation of cytosine methylation in ribosomal DNA and nucleolus organizer expression in wheat. J Mol Biol. 1988;204:523–534. doi: 10.1016/0022-2836(88)90352-x. [DOI] [PubMed] [Google Scholar]
- 34.Sardana R, O’Dell M, Flavell R. Correlation between the size of the intergenic regulatory region, the status of cytosine methylation of rRNA genes and nucleolar expression in wheat. Mol Gen Genet. 1993;236:155–162. doi: 10.1007/BF00277107. [DOI] [PubMed] [Google Scholar]
- 35.Houchins K, O’Dell M, Flavell RB, Gustafson JP. Cytosine methylation and nucleolar dominance in cereal hybrids. Mol Gen Genet. 1997;255:294–301. doi: 10.1007/s004380050500. [DOI] [PubMed] [Google Scholar]
- 36.Neves N, Heslop-Harrison JS, Viegas W. rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat x rye hybrids. Theor Appl Genet. 1995;91:529–533. doi: 10.1007/BF00222984. [DOI] [PubMed] [Google Scholar]
- 37.Macleod D, Bird A. DNase I sensitivity and methylation of active versus inactive rRNA genes in Xenopus species hybrids. Cell. 1982;29:211–218. doi: 10.1016/0092-8674(82)90105-2. [DOI] [PubMed] [Google Scholar]
- 38.Amado L, Abranches R, Neves N, Viegas W. Development-dependent inheritance of 5-azacytidine-induced epimutations in triticale: analysis of rDNA expression patterns. Chromosome Res. 1997;5:445–450. doi: 10.1023/a:1018460828720. [DOI] [PubMed] [Google Scholar]
- 39.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 41.Durica DS, Krider HM. Studies on the ribosomal RNA cistrons in Drosophila hybrids. II. Heterochromatic regions mediating nucleolar dominance. Genetics. 1978;89:37–64. doi: 10.1093/genetics/89.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tweedie S, Ng HH, Barlow AL, Turner BM, Hendrich B, Bird A. Vestiges of a DNA methylation system in Drosophila melanogaster? Nat Genet. 1999;23:389–390. doi: 10.1038/70490. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental rRNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci USA. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 45.Dryden DT, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Scheid OM. Arabidopsis Histone Deacetylase HDA6 Is Required for Maintenance of Transcriptional Gene Silencing and Determines Nuclear Organization of rDNA Repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moss T, Stefanovsky VY. At the center of eukaryotic life. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 50.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 51.Grummt I, Pikaard CS. Epigenetic mechanisms controlling RNA polymerase I transcription. Nature Rev Mol Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- 52.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 53.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dammann R, Lucchini R, Koller T, Sogo JM. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller OL, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- 56.McKnight SL, Miller OL. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- 57.Milkereit P, Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. Embo J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 60.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavanaugh AH, Hirschler-Laszkiewicz I, Hu Q, Dundr M, Smink T, Misteli T, Rothblum LI. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J Biol Chem. 2002;277:27423–27432. doi: 10.1074/jbc.M201232200. [DOI] [PubMed] [Google Scholar]
- 62.Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Cote J, Moss T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 63.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. Embo J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartsch I, Schoneberg C, Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988;8:3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langst G, Blank TA, Becker PB, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. Embo J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. Embo J. 1998;17:3135–3145. doi: 10.1093/emboj/17.11.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. Embo J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sleutels F, Barlow DP, Lyle R. The uniqueness of the imprinting mechanism. Curr Opin Genet Dev. 2000;10:229–233. doi: 10.1016/s0959-437x(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 70.Brannan CI, Bartolomei MS. Mechanisms of genomic imprinting. Curr Opin Genet Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 71.Huynh KD, Lee JT. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr Opin Cell Biol. 2001;13:690–697. doi: 10.1016/s0955-0674(00)00272-6. [DOI] [PubMed] [Google Scholar]
- 72.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 73.Lee JT, Jaenisch R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr Opin Genet Dev. 1997;7:274–280. doi: 10.1016/s0959-437x(97)80138-4. [DOI] [PubMed] [Google Scholar]
- 74.Flavell RB, O’Dell M. The genetic control of nucleolus formation in wheat. Chromosoma. 1979;71:135–152. [Google Scholar]
- 75.Flavell RB. The structure and control of expression of ribosomal RNA genes. Oxford Surveys Plant Mol Cell Biol. 1986;3:252–274. [Google Scholar]
- 76.Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997;94:3442–3447. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reeder RH. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reeder RH, Roan JG. The mechanism of nucleolar dominance in Xenopus hybrids. Cell. 1984;38:39–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- 79.Martini G, O’Dell M, Flavell RB. Partial inactivation of wheat nucleolus organizers by the nucleolus organizer chromosomes from Aegilops umbellulata. Chromosoma. 1982;84:687–700. [Google Scholar]
- 80.Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA. Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol. 2004;56:439–463. doi: 10.1007/s11103-004-4678-x. [DOI] [PubMed] [Google Scholar]
- 81.Caudy AA, Pikaard CS. Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J Biol Chem. 2002;277:31577–31584. doi: 10.1074/jbc.M202737200. [DOI] [PubMed] [Google Scholar]
- 82.Busby SJ, Reeder RH. Spacer sequences regulate transcription of ribosomal gene plasmids injected into Xenopus embryos. Cell. 1983;34:989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- 83.Labhart P, Reeder RH. Enhancer-like Properties of the 60/81 bp Elements in the Ribosomal Gene Spacer of Xenopus laevis. Cell. 1984;37:285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- 84.Pikaard CS. Ribosomal gene promoter domains can function as artificial enhancers of RNA polymerase I transcription, supporting a promoter origin for natural enhancers. Proc Natl Acad Sci. 1994;91:464–468. doi: 10.1073/pnas.91.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 86.Frieman M, Chen ZJ, Saez-Vasquez J, Shen LA, Pikaard CS. RNA polymerase I transcription in a Brassica interspecific hybrid and its progenitors: tests of transcription factor involvement in nucleolar dominance. Genetics. 1999;152:451–460. doi: 10.1093/genetics/152.1.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flowers JM, Burton RS. Ribosomal RNA gene silencing in interpopulation hybrids of Tigriopus californicus: nucleolar dominance in the absence of intergenic spacer subrepeats. Genetics. 2006;173:1479–1486. doi: 10.1534/genetics.106.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Copenhaver GP, Browne WE, Preuss D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc Natl Acad Sci U S A. 1998;95:247–252. doi: 10.1073/pnas.95.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis MS, Cheverud JM, Pikaard CS. Evidence for nucleolus organizer regions as the units of regulation in nucleolar dominance in Arabidopsis thaliana interecotype hybrids. Genetics. 2004;167:931–939. doi: 10.1534/genetics.103.026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Copenhaver GP, Pikaard CS. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 1996;9:259–272. doi: 10.1046/j.1365-313x.1996.09020259.x. [DOI] [PubMed] [Google Scholar]
- 91.Dover GA, Flavell RB. Molecular co-evolution: rDNA divergence and the maintenance of function. Cell. 1984;38:622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 92.Gerbi SA. Evolution of ribosomal DNA. In: McIntyre RJ, editor. Molecular Evolutionary Genetics. Plenum Press; New York: 1985. pp. 419–517. [Google Scholar]


