Abstract
The Ig superfamily (IgSF) intercellular adhesion molecule-1 (ICAM-1) equilibrates between monomeric and dimeric forms on the cell surface, and dimerization enhances cell adhesion. A crystal structure of ICAM-1 IgSF domains (D) 3–5 revealed a unique dimerization interface in which D4s of two protomers fuse through edge β-strands to form a single super β-sandwich domain. Here, we describe a crystal structure at 2.7-Å resolution of monomeric ICAM-1 D3–D5, stabilized by the monomer-specific Fab CA7. CA7 binds to D5 in a region that is buried in the dimeric interface and is distal from the dimerization site in D4. In monomeric ICAM-1 D3–D5, a 16-residue loop in D4 that is disordered in the dimeric structure could clearly be traced as a BC loop, a short C strand, and a CE meander with a cis-Pro followed by a solvent-exposed, flexible four-residue region. Deletions of 6 or 10 residues showed that the C-strand is essential for monomer stability, whereas a distinct six-residue deletion showed little contribution of the CE meander. Mutation of two inward-pointing Leu residues in edge β-strand E to Lys increased monomer stability, confirming the hypothesis that inward-pointing charged side chains on edge β-strands are an important design feature to prevent β-supersheet formation. Overall, the studies reveal that monomer–dimer transition is associated with a surprisingly large, physiologically relevant, IgSF domain rearrangement.
Keywords: crystal structure, leukocytes, flow cytometry, mutagenesis
Intercellular adhesion molecule-1 (ICAM-1; CD54) is perhaps the most important member of a family of related Ig superfamily (IgSF) molecules that serve as ligands for the integrins αLβ2, αMβ2, and αXβ2 (1). ICAM-1 is expressed on the surface of cells important in immune responses. Inflammatory mediators further enhance expression of ICAM-1 on these cells and induce it on other cell types, including endothelial, epithelial, and fibroblastic cells. Increased ICAM-1 expression augments immune responses and leukocyte accumulation in inflamed tissues.
ICAM-1 consists of five extracellular IgSF domains (D1–D5), a hydrophobic transmembrane domain, and a short cytoplasmic domain (1). In its native state on the cell surface ICAM-1 is in equilibrium between a monomeric and dimeric state (2, 3). Chemical cross-linking reveals a substantial proportion of dimeric material (2, 3). The fraction of monomeric cell-surface ICAM-1 can be estimated by using a mAb termed CA7 that binds to D5 of ICAM-1 (4) and binds much better to monomeric than dimeric ICAM-1 (2). The transmembrane domain of ICAM-1 stabilizes dimerization. ICAM-1 with the transmembrane domain replaced with a glycosylphosphatidylinositol (GPI) anchor is largely monomeric, whereas wild-type ICAM-1 is largely dimeric (2, 3). Recombinant soluble ICAM-1, lacking the transmembrane and cytoplasmic domains, exists as a monomer in solution (2, 4, 5).
Crystal structures of D1–D2 and D3–D5 fragments of ICAM-1 have revealed two sites for dimerization, in D1 and D4. A dimerization site on the BED sheet of D1 buries 450 Å2 per monomer (6). Dimerization in D1 is compatible with binding of the integrin αL I domain to D1 of ICAM-1, as shown by a cocrystal structure (7). A dimerization interface in D4 buries 980 Å2 per monomer (8). The two protomers come into such intimate contact in D4 that the two D4s merge into an integrated structure with two super β-sheets each containing the ABE strands from one molecule and A′GF strands from the other molecule, with 12 main-chain hydrogen bonds across the pseudodyad axis.
Models of intact ICAM-1 molecules constructed from the ICAM-1 D1–D2 crystal structure in complex with the αI domain of integrin αLβ2, and from the ICAM-1 D3–D5 crystal structure, suggest that dimerization on the cell surface is important for optimally orienting for cell adhesion the binding site in D1 for αLβ2 (5) and in D3 for αMβ2 (9). Furthermore, one monomer is likely to dimerize with distinct monomers at D1 and D4, in agreement with visualization of ICAM-1 tetramers in electron microscopy (10). Thus, chain-like 1D clusters of ICAM-1 molecules may form on the cell surface (8).
Dynamic equilibration of ICAM-1 between monomeric and dimeric states may be important during diffusion on the cell surface and formation of organized structures such as the immunological synapse in which ICAM-1 has a ring-shaped distribution (11). Remarkably, in the dimeric D3–D5 ICAM-1 crystal structure, a 16-residue-long region between the B and E strands of D4 at the dimerization interface in each protomer is disordered and not resolved by x-ray diffraction (8). We hypothesized that in the monomer structure this disordered loop may fold to provide additional edge β-strand(s) and cap the hydrophobic super-β -sheet dimerization interface. Such a large, physiologically relevant structural transition in IgSF domains or related β-sandwich folds is unprecedented, although smaller structural changes occur in cadherins (12–15). To test the hypothesis of a structural transition in D4 between monomeric and dimeric ICAM-1, we determined a crystal structure of monomeric ICAM-1 D3–D5 in complex with CA7 Fab. Through structural comparison with dimeric ICAM-1, coupled with mutagenesis studies, we demonstrate an IgSF domain rearrangement responsible for monomer–dimer transition.
Results
The Structure of D3–D5/CA7 Fab Complex.
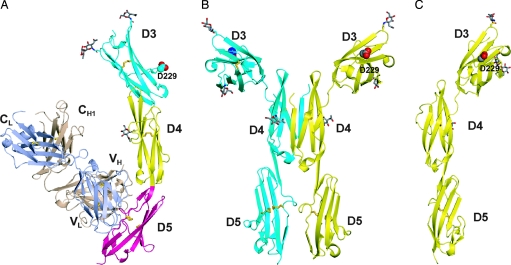
The crystal structure of ICAM-1 D3–D5 in complex with CA7 Fab at 2.7-Å resolution (Table 1) shows that CA7 Fab binds to D5 of ICAM-1 (Fig. 1A). D3–D5 are in an overall extended conformation with an obtuse twist angle between successive domains. In the D3–D5 dimer structure (8), there is a large bend between D3 and D4. Assuming that the dimer dyad axis is normal to the plasma membrane, this bend orients the αMβ2 binding site in D3 that includes Asp-229 optimally for cell adhesion (Fig. 1B). In the D3–D5 monomer structure, the angle between D3 and D4 decreases by 18° and 16° compared with monomers A and B of the dimer, respectively; the key αMβ2 binding residue, Asp-229 (9), still adopts an orientation easily accessible to an integrin (Fig. 1C).
Table 1.
Data collection and refinement statistics
| Data collection statistics | |
| Space group | C2 |
| Unit cell | a = 185.4 Å, b = 69.3 Å, c = 88.2 Å |
| α =90°, β =112.76°, γ = 90° | |
| Resolution, Å | 28.5–2.7 |
| Outer resolution shell, Å | 2.77–2.70 |
| No. of observations | 90,579 |
| Unique reflections | 26,015 |
| Completeness, % | 90.6 (58.1) |
| Rsym, % | 8.7 (54.3) |
| Refinement statistics | |
| Refinement range, Å | 28.5–2.70 |
| Data cutoff (|F|) | 0.0 |
| No. of reflections (work/test) | 23,400/2,559 |
| Rwork (Rfree), % | 20.4 (25.3) |
| rmsd bond lengths, Å | 0.005 |
| rmsd bond angles, ° | 0.91 |
| Ramachandran statistics, % (favored/allowed/outlier)* | 96.5/3.4/0.1 |
*Determined with RAMPAGE (28).
Fig. 1.
Ribbon diagrams of ICAM D3–D5 crystal structures. (A) Monomeric ICAM-1 D3–D5 complexed with CA7 Fab. (B) The dimeric ICAM-1 D3–D5 structure (8). (C) The monomeric ICAM-1 D3–D5 structure with CA7 Fab omitted, shown in the same orientation as the right monomer of dimeric D3–D5 in B, after superposition on D4. The side chain of the key αMβ2 binding residue in D3, Asp-229, is shown with large spheres. The Fab light and heavy chains are shown in light blue and wheat, respectively. Glycans are drawn in stick with red oxygens and gray carbons. Disulfide bonds are drawn as gold sticks.
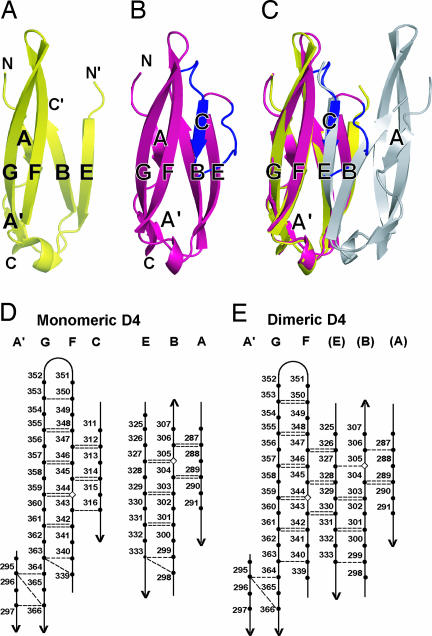
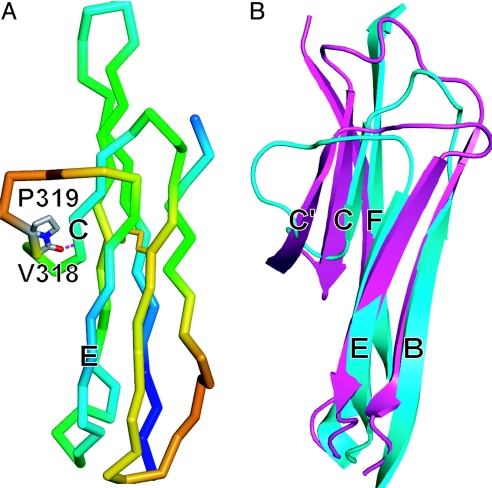
Sixteen residues in D4 that were disordered in the dimeric D3–D5 structure (Fig. 2A) could be clearly traced in the electron density of the monomeric structure (Fig. 2B). These residues, 308–323, form four residues of the BC loop, a C strand of four residues, and a meander of eight residues that connects to the E strand (Fig. 2B). Thus, monomeric D4 is made up of two β-sheets containing the ABE and A′GFC β strands, respectively (Fig. 2D). Residues 318 and 319 occupy a position similar to a C′ strand, and residue 318 forms a hairpin turn backbone hydrogen bond to the C strand (Fig. 3A); however, the cis configuration of Pro-319 kinks the backbone out of the plane occupied by the A′GFC sheet in the direction away from the ABE sheet (Fig. 3B). These results, together with the presence of an A′ strand and lack of a D strand show that D4 belongs to the I2 subset of IgSF domains (6, 16).
Fig. 2.
Structural differences between monomeric and dimeric D4. (A–C) Ribbon diagrams of D4 after superposition on D4 of one monomer of dimeric ICAM-1. (A) One monomer of dimeric D4. (B) Monomeric D4. (C) Superposition of monomeric and dimeric D4. The 16 residues that become ordered in monomeric D4 are shown in blue, and the residues preceding and following the disordered region in dimeric ICAM-1 are marked C′ and N′, respectively. (D and E) β-sheet hydrogen bonds in monomeric (D) and dimeric (E) D4. Backbone hydrogen bonds of ≥1.0 kcal/mol as determined with DSSP (29) are shown as dashed lines. The disulfide-bonded cysteines in β-strands B and F are shown as diamonds.
Fig. 3.
Structural properties of D4. (A) Backbone Cα trace of D4 colored in rainbow from highest (red) to lowest (blue) B factor. Atoms of cis-Pro-319 and atoms C and O of Val-318 are represented with sticks, and the hydrogen bond in the turn between the C-strand and CE meander is dashed. The disulfide bond is shown in yellow. (B) Comparison of the CE edges of D2 (magenta) and monomeric D4 (cyan) of ICAM-1. Superposition is on β-strands B, C, E, and F and the region containing β-strands A, A′, and G is omitted for clarity.
The CA7 Fab Binding Site on ICAM-1 D5.
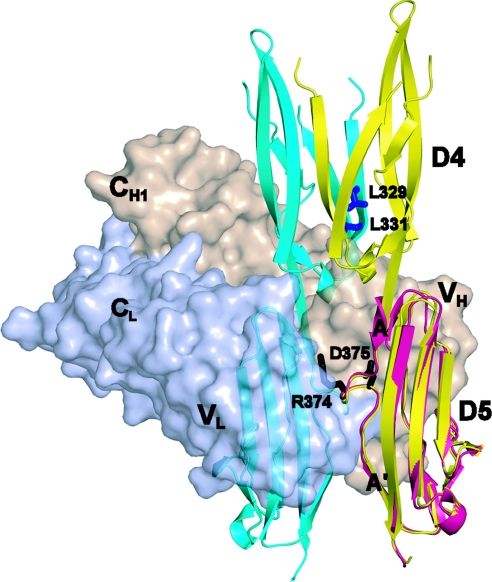
The binding site for CA7 Fab is centered on Asp-375 in a highly charged loop in the meander between the A and A′ strands in D5 of ICAM-1 (Fig. 4). This loop protrudes into the antigen-binding cavity between VH and VL, which is recessed because of the unusually short heavy chain CDR3 loop at its center. Contributing to the interaction are an extensive hydrogen bond network including D5 residues Arg-374 and Asp-375, two Trp residues in D5, and aromatic residues in the Fab. The CA7 Fab buries 1,070 Å2 of solvent-accessible surface area on D5 and 80 Å2 on D4 at its interface with D5. The interface between D4 and D5 changes by 29° and 35° compared with monomers A and B of the ICAM-1 D3–D5 dimer, respectively, to accommodate contacts of the Fab with Glu 336 of the EF loop of D4 and the first residue of D5, Tyr-367. Antibody binding also induces a flip in the side chain of the nearby Trp-395 residue of D5.
Fig. 4.
The CA7 Fab binding site in D5. CA7 Fab is shown as a surface representation colored in wheat (heavy chain) and light blue (light chain) bound to monomeric D5 shown as a magenta ribbon, with indicated side chains in the AA loop at the center of the epitope as black sticks. Dimeric D4 and D5 are shown as ribbons with a cyan monomer and a yellow monomer superimposed on monomeric D5. The inward-pointing Leu residues of β-strand E of the cyan monomer are shown as blue sticks.
In the dimer structure, the protruding AA′ meanders of D5 of the two monomers face one another and are in close proximity (Fig. 4), although their contact contributes only a small amount (310 Å2) to the total buried solvent-accessible surface area comared with the contacts in D4 (1,950 Å2). It is interesting that the same loop in the D5 AA′ meander that binds to CA7 Fab mediates the D5 contact in dimeric ICAM-1 (Fig. 4). Superposition of the D3–D5 dimer structure onto the Fab complex shows that the Fab completely occludes the position of the cognate D5 in a dimer (Fig. 4), explaining why CA7 antibody binds to monomeric but not dimeric ICAM-1 (2). Importantly, the Fab binding site is distal from, and has no direct influence on, the structurally plastic region in D4 involved in dimerization.
Structural Plasticity in Monomer/Dimer Transition.
In the ICAM-1 D3–D5 dimer structure, the two D4s merge, through a largely hydrophobic interface, into two super β-sheets that are formed by fusion of A′GF sheets with EBA sheets by hydrogen-bonding across their F strand and E strand edges (Fig. 2E). Strikingly, superposing monomeric D4 onto one monomer of dimeric D4 reveals that the C strand of monomeric D4 occupies the same position as the upper portion of strand E of the other monomer of dimeric D4 (Fig. 2C). All three residues in strand F that hydrogen-bond across the dyad axis to strand E in the dimer, hydrogen-bond to strand C in the monomer (Fig. 2 D and E). Notably, to form the supersheet in the dimer, the N-terminal, upper portion of β-strand E splays away from β-strand B and toward β-strand F (Fig. 2 A and B). Thus, the formation of six hydrogen bonds between β-strands F and E across the dyad axis of each supersheet is accompanied by the loss of three hydrogen bonds between the upper portions of β-strands B and E that are present in monomeric D4 (Fig. 2 D and E). This interchangeable role of β-strand C in the monomeric structure and the upper portion of β-strand E in the dimeric structure demonstrates the pivotal role of β-strand C in monomer–dimer transition in ICAM-1.
The C strand and CE loop in monomeric D4 must become unstructured to enable dimerization of D4. B factors suggest that the meander between the C and E strands in D4 is the most flexible region in monomeric D3–D5, and that the segment P319AQP322, which makes a solvent-exposed 180° turn is especially flexible (Fig. 3A). Comparison to other IgSF domains, including D2 of ICAM-1, which is also a member of the I2 set, shows that in addition to lack of a C′ strand D4 of ICAM-1 contains an unusually short C strand (Fig. 3B). The inherent flexibility of the CE meander and the shortness of the C strand are undoubtedly important for unfolding of this region, which would have to occur before dimerization of D4.
Mutational Studies of the D4 Dimer Interface.
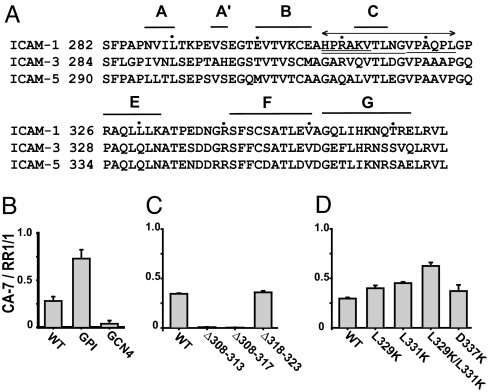
We studied by mutagenesis the importance to the monomer–dimer equilibrium of residues in the BC loop, C strand, and CE meander of D4. As standards, we first compared wild-type ICAM-1, GCN4–ICAM-1, and GPI-anchored ICAM-1 (2). GCN4–ICAM-1 contains, fused C-terminally to its transmembrane domain, a GCN4 peptide that forms a dimeric α-helical coiled-coil and, as shown below, further stabilizes dimerization. In contrast, GPI–ICAM-1 is predominantly monomeric (2). Cell surface dimerization was assessed by staining with CA7 antibody, which is specific for monomeric ICAM-1, or staining with RR1/1 antibody, which binds to D1 (5). Whereas ≈25% of wild-type ICAM-1 was monomeric, ≈75% of GPI-linked ICAM-1 was monomeric and only ≈5% of GCN4-stabilized ICAM-1 was monomeric, demonstrating the validity and dynamic range of this assay (Fig. 5B).
Fig. 5.
Secondary structure of D4 and the effect of mutations on monomer–dimer equilibrium on the cell surface. (A) Secondary structure of D4 of ICAM-1 and alignment with D4 of ICAM-3 and ICAM-5. Every 10th residue is marked with a dot. The 16 residues disordered in dimeric ICAM-1 are indicated by a line with two arrowheads. The three deleted regions are underlined. (B–D) The proportion of monomeric ICAM-1 on wild-type ICAM-1, GPI–ICAM-1, and GCN4–293T transfectants as measured by immunofluorescent flow cytometry and the ratio of CA7 (monomer specific) to RR1/1 (conformation-independent) antibody staining. (B) Comparisons of GPI–ICAM-1 and GCN4–ICAM-1 mutants. (C) Comparison of deletion mutants. (D) Comparison of point mutants.
Our structure predicts that the presence of a C strand in D4 should be sufficient to inhibit dimerization. To test this hypothesis, we deleted a portion of the BC loop and a portion (Δ308–313) or all (Δ308–317) of the C strand. Alternatively, we deleted a portion of the CE meander (Δ318–323) (Fig. 5A). The Δ308–313 and Δ308–317 mutations completely abolished CA7 binding, showing that deletion of β-strand C profoundly increases the stability of the dimer relative to the monomer (Fig. 5C). In contrast, deletion of residues 318–323 in the CE meander had little effect on CA7 binding (Fig. 5C). Furthermore, the D337K mutant, which breaks a salt bridge across the dyad axis in dimeric ICAM-1, had little effect on the CA7 to RR 1/1 ratio (Fig. 5D).
Uncontrolled, promiscuous formation of supersheets by β-sheets would have pathologic consequences. The Richardsons (17) found that inward-pointing, charged side chains are the predominant negative design principle used in edge β-strands of β-sandwich domains to avoid edge-to-edge aggregation. Inward-pointing residues point toward the opposite β-sheet of the β-sandwich and would occupy the hydrophobic interior after dimerization. In ICAM-1 D4, β-strand E is the only edge strand in the monomer that is involved in aggregation into the dimer (Fig. 2 D and E). It has no inward-pointing, charged residues, in agreement with its propensity for dimerization. However, it has two inward-pointing Leu residues that are exposed to solvent in monomeric D4 and are buried in dimeric D4 (Fig. 4). These residues, Leu-329 and Leu-331, were mutated to lysine, which is the most frequently occurring inward-pointing charged side chain in β-sandwich edge strands (17). Indeed, as predicted, the ratio of CA7-to-RR 1/1 binding, and hence the proportion of monomer, was significantly increased for both L329K and L331K mutants. Furthermore, these effects were additive, because the double mutant L329K/L331K had a markedly higher CA7-to-RR 1/1 ratio (Fig. 5D).
Discussion
The monomeric structure of D3–D5 of ICAM-1 determined here, and the previously determined dimeric structure (8), define two states that appear to coexist in equilibrium with each other and dynamically interconvert on the cell surface. The 16 residues in D4 that are disordered in dimeric ICAM-1 fold in monomeric ICAM-1 into a C β-strand and a CE meander, revealing an I2-set IgSF fold. Thus D1, D2, D3, D4, and D5 of ICAM-1 have I1, I2, I1, I2, and distorted-I2 set folds, respectively (6, 8).
The CA7 Fab binds to a loop in the AA′ meander of D5 that also participates in a minor (310 Å2 total buried solvent-accessible surface area) monomer–monomer contact in dimeric ICAM-1 (8). The predominant dimerization site in D4 (which buries 1,950 Å2), where the 16-residue folding–unfolding transition occurs, is distal from the CA7 binding site. CA7 Fab binding to D5 prevents close approach of two monomers, both directly at the D5 interface, and indirectly at the D4 interface, i.e., dimerization of D4 is sterically hindered. Thus, we believe that the completely folded state of D4 seen here represents the native fold for monomeric ICAM-1.
In the monomer–dimer equilibrium on the cell surface, the overall stability of the dimer is modest. The transmembrane domain contributes to dimerization at D4 and D5, as shown previously (2) and here by the marked increase in CA7 staining after substitution with a GPI anchor. This finding is consistent with the proximity of the C termini of the two D5s in dimeric D3–D5, and the prediction that the transmembrane domain of ICAM-1 has amphipathic and glycophorin-like features that contribute to dimerization (3). In solution, ICAM-1 D1–D5 is monomeric at >20 mg/ml; however, addition of a short, C-terminal hemagglutinin tag is sufficient to drive dimerization at 15 mg/ml as shown by physicochemical techniques and loss of CA7 antibody binding (2). Dimerization of D3–D5 is also driven by the high protein concentrations present in crystal lattices (8), which is why we used CA7 in this study to obtain the monomeric form of D3–D5.
The structure of the monomer and the mutational studies described here provide insights into the features that affect the monomer–dimer equilibrium. Among the 16 residues that become folded in monomeric D4, those in the BC loop, in strand C, and up to the backbone H-bonded residue Val-318 in the CE meander have moderate B factors and good electron density. By contrast, much of the CE meander is highly solvent exposed and has high B factors, especially at residues 320–323. Interestingly, deletion of residues 318–323 had no effect on the stability of cell surface ICAM-1 dimers, which suggests that folding of residues 320–325 contributes little to thermostability of the monomer, consistent with solvent exposure and high B factors in this region. The lack of effect of this deletion is also consistent with the observation that the Cα atoms of residues 318 and 327 are only 8 Å apart, enabling the connection between residues 317 and 324 in the deletion mutant to be formed with few backbone rearrangements. By contrast, partial or complete deletion of β-strand C almost completely abolished detection of monomer on the cell surface, demonstrating a crucial role for β-strand C in monomer stability.
β-Strand E in D4, which is an edge strand in the monomer, hydrogen-bonds to β-strand F in another monomer to form the dimer. The Richardsons (17) found two different types of negative design features in β-sandwich edge strands that prevent edge-to-edge dimerization or aggregation of β-sheets: (i) β-bulges and (ii) inward-pointing, charged side chains. Neither feature is present in β-strand E in D4. Instead, it has two inward-pointing Leu residues that contribute to the hydrophobic core of the dimerization interface in D4. Single or double mutation of these Leu residues to Lys markedly shifted the equilibrium toward monomer, validating the importance of the Leu residues in the hydrophobic core and the use of charged residues in negative design of edge β-strands. However, a portion of cell surface ICAM-1 remained dimeric, even with the double Leu → Lys mutant, suggesting a strong disposition toward dimer formation, and the possibility that the long Lys side chains might be able to arrange with the ε-amino groups escaping burial in the hydrophobic core.
What features of D4 of ICAM-1 favor unfolding of the C strand and CE meander? This region, between the B and E strands, is short in D4 of ICAM-1 at 19 residues. By contrast, in other I2 set domains, the corresponding regions of D2 of ICAM-1 [Protein Data Bank (PDB) code 1IC1], D2 of ICAM-2 (PDB code 1ZXQ), D2 of CD2 (PDB code 1HNF), D2 of CD4 (PDB code 3CD4), D4 of CD4 (PDB code 1CID), D2 of LFA-3 (PDB code 1CCZ), D2 of VCAM-1 (PDB code 1VCA), and D2 of SLAM-6 (PDB code 2IF7) are 21–38 (mean = 29) residues. Furthermore, the two-residue region corresponding in ICAM-1 D4 to that between residue 315, the last C-strand residue, and residue 318, which occupies a position similar to a C′-strand residue, was seven to eight residues in the other domains. In other words, other I2 set domains not only have edge C′ strands, which protect the C strand, but also have either longer C strands, or longer CC′ loops, which project further toward the C-terminal end of the domain, as seen in D2 of ICAM-1 (Fig. 3B). D4 is >50% identical between ICAM-1, ICAM-3, and ICAM-5 (Fig. 5A). Furthermore, the length of the region that unfolds in dimeric ICAM-1 is conserved, as is the cis-Pro 319 in ICAM-1 that is responsible for the unusual course of the CE meander and may contribute to the lack of a C′ strand. Therefore, we propose that ICAM-3 and ICAM-5 also equilibrate between monomeric and dimeric species on the cell surface (8).
This study reveals a remarkable amount of physiologically relevant structural plasticity in an IgSF domain. Not only do 16 residues undergo a folding/unfolding transition, but another four residues near the beginning of β-strand E move >4 Å in the monomer/dimer transition. Although D5 of Trk receptors expressed in Escherichia coli can undergo a swap of β-strand A to form a dimer, it is reported to be an artifact because it does not occur when Trk D5 binds ligand (18) and is prevented in intact Trk by the interface with D4 and N-glycosylation (19). A physiologically relevant swap occurs in D1 of cadherins, which have an Ig-like β-sandwich fold. N-terminal residues 1–3 move 3–10 Å, and Trp-2 and Val-3 reverse their side-chain orientations reciprocally to engage D1 of another cadherin molecule, which is thought to occur in trans between cadherins on different cells to mediate cell adhesion (12–14). Type II cadherins undergo a similar swap, except a longer four-residue segment containing two Trp residues is exchanged (20). Cadherin crystal structures and mutagenesis also suggest a significant front-to-back interaction in cis between D1 and D2 in a line of molecules on each cell (14), in contrast to the dimeric, side-to-side interaction in cis with rearrangements in D4 in ICAM-1. Dimerization in D4 is hypothesized to help orient the dimer dyad axis normal to the cell surface so that the binding sites in D1 and D3 for integrins αLβ2 and αMβ2 are optimally oriented (8) and is known to enhance adhesion to integrin αLβ2 (2). The disorder/order rearrangement of 16 residues and significant movement of four other residues that are involved in mediating dimerization is surprisingly large, and to our knowledge, unprecedented for a β-sandwich domain rearrangement that occurs physiologically.
Materials and Methods
Protein Preparation and Crystallization.
ICAM-1 domain 3–5 fragment (Phe-185–Pro-450) was purified as described (8) from CHO Lec 3.2.8.1 cell supernatants with CBR IC1/11 mAb affinity chromatography. The purified protein was deglycosylated with endoglycosidase H and further purified by Resource Q ion-exchange chromatography in 10 mM Tris·HCl, pH 8.0 using a gradient of 50–600 mM NaCl.
The CA7 hybridoma (4) was kindly provided by P. Giblin (Boehringer Ingelheim, Ridgefield, CT). The antibody was purified from culture supernatant with a protein A column followed by Superdex 200 (Amersham Pharmacia, Piscataway, NJ) gel filtration. The Fab was generated by immobilized papain (Pierce, Rockford, IL) digestion following the manufacturer's instruction. Undigested IgG and Fc fragments were removed by reloading onto the Protein A column, and the Fab was purified further with Resource S ion exchange chromatography in 20 mM sodium acetate, pH 5 with a gradient of 0–300 mM NaCl. Purified ICAM-1 D3–D5 fragment was incubated with CA7 Fab at a 1:2 molar ratio at room temperature for 30 min. The D3–D5 complex with Fab was separated from excess, free Fab using Superdex 200 in 10 mM Tris (pH 8.0) and 150 mM NaCl and concentrated for crystallization.
Crystals were grown by using vapor diffusion in hanging drops at room temperature with equal volumes of 12 mg/ml protein solution and the reservoir solution of 1.5 M Li2SO4/0.1M Hepes (pH 7.5). Crystals were cryo-protected by transfer to 1.9 M Li2SO4/0.1 M Hepes (pH 7.5). The sequence of the CA7 Fab was determined by hybridoma cDNA sequencing exactly as described (21).
Structure Determination and Refinement.
The diffraction data were collected at the 19-ID and 24-ID stations of the Advanced Photon Source at the Argonne National Laboratory (Argonne, IL) and processed with the program suite HKL2000 (22). The program molrep (23) was used for molecular replacement. The models used were the Fab fragment from F124 (PDB code 1F11), and D3 and D5 of ICAM-1 D3–D5 (PDB code 1P53). The solutions from molecular replacement were subjected to iterative cycles of model rebuilding in COOT and refinement using CNS (version 1.1) (24). Sigma A weighted 2 Fo − Fc and Fo − Fc maps were computed during rebuilding of ICAM-1 D4, and refinement was monitored by decrease of Rfree. Composite omit maps were calculated during of refinement to reduce model bias. The final rounds of refinement were performed with REFMAC (25) using TLS and maximum-likelihood refinement.
The final model, refined to 2.7-Å resolution, contains amino acid residues 186–450 of ICAM-1 D3–D5, residues 1–214 of the Fab κ-light chain, residues 1–127 and 134–214 of the Fab heavy chain, and four carbohydrate residues. Backbone density is poor at ICAM-1 residue 320, and only backbone density is evident for residues 321–323 at 1 σ (residue 321 is the one Ramachandran outlier). Two densities that show (i) coordination to Asp-213 and Asp-241 of ICAM-1 and the backbone of Asp-1 of the light chain and (ii) to Glu-185 and His-189 of symmetry-related light chains were modeled as Zn atoms. An x-ray fluorescence spectrum at ID-23 at the Advanced Photon Source suggested the presence of zinc and arsenic. A fluorescence scan at the K edge confirmed the presence of zinc. The data were reprocessed with HKL2000, and weak anomalous signals were seen to 5 Å. The anomalous signals were combined with the phases from a model lacking zinc to calculate an anomalous difference Fourier map. Only two peaks stood out from the background, which corresponded to the positions of the Zn atoms. Final statistics are shown in Table 1. Figures were prepared with PYMOL (www.pymol.org).
Flow Cytometry Studies.
Wild-type and GPI-ICAM-1 cDNA constructs have been described (5). For GCN4–ICAM-1, the ICAM-1 sequence after L475 at the end of the transmembrane domain was fused to two Gly residues and residues R249 to G279 of the yeast transcription factor GCN4 α-helical coiled-coil domain, containing a Cys residue inserted between GCN4 residues Q252 and L253. Plasmid constructs expressing ICAM-1 fusions with monomeric cyan fluorescent protein (mCFP) (ICAM–1-mCFP) were generated by ligation of 1.8-kb fragments encoding ICAM-1 to the AgeI–NotI sites of αL-mCFP (26). Mutants were made by using site-directed mutagenesis with the QuikChange kit (Stratagene, La Jolla, CA). Human embryonic kidney 293T cells were cultured in DMEM supplemented with 10% FBS and 2 mM glutamine. Human embryonic kidney 293T cells were transfected by calcium phosphate precipitation (27). In brief, 12 μg of wild-type or mutant ICAM-1 cDNA was used to transfect one 6-cm plate of 70–80% confluent cells. Two days after transfection, cells were harvested with Hanks' balanced salt solution containing 5 mM EDTA and washed twice with PBS. Cells (105) were incubated with 20 μg/ml (1/10 dilution) of primary antibodies (RR1/1 and CA7) in 100 μl PBS for 30 min on ice. Cells were then washed twice with PBS and incubated with R-phycoerythrin-conjugated goat anti-mouse IgG (Jackson Laboratories, San Francisco, CA) for 30 min on ice. After washing with PBS, cells were resuspended in cold PBS and analyzed with a FACScan (BD Biosciences, San Jose, CA).
To calculate the CA7/RR1/1 ratio, the mean fluorescence intensity (MFI) of untransfected cells was subtracted from that of transfected cells. For example, the MFI ratio of CA7/RR1/1 of wild-type ICAM-1 is computed as: [MFI CA7 of transfected cell − MFI CA7 of untransfected cell]/[MFI RR1/1 of transfected cell − MFI RR1/1 of untransfected cell]. For mutants, similar level of surface expression to wild-type ICAM-1 was verified by RR1/1 mAb staining.
Acknowledgments
We thank Drs. Jia-huai Wang, Larry Shapiro, and Mike Dustin for reviewing this manuscript. This work was supported by National Institutes of Health Grant CA31798 (to T.A.S.) and the Arthritis Foundation (C.V.C.).
Abbreviations
- IgSF
Ig superfamily
- ICAM
intercellular adhesion molecule
- D(n)
domain (n)
- GPI
glycosylphosphatidylinositol
- PDB
Protein Data Bank
- MFI
mean fluorescence intensity.
Footnotes
The authors declare no conflict of interest.
References
- 1.Dustin ML, Springer TA. In: Guidebook to the Extracellular Matrix and Adhesion Proteins. Kreis T, Vale R, editors. New York: Sambrook and Tooze; 1999. pp. 216–220. [Google Scholar]
- 2.Miller J, Knorr R, Ferrone M, Houdei R, Carron CP, Dustin ML. J Exp Med. 1995;182:1231–1241. doi: 10.1084/jem.182.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reilly PL, Woska JR, Jr, Jeanfavre DD, McNally E, Rothlein R, Bormann B-J. J Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- 4.Rothlein R, Mainolfi EA, Czajkowski M, Marlin SD. J Immunol. 1991;147:3788–3793. [PubMed] [Google Scholar]
- 5.Staunton DE, Dustin ML, Erickson HP, Springer TA. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 6.Casasnovas JM, Stehle T, Liu J-h, Wang J-h, Springer TA. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimaoka M, Xiao T, Liu J-H, Yang Y, Dong Y, Jun C-D, McCormack A, Zhang R, Joachimiak A, Takagi J, et al. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Jun C-D, Liu J-h, Zhang R-G, Jochimiak A, Springer TA, Wang J-h. Mol Cell. 2004;14:269–276. doi: 10.1016/s1097-2765(04)00204-7. [DOI] [PubMed] [Google Scholar]
- 9.Diamond MS, Staunton DE, Marlin SD, Springer TA. Cell. 1991;65:961–971. doi: 10.1016/0092-8674(91)90548-d. [DOI] [PubMed] [Google Scholar]
- 10.Jun C-D, Carman CV, Redick SD, Shimaoka M, Erickson HP, Springer TA. J Biol Chem. 2001;276:29019–29027. doi: 10.1074/jbc.M103394200. [DOI] [PubMed] [Google Scholar]
- 11.Carman CV, Springer TA. Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 14.Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 15.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Proc Natl Acad Sci USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J-h, Springer TA. Immunol Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 17.Richardson JS, Richardson DC. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ultsch MH, Wiesmann C, Simmons LC, Henrich J, Yang M, Reilly D, Bass SH, de Vos AM. J Mol Biol. 1999;290:149–159. doi: 10.1006/jmbi.1999.2816. [DOI] [PubMed] [Google Scholar]
- 19.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Proc Natl Acad Sci USA. 2002;99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, et al. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 21.Xiao T, Takagi J, Wang J-h, Coller BS, Springer TA. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Kim M, Carman CV, Springer TA. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 27.Heinzel SS, Krysan PJ, Calos MP, DuBridge RB. J Virol. 1988;62:3738–3746. doi: 10.1128/jvi.62.10.3738-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]