Abstract
Activation of macrophages and subsequent “killing” effector functions against infectious pathogens are essential for the establishment of protective immunity. NF-IL6 is a transcription factor downstream of IFN-γ and TNF in the macrophage activation pathway required for bacterial killing. Comparison of microarray expression profiles of Listeria monocytogenes (LM)-infected macrophages from WT and NF-IL6-deficient mice enabled us to identify candidate genes downstream of NF-IL6 involved in the unknown pathways of LM killing independent of reactive oxygen intermediates and reactive nitrogen intermediates. One differentially expressed gene, PKCδ, had higher mRNA levels in the LM-infected NF-IL6-deficient macrophages as compared with WT. To define the role of PKCδ during listeriosis, we infected PKCδ-deficient mice with LM. PKCδ-deficient mice were highly susceptible to LM infection with increased bacterial burden and enhanced histopathology despite enhanced NF-IL6 mRNA expression. Subsequent studies in PKCδ-deficient macrophages demonstrated that, despite elevated levels of proinflammatory cytokines and NO production, increased escape of LM from the phagosome into the cytoplasm and uncontrolled bacterial growth occurred. Taken together these data identified PKCδ as a critical factor for confinement of LM within macrophage phagosomes.
Keywords: microarray, phagosomal escape, bacterial killing
Listeria monocytogenes (LM) is a Gram-positive, facultative intracellular bacterium responsible for disseminated infections in immunocompromised individuals that can result in septicemia and meningitis (1). Effective control of listeriosis requires both innate and adaptive immune responses with the principal mediators of bacterial killing being neutrophils and macrophages (2). Key cytokines driving the innate immune response are IFN-γ (3) and TNF (4), which promote macrophage activation and drive the production of antimicrobial mediators such as reactive oxygen intermediates (ROIs) and reactive nitrogen intermediates (RNIs). Several studies using gene-deficient mice demonstrated that ROI and RNI are required for optimal pathogen killing (5–8) but that macrophages also utilize an alternative, unknown mechanism besides ROI and RNI to efficiently control bacterial infection.
Mice deficient for the transcription factor NF-IL6 are highly susceptible to a number of intracellular bacterial and fungal infections, including LM (9–12). NF-IL6-deficient mice infected with LM were unable to prevent bacterial dissemination despite production of NO, IFN-γ, and TNF being equivalent to WT controls (12). From these results, we hypothesized that NF-IL6 acts downstream from IFN-γ and TNF during the activation of macrophage effector functions against LM (2). To facilitate the identification of putative effector genes in this unknown pathway, we compared gene expression profiles of WT and NF-IL6-deficient LM-infected macrophages by differential microarray. One differentially expressed (DE) gene, PKCδ, had higher mRNA levels in the LM-infected NF-IL6−/− macrophages as compared with WT controls.
PKCδ belongs to the PKC family of diacylglycerol-activated serine threonine kinases (13) and is widely expressed in many cell types, including macrophages (14). PKCδ exerts its influence on a broad range of cellular responses such as cell growth, differentiation, apoptosis, and phagocytosis (15). In addition, PKCδ has been shown to regulate NF-IL6 activity through direct phosphorylation (16, 17, 29). However, the role of PKCδ in infectious diseases has not previously been investigated in vivo. Here we demonstrate a critical role for PKCδ in controlling LM infection. We report that, in the absence of PKCδ, mice were highly susceptible to LM and died before the onset of a protective adaptive immune response. This heightened susceptibility was driven by increased phagosomal escape of LM in PKCδ−/− macrophages, resulting in uncontrolled listerial growth despite increased proinflammatory cytokine and NO response. Furthermore, levels of NF-IL6 were markedly enhanced in PKCδ−/− mice after infection. Taken together the results clearly demonstrate that PKCδ is a critical factor for confinement of LM within macrophage phagosomes.
Results
Identification of PKCδ by Differential Microarray.
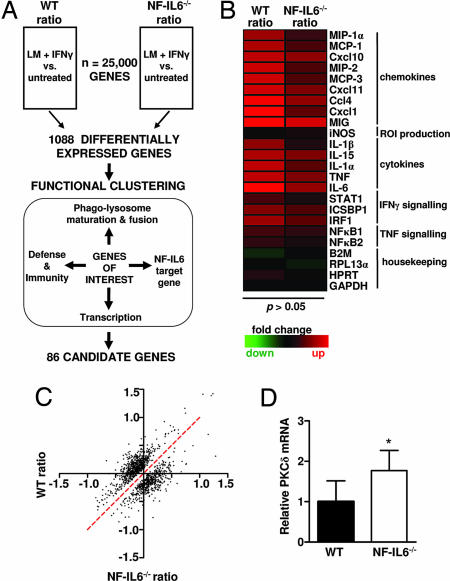
A differential microarray strategy combined with functional clustering was used to identify and select genes involved in listerial killing independent of RNI and ROI (Fig. 1A). Here, bone marrow-derived macrophages (BMDMs) from WT and NF-IL6−/− mice were simultaneously activated with IFN-γ and infected with LM (LM plus IFN-γ) or were left untreated. Total RNA isolated from untreated and infected samples 4 h postinfection (p.i.) was used to generate cDNA probes that were hybridized to microarrays. As expected from the literature (12) and predicted from our hypothesis, we found proinflammatory cytokines to be similarly induced in both LM-infected WT and NF-IL6−/− macrophages (Fig. 1B) (P > 0.05). Indeed, equivalent levels of secretion of these proteins from LM-infected WT and NF-IL6−/− macrophage cultures was confirmed by ELISA (data not shown). The impaired induction of granulocyte colony-stimulating factor (12), Clecsf9 (18), IL-12p35 (10), and ISGF3-γ (19) mRNAs, which are induced by NF-IL6, were not detectable by microarray. However, impaired transcription in LM-infected NF-IL6−/− macrophages was confirmed by quantitative RT-PCR (data not shown). Together, these data demonstrated the biological relevance of the microarray data. In an exploratory statistical approach, the top 10% of genes with the best evidence of differential expression between WT and NF-IL6−/−-infected vs. untreated gene expression ratios were selected. Of the whole mouse genome comprising ≈25,000 genes, 1,088 DE genes were selected. The fold induction or repression of DE genes in the infected vs. the untreated samples ranged from −1.5 to 1.5 on a log2 scale (Fig. 1C). These relatively small, yet significant, changes are a result of the two mandatory sequential normalization procedures that enable comparison of data between multiple microarrays. Moreover, because the macrophages were simultaneously infected and stimulated with IFN-γ, fewer genes may have been fully induced after 4 h of infection. Functional clustering of the DE genes revealed that 11% were involved in regulation of transcription. Using a set of predicted cis-regulatory modules (CRMs) that contain several phylogenetically conserved binding sites for different transcription factors (20), we found upstream and downstream promoter regions of the DE genes enriched for CRMs containing putative binding sites for NF-IL6 as compared with the CRMs for the remaining mouse genes (15% vs. 12%; P < 0.05). Among the DE genes, 3% were involved in host immunity and defense, 6% in the production of ROI, 18% in phagosome maturation and phagolysosome fusion, and 19% in signaling. Thirty-two percent of the candidate genes did not have any functional annotation and were classified as “unknown.” DE genes that belonged to all or several of the functional groups were selected for ongoing studies. One of these genes, PKCδ, had significantly higher mRNA levels in the infected NF-IL6−/− macrophages, and quantitative RT-PCR confirmed the PKCδ mRNA to be 1.77 times higher in the infected NF-IL6 macrophages as compared with the WT controls (Fig. 1D) (P < 0.05). PKCδ was selected for further analysis based on this differential expression and functional clustering. Furthermore, PKCδ regulatory roles in cellular responses relevant to the control of bacilli within the macrophage, including production of superoxide (21, 22) and proinflammatory signaling pathways mediated by NF-κB (23), TNF (24, 25), IL-6 (26), 15-lipoxygenase (27), and IFN-γ (28), are established. Additionally, PKCδ has been shown to regulate the transcriptional activity of NF-IL6 (16, 17, 29). This body of work is suggestive of a role for PKCδ in regulating host responses to infectious diseases. Surprisingly, such a role of PKCδ in infectious immunity has not been investigated to date; as such, we decided to examine its role in innate immunity using a PKCδ gene-deficient mouse model.
Fig. 1.
Identification of PKCδ by microarray. (A) Differential microarray and functional clustering to identify candidate genes. BMDMs from WT and NF-IL6−/− mice were untreated or were simultaneously activated with IFN-γ and infected with LM (LM + IFN-γ). RNA was isolated from untreated and infected samples at 4 h p.i. and used to generate labeled cDNA probes, which were hybridized to the microarray. DE genes were identified by comparing the log2-transformed ratio (LM + IFN-γ vs. untreated) for each gene between all four WT and NF-IL6−/− microarrays. DE genes were functionally clustered, and genes belonging to multiple functional categories were selected for further study. (B) Heat map showing up-regulation of proinflammatory mediators in both WT and NF-IL6−/−-infected BMDMs. Rows represent individual genes, and columns show the log2 transformed gene expression ratio. Up- or down-regulated genes in the LM plus IFN-γ sample, as compared with the untreated, are shaded red or green, respectively. (C) Scatter plot of the DE genes. Plotted on the x and y axes are the gene expression ratios for WT and NF-IL6−/− mice. The red dotted line represents non-DE ratios. (D) Quantitative RT-PCR confirming higher PKCδ mRNA levels in infected NF-IL6−/− BMDMs as compared with WT. Data shown are means ± SEM (*, P < 0.05).
Increased Mortality in LM-Infected PKCδ−/− Mice Despite Enhanced Levels of NF-IL6 and IL-6.
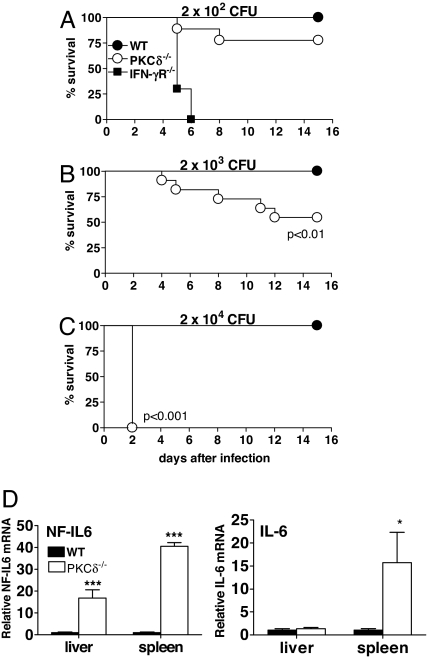
To address the possible role of PKCδ in host-protective responses in vivo, PKCδ−/− mice and their control littermates were infected i.p. with titrated doses of LM and mortality was measured (Fig. 2 A–C). WT mice were resistant to LM infection with an LD50 of 2 × 105 cfu (data not shown). No difference between PKCδ+/− heterozygous littermates and WT mice in controlling the infection was seen (data not shown). In contrast, PKCδ−/− mice were highly susceptible to LM infection with a LD50 of 2 × 103 cfu (Fig. 2B). IFN-γR−/− mice are highly susceptible to LM infection (30) and were included to assess the relative degree of susceptibility of PKCδ−/− mice to LM infection. As expected, all IFN-γR−/− mice died within 7 days of infection with 2 × 102 cfu (Fig. 2A). These results clearly demonstrated that PKCδ−/− mice had defective innate immune responses rendering them highly susceptible to LM infection. To explore a possible relationship between this PKCδ-dependent susceptibility and NF-IL6, mRNA levels of NF-IL6 were examined in the liver and spleen from WT and PKCδ−/− mice at 2 days p.i. (Fig. 2D). Levels of NF-IL6 (P < 0.0001) and IL-6 (P < 0.05) were found to be significantly higher in PKCδ−/− mice when compared with WT mice.
Fig. 2.
Enhanced mortality in PKCδ−/− mice after LM infection despite increased levels of NF-IL6 and IL-6. Mice were infected with 2 × 102 cfu (A), 2 × 103 cfu (B), or 2 × 104 cfu (C) of LM, and mortality was measured for n = 7–10 mice per group (A), n = 11–12 mice per group (B), and n = 5 mice per group (C). (D) Quantitative RT-PCR of NF-IL6 and IL-6 in liver and spleens of WT and PKCδ−/− mice at 2 days after LM infection with 2 × 104 cfu (n = 5 per group). Data shown are means ± SEM (*, P < 0.05; ***, P < 0.0001). All results are representative of two independent experiments.
Enhanced Bacterial Burden and Increased Histopathology in LM-Infected PKCδ−/− Mice.
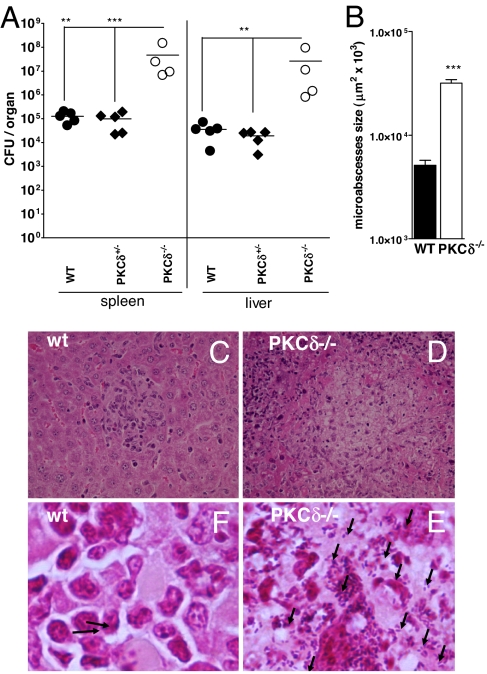
PKCδ−/−, WT, and PKCδ+/− heterozygous littermate mice were infected with 2 × 104 cfu of LM, and bacterial burdens in liver and spleen were measured at 2 days p.i. (Fig. 3A). Spleen and liver cfu from PKCδ−/− mice were significantly increased when compared with WT and PKCδ+/− mice (P < 0.01). Protective immune responses against LM involve the formation of small microabscesses consisting of infiltrating immune cells that confine and control bacterial growth. At 2 days p.i. liver microabscesses from WT mice were small (Fig. 3B) and well defined (Fig. 3C). In contrast, PKCδ−/− microabscesses (Fig. 3D) were larger (P < 0.0001) and more abundant and contained many polymorphonuclear cells and zones of hepatocellular necrosis. This elevated histopathology was accompanied by higher bacilli counts in PKCδ−/− microabscesses (Fig. 3E) than WT microabscesses (Fig. 3F). This failure to control bacterial growth resulted in necrotic lesions and liver destruction in the PKCδ−/− mice. Taken together, histopathology from LM-infected PKCδ−/− mice showed significantly more and larger microabscesses in PKCδ−/− mice than were seen in WT mice, which would drive the increased mortality we described.
Fig. 3.
Increased bacterial burden and enhanced histopathology in PKCδ−/− mice. WT, PKCδ+/− heterozygous littermates, and PKCδ−/− mice were infected i.p. with 2 × 104 cfu of LM (n = 4–5 per group). (A) Bacterial load in spleen and liver was determined at 2 days p.i. (**, P < 0.01; ***, P < 0.001). (B) Size quantification of liver microabscesses in WT and PKCδ−/− mice. Data correspond to the mean of 80 microabscesses measured in three mice per group (***, P < 0.0001). Liver sections were stained with hematoxylin/eosin (C and D) and Gram-positive stain (E and F), which colors LM in blue (arrow). [Magnifications: ×400 (C and D) and ×1,000 (E and F).] All data are representative of two independent experiments.
Efficient Activation of PKCδ−/− Macrophages During LM Infection.
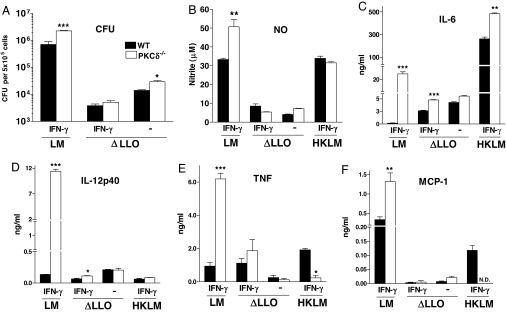
To determine whether increased susceptibility of the PKCδ−/− mice to LM is due to impaired macrophage activation, we measured bacterial growth along with NO, proinflammatory cytokines, and a chemokine in supernatants from LM-infected macrophages. Bacterial growth of live virulent LM was significantly increased in PKCδ−/− compared with WT macrophages at 12 h p.i. (Fig. 4A). Nitrite production, representative of the bactericidal mediator NO, was significantly enhanced in LM-infected PKCδ−/− macrophages (Fig. 4B). Cytokine levels of IL-6, IL-12p40, TNF, and monocyte chemoattractant protein 1 (MCP-1) were also increased in LM-infected PKCδ−/− macrophages (Fig. 4 C–F). These results demonstrated that, during LM infection, macrophage activation in the absence of PKCδ was not abrogated. To determine whether increased proinflammatory responses in PKCδ−/− macrophages were a consequence of a higher bacterial burden, macrophages were infected with heat-killed LM (HKLM) or listeriolysin O (LLO)-deficient LM mutant strain (ΔLLO), which are unable to escape from the phagosome. Levels of nitrite, TNF, and MCP-1 were similar between WT and PKCδ−/− macrophages infected with ΔLLO mutant LM, suggesting that the increased levels of these cytokines in LM-infected PKCδ−/− macrophages was due to a higher bacterial load.
Fig. 4.
Efficient activation of PKCδ−/− macrophages. Bacterial growth (A) and secretion of nitrite (B), cytokines (C–E), and chemokine (F) into the culture supernatant by WT and PKCδ−/− BMDMs 12 h p.i. with live LM, LLO-deficient LM mutant strain (ΔLLO), or heat-killed LM (HKLM). Macrophages were either activated overnight with IFN-γ or not stimulated. Data shown are means ± SEM of triplicate samples. LM results are representative of three independent experiments, and ΔLLO and HKLM results are from one experiment (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Small, yet significantly increased, levels of IL-6 were seen in ΔLLO mutant-infected PKCδ−/− macrophages when compared with WT controls. Such an increase occurred despite both strains having equivalent bacterial burdens, suggesting that this increased level of IL-6 in LM-infected PKCδ−/− macrophages was due to higher bacterial loads combined with alleviation of residual PKCδ-dependent transcriptional repression (29). In contrast, HKLM activation in PKCδ−/− macrophages induced increased levels of IL-6 but reduced levels of TNF and MCP-1, suggesting that different signaling pathways are induced during infection with ΔLLO mutant and HKLM in the absence of PKCδ.
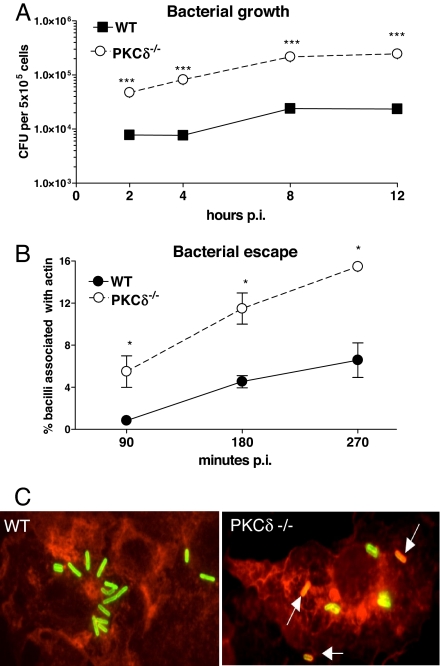
LM-Infected PKCδ−/− Macrophages Have Enhanced Bacterial Growth and Increased Bacterial Escape from Phagosomes.
To address effector functions of PKCδ, we infected PKCδ−/− BMDMs with LM and measured bacilli growth at 2, 4, 8, and 12 h p.i. (Fig. 5A). Macrophages from WT mice were able to restrict initial bacterial growth and reached a steady-state plateau at 8 and 12 h p.i. In contrast, PKCδ−/− macrophages were unable to control bacterial growth and had dramatically increased bacterial counts at all time points (P < 0.0001). To determine whether this increased bacterial growth was due to enhanced bacterial escape from phagosomes, BMDMs were infected with LM and bacterial escape was measured by fluorescent microscopy (Fig. 5 B and C). WT and PKCδ−/− macrophages contained equal numbers of bacteria per cell at 90 min p.i. (data not shown), indicating that bacterial phagocytosis was similar in all groups. The percentage of bacteria in the cytoplasm associated with actin and the number of bacteria trapped in phagosomes were scored, and the percentage bacterial escape in PKCδ−/− macrophages was significantly higher (P < 0.05) than in WT controls at 90, 180, and 270 min p.i. (Fig. 5B). These results clearly show that impaired listericidal activity in PKCδ−/− macrophages was due to a striking inability in phagosomal confinement of LM, resulting in enhanced listerial escape and subsequent uncontrolled bacterial growth.
Fig. 5.
Enhanced bacterial growth and increased bacterial escape from PKCδ−/− phagosomes. WT and PKCδ−/− BMDMs were activated overnight with IFN-γ and infected with LM. (A) Bacterial load was determined 2, 4, 8, and 12 h p.i. (***, P < 0.0001). (B) Quantification of LM escape from phagosomes at 90, 180, and 270 min p.i. The number of bacteria in the cytoplasm associated with actin (red) and trapped in the phagosome (green) was scored. Data shown are averages ± SEM of the means for three independent experiments (*, P < 0.05). (C) Deconvoluting fluorescent microscopy of BMDMs infected at 90 min with LM. Escaped bacteria (orange/yellow) are indicated by arrows. (Magnification: ×1,000.) All results are representative of three independent experiments.
Discussion
Early control of listeriosis requires an efficient innate immune response dominated by neutrophils and macrophages, the major effector cells responsible for bacilli killing (2). Full macrophage activation to LM requires IFN-γ and TNF driving the relevant production of cytokines, chemokines, and bactericidal mediators such as ROI and RNI (3, 4). Several in vivo infection studies using inducible NO synthase-deficient (7), gp47phox-deficient (6), and gp91phox/inducible NO synthase-deficient (8) mice have shown their bactericidal activity to be impaired. However, these macrophages could still kill a proportion of bacilli. Mice deficient in genes such as IFN-γR (30), TNFRp55 (6), ICSBP (31), IRF2 (31), RelB (32), LRG-47 (33), and NF-IL6 (12) were susceptible to LM infection despite full macrophage activation and production of ROI and RNI, suggesting that additional effector mechanisms besides ROI and RNI are used in macrophage-mediated bacterial killing. We hypothesize that this or these unknown pathway(s) are most likely mediated by IFN-γ- and TNF-induced responses, with the ultimate downstream effector molecule being NF-IL6 (2).
To identify genes involved in these unknown killing pathway(s), we compared transcriptional responses of LM-infected WT and NF-IL6−/− macrophages by microarray. As expected, up-regulation of proinflammatory cytokines and killing effector genes was comparable between LM-infected WT and NF-IL6−/− macrophages, confirming the biological relevance of the microarray data. Our strategy combining differential microarray efficiently reduced the number of possible candidate genes by 23-fold from ≈25,000 mouse genes to 1,088 genes. Functional clustering further reduced the candidates down to 86. Of these candidates, PKCδ was selected for further study because functional clustering suggested that it may act as a detrimental factor by repressing NF-IL6 activity (29) and IL-12 production (34) and promoting escape of LM from the phagosome (14). In contrast, we found PKCδ-deficient mice to be highly susceptible to LM infection with enhanced bacterial burden being apparent in the early stage of infection. This phenotype suggested a defect in innate bactericidal macrophage response in the absence of PKCδ. Indeed LM-infected PKCδ−/− macrophages were unable to contain the bacilli within the phagosome, resulting in uncontrolled bacterial growth, dissemination, and heightened mortality in PKCδ−/− mice. In line with the hypothesized unknown killing pathway(s), this increased lethality was independent of macrophage activation because PKCδ−/− macrophages secreted significantly higher levels of IL-6, IL-12p40, TNF, MCP-1, and NO in response to the higher bacterial burden. Moreover, the higher levels of IL-6 in LLO-infected PKCδ−/− macrophages, despite having an equal bacterial burden, may be attributed to alleviated PKCδ-dependent transcriptional repression (29). These data clearly demonstrate a novel protective role for PKCδ during innate immune response against LM.
The key question arising from our data was how PKCδ confines LM to the phagosome. Studies using rottlerin, a putative PKCδ-specific inhibitor, have reported that PKCδ induces extracellular calcium influxes through its activation of calcium channels, resulting in PKCβII translocation to early endosomes and leading to listerial phagosomal escape (14, 35). However, we found that extracellular calcium influxes mediated by L-type channels occurred independent of PKCδ, resulting in reduced listerial growth in both WT and PKCδ−/− macrophages (data not shown). PKCδ therefore does not appear to induce calcium influxes during LM infection as previously reported (14). This discrepancy may be due to the approach to block PKCδ by rottlerin, which is now accepted as a nonspecific inhibitor of a number of kinases and other enzymes in addition to PKCδ (37). Impaired ROI production may explain the heightened susceptibility of PKCδ−/− mice to LM because PKCδ is essential for stimulating localized release of superoxide into the phagosome (21, 22). However, because PKCδ−/− mice had increased susceptibility to LM compared with mice deficient for p47phox (6) and gp91phox (5) and doubly deficient for inducible NO synthase and gp91phox (8), we suggest that PKCδ has a further protective role in infection other than stimulating superoxide production. Moreover, the striking up-regulation of NF-IL6 in LM-infected PKCδ−/− macrophages did not affect the heightened susceptibility, suggesting that PKCδ plays a protective role in mediating NF-IL6-dependent macrophage control of LM.
Inhibition of all PKC isoforms with GF109203 further enhanced the listerial proliferation in PKCδ−/− macrophages (data not shown), suggesting that additional PKC isoforms are involved in controlling listerial proliferation in macrophages. Indeed, PKCε, which localizes to perforated LM-containing phagosomes and induces RNI production near the phagosome (38), has been shown to be important for macrophage activation and clearance of bacterial infections (39). Moreover, LM has been shown to exploit host PKCβI and PKCβII activity during infection by activating the recruitment of PKCβI and PKCβII to early endosomes. Inhibition of these PKC isoforms resulted in increased phagocytosis and decreased escape of LM from macrophage phagosome (14).
To summarize, we used a differential microarray approach to facilitate the identification of genes involved in macrophage effector functions that are independent of RNO and RNI. We identified PKCδ as a critical factor for confinement and killing of LM within macrophage phagosomes. The mechanism whereby NF-IL6, PKCδ, and PKCε mediate the confinement and killing of LM within macrophage phagosomes requires further investigation.
Materials and Methods
Bacteria and Mice.
Live and heat-killed LM (EGD strain) and attenuated LM ΔLLO stocks were prepared as described (30). LM ΔLLO mutant strain was a gift from T. Chakraborty (Institute of Medical Microbiology, University of Giessen, Giessen, Germany). NF-IL6−/− (C57BL/6 × 129/SV) mice were a gift from V. Poli (40) (Department of Genetics, Biology, and Biochemistry, University of Turin, Turin, Italy). PKCδ−/− mice were on a 129/SV genetic background (41). Mice were bred in specific pathogen-free conditions and were matched for age (6–10 weeks) and sex in all experiments.
Genotyping.
NF-IL6−/− mice were genotyped by PCR using primer 5′-TGGACAAGCTGAGCGAC-3′ and primers specific for WT (5′-GGG CTG CTT GA ACA-3′) or targeted NF-IL6 locus (5′-GCC GAT TGT CTG TTG TGC CC-3′) and the following PCR program: 94°C for 3 min; 94°C for 30 sec, 59°C for 30 sec, 72°C for 30 sec for 45 cycles; 72°C for 3 min. PKCδ−/− mice were genotyped by using primer 5′-AAC AGC TGT GAT GGG ATC GAA-3′ and primers specific for WT (5′-ACC CTT CCT GCG CAT CTC CT-3′) or targeted PKCδ locus (5′-GAG GAT CTC GTC GTG ACC CA- 3′) and the following PCR program: 94°C for 3 min; 94°C for 30 sec, 55°C for 30 sec, 72°C for 45 sec for 40 cycles; 72°C for 5 min.
Infection of BMDMs with LM.
BMDMs were generated as described (42), washed three times in DMEM containing 10% FCS without antibiotics, activated overnight with 100 units/ml IFN-γ (BD Pharmingen, San Jose, CA), and infected with LM [multiplicity of infection (MOI) 10:1], ΔLLO mutant (MOI 50:1), or HKLM (MOI 200:1)]. Gentamycin was added at 50 μg/ml at 1 h p.i. to kill all extracellular bacilli.
Bacterial Load in BMDMs and Organs.
BMDMs or organs were lysed/homogenized in 0.05% Triton X-100, and 10-fold serial dilutions were plated on tryptose–soy agar plates. The plates were incubated at 37°C for 24 h, and the cfu per organ were enumerated.
Histology.
Liver and spleen sections were stained with hematoxylin and eosin, and the number and size of 80 liver microabscesses per mouse per group were measured as described (43).
RNA Isolation.
Total RNA was isolated from organs or BMDMs by using TriReagent (Molecular Research Center, Cincinnati, OH), DNaseI-treated, and “cleaned up” by using the RNeasy RNA Extraction Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Quantitative RT-PCR.
Quantitative RT-PCR was performed as described (43) by using primers for PKCδ (forward, 5′-CGG GCT ACG TTT TAT GC-3′; reverse, 5′-TCC AAC GGG GAT AG TG-3′), IL-6 (forward, 5′-GTT CTC TGG GAA ATC GTG GA-3′; reverse, 5′-TGT ACT CCA GGT AGC TAT GG-3′), NF-IL6 (forward, 5′-CCC ATG GAA GTG GCC AAC T-3′; reverse, 5′-GCG AAG AGG TCG GAG AGG AA-3′), and β2-microglobulin (forward, 5′-TGA CCG GCT TGT ATG CTA TC-3′; reverse, 5′-CAG TGT GAG CCA GGA TAT AG-3′). Relative mRNA expression values were calculated by dividing the calculated value for the gene of interest by the β2-microglobulin value.
ELISA.
Cytokines and chemokine in culture supernatants and sera were measured by sandwich ELISA as described (44).
NO Assay.
Production of nitrite in cell culture supernatants was measured by using the Griess reaction assay as described (45).
Fluorescent Microscopy.
Measurement of escape from the primary vacuole was performed as described (36) except that BMDMs were preactivated with 100 units/ml IFN-γ for 16 h before infection with LM (MOI 30:1), and LM were labeled with rabbit anti-LM serovar 1/2a (Capricorn Products, Portland, ME) and revealed with goat anti-rabbit IgG-FITC antibody (Sigma–Aldrich, Munich, Germany).
Microarray Experiment.
BMDMs from WT or NF-IL6−/− mice were untreated or simultaneously activated with 100 units/ml IFN-γ and infected with LM (MOI 10:1). Total RNA was extracted at 4 h p.i. from four repeat infection experiments, linearly amplified, labeled, and hybridized to Mouse Exonic Evidence-Based Oligonucleotide oligo arrays (Illumina, San Diego, CA). For each infection experiment, samples from untreated and infected macrophages for the same mouse group were hybridized to the same microarray. Image and data analysis was done by using Limma (46) and TIGR MeV (47). Data were normalized by using print-tip loess and quantile normalization. DE genes were identified by using a paired t test with unequal variance, pairing the relative gene expression ratio of infected vs. untreated macrophages from WT and NF-IL6−/− mice from the same experiment. The top 10% of genes with the smallest P values were selected for further analysis and were functionally clustered. Genes belonging to all or several functional groups and for which a gene-deficient mouse model was available were selected for further analyses. A one-tailed Fisher's exact test was used to test for enrichment of CRMs containing NF-IL6 close to the candidate genes compared with CRMs close to other genes on the array (20, 48).
Statistical Analysis.
RT-PCR data were analyzed by using a paired Student t test. All other data, except from microarrays, were analyzed by using a Student t test (two-tailed with unequal variance), and P < 0.05 was considered significant. Survival data were analyzed by Kaplan–Meier using the log-rank test. Each experiment was repeated at least once to ensure reproducibility.
Acknowledgments
We thank Dr. V. Poli and Prof. T. Chakraborty for providing the NF-IL6−/− mice and LM ΔLLO mutant, respectively; the University of Cape Town Animal Unit staff, Reagon Petersen, Erica Smit, Zenaria Abbas, and Wendy Green for maintaining and genotyping the mice; Lizette Fick, Marilyn Tyler, and Zoë Lotz for histology; Dinko Basich for technical assistance; and to Dr. Kevin Dennehy for critical reading of the manuscript. This work was supported by grants from the Swiss National Foundation and the Swiss Foundation for Fellowships in Biology and Medicine (to R.G.) and grants from the National Research Foundation, the Medical Research Council, and The Wellcome Trust (to F.B.).
Abbreviations
- DE
differentially expressed
- HKLM
heat-killed LM
- ROI
reactive oxygen intermediate
- RNI
reactive nitrogen intermediate
- CRM
cis-regulatory module
- BMDM
bone marrow-derived macrophage
- MCP-1
monocyte chemoattractant protein 1
- LLO
listeriolysin O
- MOI
multiplicity of infection
- p.i.
postinfection
- LM
Listeria monocytogenes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE6256).
References
- 1.Gellin BG, Broome CV. J Am Med Assoc. 1989;261:1313–1320. [PubMed] [Google Scholar]
- 2.Brombacher F, Kopf M. Res Immunol. 1996;147:505–511. doi: 10.1016/s0923-2494(97)85215-7. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier NA, Schreiber RD. Proc Natl Acad Sci USA. 1985;82:7404–7408. doi: 10.1073/pnas.82.21.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havell EA. J Immunol. 1989;143:2894–2899. [PubMed] [Google Scholar]
- 5.Dinauer MC, Deck MB, Unanue ER. J Immunol. 1997;158:5581–5583. [PubMed] [Google Scholar]
- 6.Endres R, Luz A, Schulze H, Neubauer H, Futterer A, Holland SM, Wagner H, Pfeffer K. Immunity. 1997;7:419–432. doi: 10.1016/s1074-7613(00)80363-5. [DOI] [PubMed] [Google Scholar]
- 7.MacMicking JD, Nathan C, Hom G, Chartrain N, Fletcher DS, Trumbauer M, Stevens K, Xie QW, Sokol K, Hutchinson N, et al. Cell. 1995;81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 8.Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M, Fang F, Dinauer M, Nathan C. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 9.Pizarro-Cerda J, Desjardins M, Moreno E, Akira S, Gorvel JP. J Immunol. 1999;162:3519–3526. [PubMed] [Google Scholar]
- 10.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, et al. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara I, Mizuno S, Yamada H, Matsumoto M, Akira S. Am J Pathol. 2001;158:361–366. doi: 10.1016/S0002-9440(10)63977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg SF. Biochem J. 2004;384:449–459. doi: 10.1042/BJ20040704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wadsworth SJ, Goldfine H. Infect Immun. 2002;70:4650–4660. doi: 10.1128/IAI.70.8.4650-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson DN, Foster DA. FASEB J. 2004;18:627–636. doi: 10.1096/fj.03-0979rev. [DOI] [PubMed] [Google Scholar]
- 16.Trautwein C, Caelles C, van der Geer P, Hunter T, Karin M, Chojkier M. Nature. 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- 17.Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. J Clin Invest. 1994;93:2554–2561. doi: 10.1172/JCI117266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto M, Tanaka T, Kaisho T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Akira S. J Immunol. 1999;163:5039–5048. [PubMed] [Google Scholar]
- 19.Weihua X, Hu J, Roy SK, Mannino SB, Kalvakolanu DV. Biochim Biophys Acta. 2000;1492:163–171. doi: 10.1016/s0167-4781(00)00111-1. [DOI] [PubMed] [Google Scholar]
- 20.Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, et al. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bey EA, Xu B, Bhattacharjee A, Oldfield CM, Zhao X, Li Q, Subbulakshmi V, Feldman GM, Wientjes FB, Cathcart MK. J Immunol. 2004;173:5730–5738. doi: 10.4049/jimmunol.173.9.5730. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Xu B, Bhattacharjee A, Oldfield CM, Wientjes FB, Feldman GM, Cathcart MK. J Leukocyte Biol. 2005;77:414–420. doi: 10.1189/jlb.0504284. [DOI] [PubMed] [Google Scholar]
- 23.Page K, Li J, Zhou L, Iasvovskaia S, Corbit KC, Soh JW, Weinstein IB, Brasier AR, Lin A, Hershenson MB. J Immunol. 2003;170:5681–5689. doi: 10.4049/jimmunol.170.11.5681. [DOI] [PubMed] [Google Scholar]
- 24.Kilpatrick LE, Lee JY, Haines KM, Campbell DE, Sullivan KE, Korchak HM. Am J Physiol. 2002;283:C48–C57. doi: 10.1152/ajpcell.00385.2001. [DOI] [PubMed] [Google Scholar]
- 25.Woo CH, Lim JH, Kim JH. Am J Physiol. 2005;288:L307–L316. doi: 10.1152/ajplung.00105.2004. [DOI] [PubMed] [Google Scholar]
- 26.Novotny-Diermayr V, Zhang T, Gu L, Cao X. J Biol Chem. 2002;277:49134–49142. doi: 10.1074/jbc.M206727200. [DOI] [PubMed] [Google Scholar]
- 27.Xu B, Bhattacharjee A, Roy B, Feldman GM, Cathcart MK. J Biol Chem. 2004;279:15954–15960. doi: 10.1074/jbc.M400413200. [DOI] [PubMed] [Google Scholar]
- 28.Deb DK, Sassano A, Lekmine F, Majchrzak B, Verma A, Kambhampati S, Uddin S, Rahman A, Fish EN, Platanias LC. J Immunol. 2003;171:267–273. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, et al. Nature. 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 30.Dai WJ, Bartens W, Kohler G, Hufnagel M, Kopf M, Brombacher F. J Immunol. 1997;158:5297–5304. [PubMed] [Google Scholar]
- 31.Fehr T, Schoedon G, Odermatt B, Holtschke T, Schneemann M, Bachmann MF, Mak TW, Horak I, Zinkernagel RM. J Exp Med. 1997;185:921–931. doi: 10.1084/jem.185.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weih F, Warr G, Yang H, Bravo R. J Immunol. 1997;158:5211–5218. [PubMed] [Google Scholar]
- 33.Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, Sher A, Taylor GA. J Exp Med. 2001;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fronhofer V, Lennartz MR, Loegering DJ. J Leukocyte Biol. 2006;79:408–415. doi: 10.1189/jlb.0805438. [DOI] [PubMed] [Google Scholar]
- 35.Goldfine H, Wadsworth SJ, Johnston NC. Infect Immun. 2000;68:5735–5741. doi: 10.1128/iai.68.10.5735-5741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wadsworth SJ, Goldfine H. Infect Immun. 1999;67:1770–1778. doi: 10.1128/iai.67.4.1770-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tapia JA, Jensen RT, Garcia-Marin LJ. Biochim Biophys Acta. 2006;1763:25–38. doi: 10.1016/j.bbamcr.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Shaughnessy LM, Lipp P, Lee KD, Swanson JA. Cell Microbiol. 2007;9:1695–1704. doi: 10.1111/j.1462-5822.2007.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castrillo A, Pennington DJ, Otto F, Parker PJ, Owen MJ, Bosca L. J Exp Med. 2001;194:1231–1242. doi: 10.1084/jem.194.9.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. J Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 41.Leitges M, Mayr M, Braun U, Mayr U, Li C, Pfister G, Ghaffari-Tabrizi N, Baier G, Hu Y, Xu Q. J Clin Invest. 2001;108:1505–1512. doi: 10.1172/JCI12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holscher C, Arendse B, Schwegmann A, Myburgh E, Brombacher F. J Immunol. 2006;176:1115–1121. doi: 10.4049/jimmunol.176.2.1115. [DOI] [PubMed] [Google Scholar]
- 43.Herbert DR, Holscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, Kirsch R, Hall P, Mossmann H, et al. Immunity. 2004;20:623–635. doi: 10.1016/s1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- 44.Mohrs M, Ledermann B, Kohler G, Dorfmuller A, Gessner A, Brombacher F. J Immunol. 1999;162:7302–7308. [PubMed] [Google Scholar]
- 45.Nickdel MB, Roberts F, Brombacher F, Alexander J, Roberts CW. Infect Immun. 2001;69:1044–1052. doi: 10.1128/IAI.69.2.1044-1052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wettenhall JM, Smyth GK. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 47.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 48.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]