Abstract
In all land plants, cellulose is synthesized from hexameric plasma membrane complexes. Indirect evidence suggests that in vascular plants the complexes involved in primary wall synthesis contain three distinct cellulose synthase catalytic subunits (CESAs). In this study, we show that CESA3 and CESA6 fused to GFP are expressed in the same cells and at the same time in the hypocotyl of etiolated seedlings and migrate with comparable velocities along linear trajectories at the cell surface. We also show that CESA3 and CESA6 can be coimmunoprecipitated from detergent-solubilized extracts, their protein levels decrease in mutants for either CESA3, CESA6, or CESA1 and CESA3, CESA6 and also CESA1 can physically interact in vivo as shown by bimolecular fluorescence complementation. We also demonstrate that CESA6-related CESA5 and CESA2 are partially, but not completely, redundant with CESA6 and most likely compete with CESA6 for the same position in the cellulose synthesis complex. Using promoter-β-glucuronidase fusions we show that CESA5, CESA6, and CESA2 have distinct overlapping expression patterns in hypocotyl and root corresponding to different stages of cellular development. Together, these data provide evidence for the existence of binding sites for three distinct CESA subunits in primary wall cellulose synthase complexes, with two positions being invariably occupied by CESA1 and CESA3, whereas at least three isoforms compete for the third position. Participation of the latter three isoforms might fine-tune the CESA complexes for the deposition of microfibrils at distinct cellular growth stages.
Cellulose microfibrils are synthesized from a multiprotein complex inserted into the plasma membrane. These “rosette” complexes consist of six globules, each of which contains multiple cellulose synthase catalytic subunits (CESAs). These complexes migrate in the plasma membrane along microtubules, propelled by the polymerization of the β-1,4-glucan chains (1).
Plant CESA genes are members of multigene families. Arabidopsis has 10 CESA isoforms that, based on sequence comparison with other plant species, can be classified into six orthologous groups (2). Mutational analysis shows that these six groups of isoforms have nonredundant functions in cellulose synthesis. Mutants for three isoforms (CESA4, CESA7, and CESA8) show defects in cellulose synthesis specifically in secondary walls (3–5). Microarray data show that the mRNAs for the three genes are coregulated (6, 7). The three proteins are expressed in the same cell types during secondary cell wall deposition, and co-immunoprecipitation (IP) experiments show that all three proteins interact (3). Although the interactions remain to be validated in vivo, these data strongly suggest that at least in these cells the complexes contain three isoforms. Mutants for isoforms CESA1, CESA3, and CESA6 have cellulose defects in primary cell walls (8–11). The three genes are also coregulated at the mRNA level (12). It is not known, however, whether the corresponding proteins are expressed in the same cells at the same time. It is also not known whether the three CESAs are present in different populations of complexes or are part of the same complex. Interestingly, a class of mutations in the C terminus of either CESA3 (cesa3ixr1) or CESA6 (cesa6ixr2) (isoxaben-resistant) confers increased resistance to the cellulose inhibitor isoxaben (13, 14). The simplest explanation for the existence of two nonredundant resistance loci is that isoxaben recognizes an epitope associated with the CESA3- and CESA6-containing complex. Null mutants for CESA1 and CESA3 are gametophytic lethal (39), indicating the essential nature of the genes. In contrast, CESA6 null mutants show a relatively mild phenotype, which might be explained by the existence of CESA2, CESA5, and CESA9, which are closely related to CESA6 (12). Partial redundancy between CESA6-related isoforms also might explain the lower isoxaben resistance conferred by cesa6ixr2 compared with cesa3ixr1. In this study, we initially focused on the isoxaben targets CESA3 and CESA6. We raised specific antibodies against both proteins and constructed functional GFP fusions. We show that CESA3 and CESA6 are expressed in the same cells at the same time and migrate with comparable velocities along linear trajectories at the cell surface during early dark-grown seedling development. The levels of both proteins decrease in mutants for either CESA1, CESA3, or CESA6. Using co-IP experiments, we further show that at least CESA3 and CESA6 interact with each other, and using bimolecular fluorescence complementation (BiFC) experiments, we show that CESA1, CESA3, and CESA6 can interact in vivo. Through the analysis of single, double, and triple mutants, we also show that CESA5 and CESA2 are partially redundant with CESA6 and most likely compete with CESA6 for the same binding site in the complex. Finally, we show that CESA6-related isoforms display distinct expression patterns during cellular differentiation.
Results
Simultaneous Accumulation and Similar Subcellular Localization of CESA3 and CESA6 in Developing Dark-Grown Seedlings.
The existence of two nonredundant isoxaben-resistant loci suggested that CESA3 and CESA6 are part of the same complex. To investigate this idea, we raised isoform-specific antibodies against the N-terminal predicted cytoplasmic domain of each protein. The antibodies were enriched by using a two-step immunopurification procedure, and their specificity was shown on immunoblots with the corresponding N-terminal fragments of CESA3, CESA6, and CESA1 produced in Escherichia coli (see Methods). The absence of cross-reactivity was also shown in the co-IP experiments described below. It is not excluded, however, that anti-CESA6 antibodies cross-reacted with more closely related isoforms CESA2, CESA5, or CESA9.
CESA3 and CESA6 were undetectable in imbibed seeds and appeared after germination [supporting information (SI) Fig. 5]. The immunostained bands showed a size of ≈120 kDa, which is the predicted molecular mass for the CESA proteins. The amount of both proteins increased simultaneously in growing seedlings to reach a maximum at 96 h postimbibition, suggesting a coregulation at the protein level in developing seedlings.
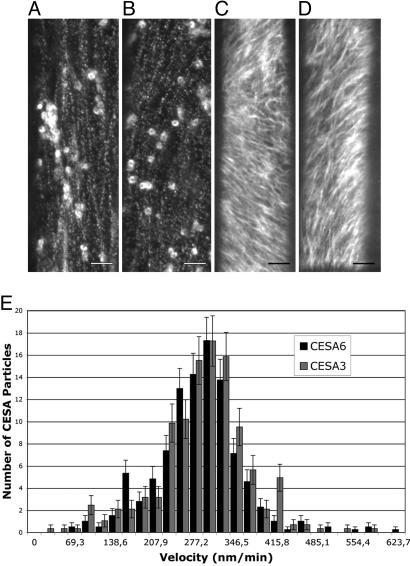
We next investigated the cellular and subcellular localization of CESA3 and CESA6. GFP-CESA3 and GFP-CESA6 were expressed from their own promoters in the respective cesa3je5 and cesa6prc1–1 mutant backgrounds. These constructs were biologically functional as shown by the complementation of the short hypocotyl phenotype of the mutants (SI Fig. 6). Both GFP-CESA3 and GFP-CESA6 could be observed in epidermal and cortical cells of 3-day-old dark-grown hypocotyls (Fig. 1 A–D). In addition, GFP-CESA3 and GFP-CESA6 fluorescence showed a similar subcellular distribution. Both proteins accumulated in similar intracellular compartments and formed rows of fluorescent particles at the cell surface as described for yellow fluorescent protein (YFP)-CESA6 (1). Among these compartments one could recognize 1.3-μm doughnut-shaped compartments as described for YFP-CESA6 (1) and smaller highly motile compartments. Time-lapse spinning disk microscopy showed bidirectional migration of surface particles for both fusion proteins. The average velocity of the particles in elongating epidermal cells was 272 nm/min (range: 53 to 622 nm/min) and 277 nm/min (range: 40 to 566 nm/min) for GFP-CESA6 and GFP-CESA3, respectively (Fig. 1E). These velocities are similar to those reported for YFP-CESA6 (1, 15).
Fig. 1.
Visualization of similar distribution and dynamics of GFP-CESA3 and GFP-CESA6. (A–D) Functional GFP-CESA3 and GFP-CESA6 fusion proteins expressed, respectively, in mutant backgrounds cesa3je5 and cesa6prc1–1 label similar intracellular compartments and complexes that migrate with comparable velocities at the cell surface. Spinning disk microscope images (A and B) and projection of a time series (10 min, 30 s, C; 10 min, D) of epidermal cells at the top of a dark-grown hypocotyl expressing GFP-CESA3 (A and C) or GFP-CESA6 (B and D). (Scale bars: 4 μm.) (E) Distribution of the velocities of the surface particles.
CESA3 and CESA6 Protein Levels Are Reduced in cesa1, cesa3, and cesa6 Mutants.
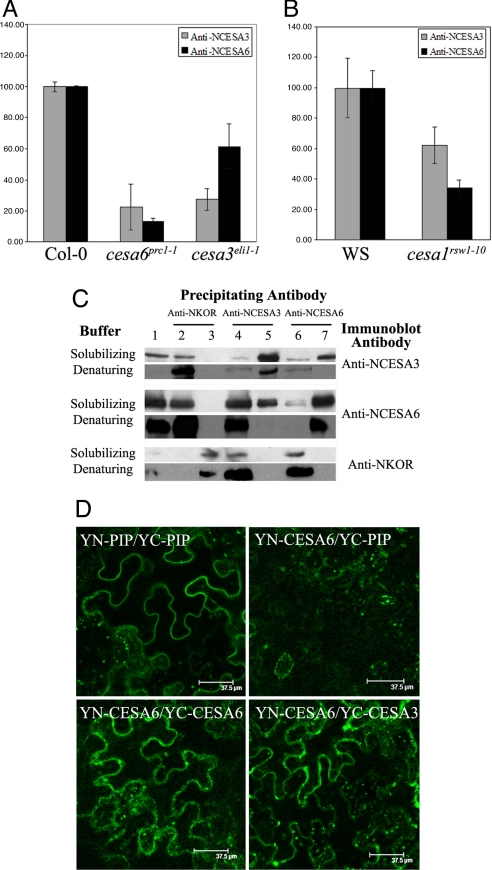
If distinct CESA isoforms are present in the same complex, one can expect that reducing the protein levels of one isoform will also affect the levels of the other isoforms. To investigate this notion, we quantified the relative abundance of CESA3 and CESA6 in 4-day-old dark-grown seedlings in cesa1, cesa3, and cesa6 mutants. cesa6prc1–1 is a null allele (10), whereas cesa3eli1–1 (11) and cesa1rsw1–10 (16) are both leaky alleles. All three mutants show a short hypocotyl and are deficient for cellulose in their primary cell walls (SI Fig. 7) (17). CESA3 and CESA6 proteins accumulated to similar levels either in Col-0 or WS backgrounds (Fig. 2 A and B). As expected, in cesa6prc1–1 the anti-CESA6 antibody detected only a few percent of the wild-type levels (Fig. 2A). The residual signal may be related to cross-reaction with related CESA subunits, such as CESA2 or CESA5. Interestingly, CESA3 protein levels were also significantly reduced in this mutant. In a similar way, in cesa3eli1–1 not only the level of CESA3 was reduced but also that of CESA6 (Fig. 2A). The less severe reduction of CESA6 compared with CESA3 in this mutant may be related to the altered properties of the remaining mutant CESA3 protein. Because we did not dispose of specific antibodies for CESA1, we could not analyze directly the CESA1 protein levels in the three mutants. However, in cesa1rsw1–10 we observed that levels of CESA3 and CESA6 were also diminished (Fig. 2B). Together, these data show that the normal accumulation of each of the CESA3 and CESA6 proteins requires the presence of normal levels of both CESA1, CESA3, and CESA6, which is consistent with the presence of these three proteins in the same complex.
Fig. 2.
CESA3 and CESA6 are in the same cellulose synthase complex. (A and B) CESA3 and CESA6 protein levels, measured in dark-grown seedlings by quantitative immunoblotting, are reduced in mutants cesa6prc1–1, cesa3eli1–1 (A) and cesa1rsw1–10 (B). Protein levels are expressed as percentage of wild-type levels, and error bars are SDs of three biological repeats. (C) CESA3 coimmunoprecipitates with CESA6 and vice versa, but not with membrane-bound cellulase KOR1. IP was carried out in nondenaturing or denaturing conditions. Shown are total Triton X-100-solubilized extracts (lane 1), IP with anti-NKOR, unbound (lane 2) and bound (lane 3) fractions; with anti-NCESA3, unbound (lane 4) and bound (lane 5) fractions or with anti-NCESA6, unbound (lane 6) and bound (lane 7) fractions. Immunoblotting was carried out with anti-NCESA3 anti-NCESA6 or anti-NKOR antibodies. (D) BiFC in N. benthamiana leaf epidermal cells shows in vivo CESA6 homodimer and CESA3/CESA6 heterodimer formation. (Upper Left) YN-PIP/YC-PIP-positive control. (Upper Right) YN-CESA6/YC-PIP. (Lower Left) YN-CESA6/YC-CESA6. (Lower Right) YN-CESA6/YC-CESA3. Other CESA combinations are shown in SI Fig. 8.
CESA3 Coimmunoprecipitates with CESA6 but Not with KOR1.
To investigate whether CESA3 and CESA6 physically interact, we carried out co-IP experiments on detergent-solubilized protein extracts from 4-day-old dark-grown seedlings (Fig. 2C). As expected, anti-CESA3 antibodies precipitated CESA3 both in nondenaturing and denaturing conditions (Fig. 2C, lane 5). Interestingly, CESA6 also coimmunoprecipitated with CESA3 in nondenaturing conditions, whereas no CESA6 signal was observed in denaturing conditions. The latter observation also confirms the absence of cross-reactivity of anti-CESA6 with CESA3. Similar results were obtained with the reciprocal experiment in which anti-CESA6 antibodies were used for the co-IP. Again, CESA6 precipitated in both nondenaturing and denaturing conditions, whereas CESA3 only coprecipitated in nondenaturing conditions. These results show that a stable interaction existed between CESA3 and CESA6 in Triton X-100-solubilized extracts and that this interaction was disrupted in denaturing buffer. None of the CESAs were immunodetected in the absence of the primary antibody (CESA3 or CESA6) (data not shown). Using the anti-CESA6 antibody, we observed a second band on the immunoblot. This band may correspond to a truncated CESA6 isoform or a CESA6-related isoform. We also investigated the interaction of CESA3 and CESA6 with the membrane-bound cellulase KOR1, which is also required for normal cellulose synthesis (18). KOR1 protein was detected only in the unbound fraction and not in the immunoprecipitates with anti-CESA3 or CESA6 antibodies. These results indicate that KOR1 did not stably interact with CESA3 nor with CESA6 in detergent-solubilized extracts and underscore the specificity of the observed CESA3–CESA6 interaction.
Visualization of CESA Homodimers and Heterodimers in Planta Using BiFC.
To confirm the co-IP results, we studied the capacity of CESA isoforms to interact in vivo by using the BiFC technique (19). The N-terminal and the C-terminal fragments of YFP were both fused to the N terminus of the coding sequences of CESA1, CESA3, and CESA6. As shown in SI Fig. 6, GFP fusions at this position do not interfere with the functionality of at least CESA3 and CESA6. Combinations of chimeric genes expressed from the cauliflower mosaic virus 35S promoter were transiently expressed in Nicotiana benthamiana leaves using Agrobacterium infiltration. As a positive control, we used the aquaporin PIP2–1 (20). Aquaporins are known to form homotetramers in the plasma membrane (21). As expected, coexpression of YN-PIP2-1 and YC-PIP2-1 yielded a YFP signal (Fig. 2D), whereas no signal was detected by expression of either YN-PIP2-1 or YC-PIP2–1 alone (data not shown). The fluorescence emission spectrum of the signal indeed confirmed that the signal was derived from a functional YFP (SI Fig. 8B). We next investigated whether the fusion proteins derived from the three CESA proteins could homodimerize. As shown in Fig. 2D and SI Fig. 8, a fluorescent signal was observed for YN-CESA3/YC-CESA3 and YN-CESA6/YC-CESA6 combinations. Fluorescent signals were observed both at the cell surface and in intracellular compartments, similar to that of CESA3 or CESA6 fused to intact YFP (SI Fig. 8C). For YN-CESA1/YC-CESA3 and YN-CESA3/YC-CESA3, the signal was always weaker, suggesting that this protein homodimerizes less efficiently (SI Fig. 8E). We also analyzed the YN-CESA1/YC-CESA1 combination, which also reconstituted the YFP fluorescence (SI Fig. 8D). For all three CESA fusions, no fluorescent signal was detected by expression of either YN-CESA or YC-CESA alone (data not shown).
To investigate whether heterodimers could also be formed, we coexpressed YN-CESA3/YC-CESA1, YN-CESA1/YC-CESA3, YN-CESA6/YC-CESA1, YN-CESA1/YC-CESA6, YN-CESA3/YC-CESA6, or YN-CESA6/YC-CESA3 (SI Fig. 8 and Fig. 2D). We observed that YFP fluorescence was reconstituted for all of the combinations, suggesting that all three isoforms can form a complex. As a negative control, we coexpressed PIP2-1 chimerae with corresponding CESA constructs (Fig. 2D). No signal was detected with combinations of either YN-PIP2-1 or YC-PIP2-1 for all of the CESA fusions tested, suggesting that PIP2-1 and CESA proteins were unable to interact despite the presence of both proteins in the plasma membrane. In conclusion, all three isoforms showed interactions in vivo, confirming the co-IP results for CESA3 and CESA6.
Partial Redundancy Among CESA6-Related CESA Isoforms.
The mild phenotype of CESA6 null alleles as compared with strong mutant alleles of CESA1 or CESA3 and the lower resistance to isoxaben of cesa6ixr2–1 mutant than cesa3ixr1–1 or cesa3ixr1–2 (13, 14) might be caused by partial redundancy with other CESA isoforms.
CESA2, CESA5, and CESA9 are closely related to CESA6. CESA9 is only expressed during embryogenesis, as shown by public microarray data and promoter-β-glucuronidase (GUS) fusions (data not shown), and was not considered in this study. We first studied single T-DNA insertion mutants for CESA2 or CESA5. Mutant light-grown seedlings and adult plants were indistinguishable from the wild type (Fig. 3 A and C). However, the dark-grown hypocotyl of cesa2 was slightly shorter than that of the wild type (Fig. 3B and Table 1). The analysis of the cell wall of dark-grown mutant hypocotyls with FTIR microspectroscopy failed to detect differences with the wild type as shown by the tight clustering of the mutants with wild-type controls (SI Fig. 7). The FTIR spectra of double cesa2/cesa5 mutants also clustered with those of the wild type. Interestingly, compared with the single cesa2 mutant, the dark-grown hypocotyl of the double mutant was slightly longer. In addition, roots of light-grown cesa2/cesa5 seedlings were slightly longer, and adult plants were slightly larger than the wild type (Fig. 3 A and C). In contrast, mutant combinations with cesa6 showed a very different picture. cesa2/cesa6 double mutants showed an enhanced phenotype compared with cesa6 as shown by the reduced hypocotyl length in the dark, the reduced root length in the light, and the strongly dwarfed and bushy adult cesa2/cesa6 plants (Fig. 3). cesa5/cesa6 double homozygotes were seedling lethal. Interestingly, the cesa5 allele showed a strong dosage effect in a homozygous cesa6 background as shown by the 1:2:1 segregation of the dark-grown hypocotyl into three size classes within the F2 population of a cesa6−/−;cesa5+/− heterozygote (SI Fig. 9). To construct the triple mutant, we crossed cesa2−/− cesa5−/− with cesa2−/− cesa6−/−. Among the cesa2−/− cesa6−/− F2 seedlings only the cesa5+/+ seedlings were viable. The cesa5+/− or cesa5−/− seedlings either bleached and died or survived as very small sterile plants (data not shown).
Fig. 3.
Partial redundancy between CESA6-related CESAs. Five-day-old light-grown (A), dark-grown seedlings (B), and 32-day-old greenhouse-grown plants (C) of (from left to right) Col0, cesa2, cesa5, cesa2/cesa5, cesa6prc1–1, cesa2/cesa6prc1–1, and cesa5/cesa6prc1–1 are shown.
Table 1.
Hypocotyl lengths of 5-day-old dark-grown seedlings
| Type | Mean | SD | n | Student's t test* |
|---|---|---|---|---|
| Col-0 | 16.26 | 1.19 | 67 | |
| cesa2 | 14.24 | 1.21 | 57 | 9.33† |
| cesa5 | 15.39 | 1.06 | 70 | 1.67 |
| cesa6 | 3.41 | 0.34 | 69 | 85.6† |
| cesa2/cesa5 | 16.95 | 1.34 | 69 | 3.18† |
| cesa2/cesa6 | 2.12 | 0.18 | 71 | 96† |
*All comparisons were made to Col-0.
†Significantly different; P = 0.01.
CESA6-Related CESAs Are Targets for Isoxaben.
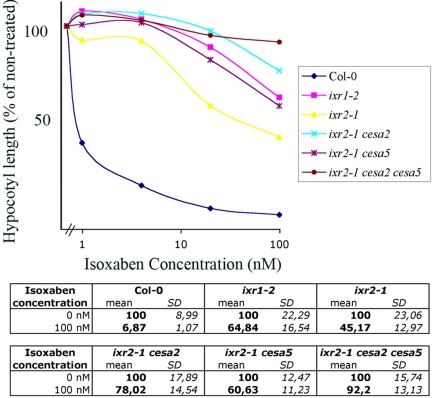
We next investigated whether the lower isoxaben resistance of cesa6ixr2–1 compared with cesa3ixr1–2 was caused by the presence of CESA6-related proteins in the cellulose synthase complexes of cesa6ixr2–1 plants. To this end, we quantified the resistance to isoxaben of all double and triple mutant combinations of cesa2 and cesa5 with cesa6ixr2–1 (Fig. 4) by measuring the hypocotyl length of 5-day-old dark-grown seedlings on increasing concentrations of isoxaben. As observed previously, cesa6ixr2–1 was ≈50-fold more resistant to isoxaben than the wild type, and cesa3ixr1–2 was 5-fold more resistant than cesa6ixr2–1. Interestingly, adding cesa5 increased the resistance of cesa6ixr2–1 to that of cesa3ixr1–2, adding cesa2 caused a >10-fold increase in resistance, and adding both cesa2 and cesa5 rendered the seedling insensitive to 100 nM isoxaben. The increased resistance was not caused by increased CESA3 or CESA6 protein levels in the mutants (data not shown). Together, these data show that all three CESA6-related isoforms are targets for isoxaben, suggesting that they compete for the same binding site within the cellulose synthase complex.
Fig. 4.
CESA6, CESA2, and CESA5 are targets for isoxaben. (Upper) The effect of isoxaben on hypocotyl length: dose–response curve. Hypocotyl length is expressed in percentage of the untreated control. Average values of ≈35 individuals are shown. (Lower) For clarity, SDs of untreated and 100 nM isoxaben-treated seedlings are shown.
Distinct Expression Patterns of CESA6-Related Isoforms During Cellular Differentiation.
Using promoter-GUS fusions we studied the expression patterns of CESA6, CESA2, and CESA5 genes in dark-grown hypocotyls and roots of 72-h-old seedlings (SI Fig. 10A). At this stage, different phases of cell expansion are represented from the top to the base of the hypocotyl or from the tip to the base of the root. Promoter CESA5-GUS was weakly expressed only in the unexpanded cells in the apical hook of the hypocotyl. Promoter CESA6-GUS expression occurred throughout the hypocotyl and root, peaking in the growth acceleration zone just below the apical hook and in the cell elongation zone in the root. Promoter CESA2-GUS expression largely overlapped with that of promoter CESA6-GUS, but in the root, peaked in more mature cells. The GUS expression patterns in the root were consistent with the microarray data for CESA2, CESA5, and CESA6 on flow-sorted root protoplasts (22). In conclusion, the three partially redundant CESA genes mark distinct cellular growth stages in hypocotyl and root.
CESA5 Is Only Partially Redundant with CESA6.
We showed that CESA5 is partially redundant with CESA6 and most likely competes for the same site in the complex. The incomplete redundancy can be caused by the partially overlapping expression patterns or intrinsic differences in the protein structure and function. To investigate this, we expressed CESA5 from the CESA6 promoter in a cesa6prc1–1 null background. Three independent transformants showed incomplete restoration of the hypocotyl growth defect (SI Fig. 10B). On the contrary, expressing CESA6 from the same promoter sequence completely restored the wild-type phenotype to cesa6prc1–1 (data not shown). The results show that CESA5 cannot entirely substitute for CESA6, suggesting that both isoforms have specialized roles in the deposition of cellulose in expanding cells.
Discussion
Several lines of evidence presented in this article show that CESA3, CESA6, and most likely CESA1, are in the same protein complex in dark-grown seedlings. This idea was already suggested by the coexpression of transcripts for CESA1, CESA3, and CESA6 (12, 23), the existence of isoxaben-resistant alleles for both CESA3 and CESA6 (13, 14), and the observation that the complexes involved in secondary wall synthesis also contain three nonredundant CESA isoforms (3). Here, we showed that CESA3 and CESA6 proteins follow the same accumulation kinetics in developing seedlings, functional GFP-fusion proteins are expressed in the same cells and occupy similar subcellular compartments, and the GFP-fusion proteins migrate at the cell surface with comparable velocities. Although we did not observe CESA1 directly, the coexpression at the mRNA level strongly suggests that CESA1 is also expressed in the same cells. We further showed that mutations in either CESA1, CESA3, or CESA6 cause a decrease in protein levels of CESA3 and CESA6, which is compatible with the idea that the assembly of the complex is prevented in the absence of one component, and that CESA3 and CESA6 coimmunoprecipitate with each other but not with KOR1. It remains possible however, that the antibodies coimmunoprecipitated not only the CESA complex but also detergent-resistant membrane (DRM) fractions that contain other membrane proteins. This is unlikely the case because KOR1, which also has been detected in DRMs (24), did not coimmunoprecipitate with CESA3 or CESA6. Similar observations have been reported for the secondary cell wall CESAs (3, 25). Finally, using BiFC we show that CESA3, CESA6, and CESA1 can interact in vivo.
One might question the validity of the BiFC technique for the study of the interaction of membrane proteins because false positives might arise as a result of proximity of noninteracting proteins confined within membranes. However, highly specific interactions between membrane proteins have been demonstrated using this technique in other systems (26–28). Also, the absence of an interaction with the plasma membrane protein PIP2-1 makes this possibility unlikely. Although we tested only pairwise combinations of CESA isoforms, a positive signal does not exclude indirect interactions, perhaps through the formation of complexes with endogenous tobacco proteins. Collectively, these results provide strong evidence for the presence of three CESA proteins in the same complex.
The strong phenotypes of cesa1 and cesa3 mutants demonstrate the absence of redundancy with other CESAs. We show in this article that the relatively mild phenotype of cesa6 instead reflects partial redundancy with two related isoforms, CESA2 and CESA5, which presumably can replace CESA6 in the complex. We also show that all three CESA6-related isoforms are isoxaben targets, which explains the lower resistance of cesa6ixr2–1 to isoxaben compared with cesa3ixr1, because CESA3 does not compete with other isoforms for its position in the complex.
The different expression patterns of CESA5, CESA6, and CESA2 during hypocotyl and root growth suggests that the composition of the cellulose synthase complex changes at different stages of cellular development. The partial complementation of the cesa6prc1–1 phenotype, through the expression of CESA5 under control of the CESA6 promoter, also shows that the isoforms are functionally specialized. Similar observations have been made for CESA2 and CESA6 (39). We previously showed that distinct stages of cellular development can be distinguished from the top to the base of the dark-grown hypocotyl (29). It is conceivable that properties of the cellulose synthase complexes change to cope with varying constraints associated with cellulose deposition at successive growth stages. It will be interesting to see to what extent the three CESA6-related isoforms confer distinct properties to the terminal complexes, such as the velocity and the density of the complexes, the orientation of the trajectories, interaction with the cytoskeleton, and coupling to cell elongation.
Methods
Plant Material, in Vitro Growth Conditions, and Genetic Analysis.
Arabidopsis wild-type Col-0 and mutant lines were grown as described (30) at 20°C without sucrose. See SI Table 2 for mutants and T-DNA insertion lines used in this study. Plants were grown in the dark as described (29) or in a 16-h light and 8-h dark cycle. Homozygous lines for cesa2 and cesa5 were screened for the simultaneous presence of a PCR product amplified from the left T-DNA border and absence of a PCR product from the genomic sequence using primers flanking the T-DNA insertion (SI Table 3). Double and triple cesa mutants were genotyped by PCR. Promoter-GUS lines for CESA2, CESA5, and CESA6 (described in ref. 31) were kindly provided by M. Doblin (University of Melbourne, Melbourne, Australia). The length of hypocotyls fixed with 0.2% formaldehyde were measured by using Optimas 4.1 software as described (29).
Production of Polyclonal Antibodies Against CESA3 and CESA6.
The N-terminal regions upstream of the first transmembrane domains of CESA3 and CESA6 were amplified (primers in SI Table 3) and cloned into the EcoR1 site of pGEX3X and pGEX2T (Amersham Biosciences, Piscataway, NJ), respectively. GST-fusion proteins were purified on GHS-Sepharose 4B resin according to the manufacturer's recommendations (Amersham Biosciences). Rabbit antibodies against the GST-fusion polypeptides were produced by BioGenes (Berlin, Germany). GST-fusion proteins were coupled to CNBr-activated Sepharose 4F according to the supplier's protocol (Amersham Biosciences). The serum was incubated with the antigen-Sepharose matrix, and the unbound fraction was extensively washed from the resin according to the manufacturer's recommendations. Antibodies were eluted with 0.2 M glycine (pH 2), 0.15 M NaCl and neutralized with 1.5 M Tris·HCl, pH 8.8. To further improve the specificity of the sera, the pooled fractions eluted from the CESA3-Sepharose resin were loaded on the CESA6-Sepharose resin, and the unbound fraction on this matrix was used as purified CESA3 antibody. The antibodies raised against CESA6 were prepared in the same manner. The specificity of the CESA3 and CESA6 antibodies was verified by immunoblotting against the GST-fusion proteins (SI Fig. 11A).
Quantification of Protein Levels.
Samples corresponding to 50 dark-grown seedlings were collected at different times and ground in 50 μl of extraction buffer (4 M urea and 100 mM DTT). Twenty-five microliters of modified Laemmli buffer (without bromophenol blue) was added, and the samples were boiled for 5 min and centrifuged at 10,000 × g for 10 min at room temperature. Protein content of the supernatant was quantified by using the RC DC Protein Assay method (Bio-Rad, Hercules, CA). Aliquots of 20 μg protein were then analyzed by 8% SDS/PAGE (32) and immunoblotted according to standard protocols. The purified anti-CESA sera were used in 1:1,000 dilution, and the signals were measured by fluorescent detection of AlexaFluor 488 goat anti-rabbit (used at 1/3,000; Molecular Probes–Invitrogen, Carlsbad, CA) and quantified with ImageGauge software (Fuji, Tokyo, Japan). Fifty Arabidopsis seedlings grown in liquid medium (30) for 48 h in the dark were labeled with a mixture of 35S-labeled methionine and [35S]cysteine (0.2 MBq; PerkinElmer Life Science, Albany, NY) for 4 h. Seedlings were then extensively washed and used for IP. Radiolabeled proteins were separated by 8% SDS/PAGE (32) and visualized by phosphorimaging (Fuji FLA 5000).
Co-IP Experiments.
Two grams of 3-day-old dark-grown seedlings was ground in liquid nitrogen, and the powder was transferred to either 6 ml of solubilization buffer [50 mM Tris·HCl, pH 8/150 mM NaCl/2% Triton X-100, containing 1% (wt/vol) polyvinylpyrrolidone (Polyclar AT), plant protease inhibitor mixture and phosphatase inhibitor mixture 1 and 2 (Sigma, St. Louis, MO) used, respectively, at a 100- and 200-fold dilution] or denaturing buffer [50 mM Tris·HCl, pH 8/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS, containing 1% (wt/vol) polyvinylpyrrolidone (Polyclar AT) and the mixture of inhibitors]. A cleared cell lysate was obtained by centrifugation at 8,000 × g for 10 min and filtration of the supernatant through Miracloth. An aliquot of 2 mg of protein was supplemented with purified polyclonal antisera at 1:50 dilutions, and IPs were carried out essentially according to established protocols (33). Immunoprecipitated proteins bound on Protein A Sepharose CL-4B beads (Amersham Biosciences) were boiled in 2× Laemmli buffer, and proteins were analyzed by 8% SDS/PAGE (32) and immunoblotted using the ECL Luminescent Detection System (Amersham Biosciences). Autoradiography of the proteins precipitated in denaturing conditions with either anti-CESA6 or anti-KOR1 out of extracts from 35S-labeled seedlings confirmed the specificity of the respective antibodies for proteins of the expected molecular masses (respectively, 120 and 72 kDa; SI Fig. 11B) also when used in IP experiments.
Plant Expression Vectors.
For plant expression vectors see SI Text.
Transient Expression in N. benthamiana.
Leaves of 3-week-old plants were transformed by infiltration as described (34). YFP fluorescence was detected 3 days after infiltration by using the 514-nm laser line of a SP2 AOBS confocal laser scanning microscope (Leica, Solms, Germany,) equiped with an argon laser. To check the YFP reconstitution, spectral analysis was performed with the 496-nm laser line. For BiFC experiments, all constructs fluorescence was detected at the same photo-multiplier tube (PMT) settings (760), except for the negative interactions for which the PMT was increased up to 880.
Imaging of GFP-CESA and Quantification of Velocity.
Seedlings expressing GFP-CESA3 and GFP-CESA6 were cultured as described (35). Three-day-old etiolated seedlings were analyzed on an Axiovert 200M confocal microscope (Zeiss, Thornwood, NY) equipped with a Yokogawa CSU22 spinning disk, Zeiss ×100/1.4 N.A. oil objective, and Andor EMCCD DV885 camera (Plateforme d'Imagerie Dynamique, Institut Pasteur, Paris). GFP was excited at 488 nm by a diode pumped solid-state laser, and fluorescence emission was collected through a 505/555-nm band-pass filter (Semrock, Rochester, NY). GFP-CESA velocities were quantified by using the manual tracking plugin (F. Cordelières) in ImageJ (W. Rasband, National Institutes of Health, Bethesda, MD) (n = 307 for CESA3; n = 393 for CESA6).
GUS Staining.
Seventy-two-hour-old dark-grown seedlings expressing promoter CESA-GUS fusion were submerged in GUS buffer as described (36), infiltrated three times (3 min) under vacuum, and incubated at 37°C for 1 or 2 h. Seedlings were washed three times with 70% ethanol.
FTIR Microspectroscopy.
Four biological replicates of wild-type or mutant dark-grown seedlings were analyzed by FTIR microscopy as described (37). The collected spectra were baselined and normalized as described, and statistical analysis was performed by using Student's t test (38).
Supplementary Material
Acknowledgments
We thank G. Mouille for assistance with FTIR analysis, A. Urbain for help with statistics, O. Grandjean for assistance with confocal laser scanning microscope analysis, C. Machu for help with spinning disk microscopy (Plateforme d'Imagerie Dynamique, Institut Pasteur, Paris), M. Doblin for promoter-GUS lines, and C. Maurel and D. T. Luu (Centre National de la Recherche Scientifique, Montpellier, France) for the PIP2–1 cDNA. M.J. was supported by French-Polish PAI-POLONIUM Contract 09195RK. E.C. was supported by National Agency for Research Project “IMACEL” ANR-06-BLAN-0262. Z.P. was supported by Marie Curie Program MEST-CT-2004-7576-VERT. Additional funding was provided by grants from the French Ministery of Research, Action Concertée Incitative Biologie du Développement, and the European Economic Community FP6 program CASPIC.
Abbreviations
- CESA
cellulose synthase catalytic subunit
- BiFC
bimolecular fluorescence complementation
- IP
immunoprecipitation
- YFP
yellow fluorescent protein
- GUS
β-glucuronidase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706569104/DC1.
References
- 1.Paredez AR, Somerville CR, Ehrhardt DW. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 2.Somerville C. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 3.Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Proc Natl Acad Sci USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor NG, Laurie S, Turner SR. Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persson S, Wei H, Milne J, Page GP, Somerville CR. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 9.Desnos T, Orbovic V, Bellini C, Kronenberger J, Caboche M, Traas J, Hofte H. Development (Cambridge, UK) 1996;122:683–693. doi: 10.1242/dev.122.2.683. [DOI] [PubMed] [Google Scholar]
- 10.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Plant J. 2003;34:351–362. doi: 10.1046/j.1365-313x.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- 12.Hématy K, Höfte H. In: The Expanding Cell. Verbelen J-P, Vissenberg K, editors. Berlin: Springer; 2006. pp. 33–56. [Google Scholar]
- 13.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Hofte H. Plant Physiol. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D. Proc Natl Acad Sci USA. 2007;104:5854–5859. doi: 10.1073/pnas.0700789104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagard M, Hofte H, Vernhettes S. Plant Physiol Biochem. 2000;38:1–11. [Google Scholar]
- 17.Robert S, Mouille G, Höfte H. Cellulose. 2004;11:351–364. [Google Scholar]
- 18.Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu CD, Chinenov Y, Kerppola TK. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 20.Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C. Plant Physiol. 2005;139:790–805. doi: 10.1104/pp.105.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 22.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 23.Manfield IW, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morel J, Claverol S, Mongrand S, Furt F, Fromentin J, Bessoule JJ, Blein JP, Simon-Plas F. Mol Cell Proteomics. 2006;5:1396–1411. doi: 10.1074/mcp.M600044-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Szyjanowicz PM, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR. Plant J. 2004;37:730–740. doi: 10.1111/j.1365-313x.2003.02000.x. [DOI] [PubMed] [Google Scholar]
- 26.Hynes TR, Tang L, Mervine SM, Sabo JL, Yost EA, Devreotes PN, Berlot CH. J Biol Chem. 2004;279:30279–30286. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 27.Zamyatnin AA, Jr, Solovyev AG, Bozhkov PV, Valkonen JP, Morozov SY, Savenkov EI. Plant J. 2006;46:145–154. doi: 10.1111/j.1365-313X.2006.02674.x. [DOI] [PubMed] [Google Scholar]
- 28.Anderie I, Schulz I, Schmid A. Cell Signal. 2007;19:582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Refregier G, Pelletier S, Jaillard D, Hofte H. Plant Physiol. 2004;135:959–968. doi: 10.1104/pp.104.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estelle MA, Somerville CR. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- 31.Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. Plant Cell Physiol. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Lane D, Harlow E. Using Antibodies: A Laboratory Manual. Cold Spring, NY: Cold Spring Harbor Lab Press; 1999. [Google Scholar]
- 34.Voinnet O, Rivas S, Mestre P, Baulcombe D. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 35.Chan J, Calder G, Fox S, Lloyd C. Plant Cell. 2005;17:1737–1748. doi: 10.1105/tpc.105.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jefferson RA, Kavanagh TA, Bevan MW. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouille G, Robin S, Lecomte M, Pagant S, Hofte H. Plant J. 2003;35:393–404. doi: 10.1046/j.1365-313x.2003.01807.x. [DOI] [PubMed] [Google Scholar]
- 38.Robin S, Lecomte M, Höfte H, Mouille G. J Applied Statist. 2003;30:669–681. [Google Scholar]
- 39.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Pointdexter P, Khitrov N, Auer M, Somerville CR. Proc Natl Acad Sci USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.