Abstract
Tenascin (TN) is an extracellular matrix glycoprotein that is expressed in a characteristic spatiotemporal pattern during development and is up-regulated in the adult during tumorigenesis, wound healing, and nerve regeneration. In previous studies, we identified a promoter within the proximal 250 bp upstream of the mouse TN gene that contains several putative regulatory elements that are conserved among vertebrate TN genes. We have identified four different DNA elements within this promoter and show that they contribute in different ways to TN gene expression in NIH 3T3 fibroblasts, C6 glioma cells, and N2A neuroblastoma cells. These elements comprise a binding site for Krox proteins, one for nuclear factor 1, an octamer motif that binds POU-homeodomain proteins, and a novel TN control element. The nuclear factor 1 and TN control element had positive effects on TN promoter activity and formed similar DNA–protein complexes with nuclear extracts from all three cell lines. The Krox element had a negative effect on TN promoter activity in N2A cells, a positive effect in C6 cells, and no effect in NIH 3T3 cells. Two DNA binding complexes, one correlated with the negative and the other with the positive activities of the Krox element, were found to contain the protein Krox24. In cotransfection experiments, the octamer motif was required for induction of TN promoter activity by the POU-homeodomain protein Brn2 in N2A cells but was inactive in C6 cells. Consistent with these findings, N2A cells transfected with Brn2 formed octamer-binding complexes containing N-Oct3, the transcriptionally active form of Brn2, whereas complexes formed in C6 cells contained only N-Oct5A and N-Oct5B. Our results provide a striking example of the diversity of regulatory mechanisms that can be called forth by combining different promoter motifs with transcriptional activators or repressors.
Keywords: extracellular matrix, gene regulation, Krox24, octamer, nuclear factor 1
A key problem of developmental biology is to understand how the expression of genes is regulated in a spatiotemporally coordinated fashion leading to place-dependent morphogenesis. Cell adhesion molecules and extracellular matrix proteins play central roles in morphogenesis through their ability to exert mechanical effects on cell aggregation and migration. One particular extracellular matrix cellular glycoprotein affecting such cellular interactions is tenascin (TN), also called cytotactin. TN has a modular structure composed of epidermal growth factor repeats, fibronectin type III repeats, and a globular domain resembling the β and γ chains of fibrinogen (1). TN has both adhesive and counteradhesive effects on cells, and its binding can alter cell shape, physiology, and migration patterns. Several of these activities have been ascribed to specific structural domains within the molecule that bind to a number of different cell surface receptors, including members of the integrin family (2, 3).
In the central and peripheral nervous system, TN is expressed by glia and regulates neuronal migration (4–7). Outside of the nervous system, TN is expressed at sites of embryonic induction, particularly where cells undergo extensive epithelial–mesenchymal transformations such as the lung, mammary gland, kidney, and teeth (8). TN is also known to be stimulated by growth factors of the transforming growth factor type β and basic fibroblast growth factor families (9, 10). In the adult, expression of TN is reinduced both during wound healing and nerve regeneration, and in a variety of tumor cells, including neuroblastomas and gliomas (8, 11–13).
As a basis for understanding the factors that control the different contexts of TN gene expression in vertebrates, we have identified the promoters for the chicken and mouse TN genes (14, 15). The chicken TN promoter contains a number of regulatory elements, including a binding site for homeodomain (HD) proteins (14). In cellular cotransfection experiments, this promoter was activated by the homeobox gene Evx-1, a homologue of the even-skipped gene from Drosophila (16). During vertebrate development, TN is expressed in a cephalocaudal pattern during somite formation (4), and recently, a TN-like pair-rule gene from Drosophila called tenm/odz has been shown to be expressed in a segmented seven-stripe pattern that overlaps with that of even-skipped (17, 18). Thus, the control of TN-like molecules during both invertebrate and vertebrate development may be regulated by related pattern-forming genes.
In our previous studies of the mouse TN promoter (15), the proximal 250 bp upstream of the RNA start site accounted for the majority of TN promoter activity in mouse NIH 3T3 cells and chicken embryo fibroblasts (15). In this study, we show that four DNA sequences contribute to different aspects of TN gene regulation in different cell lines. These sequences are conserved among the vertebrate TN proximal promoters (14, 15, 19) and contain binding sites for Krox, nuclear factor 1 (NF-1), and POU-HD proteins, and a novel TN control element (TCE).
MATERIALS AND METHODS
All molecular biological techniques were performed as described (20). The 5′ deletions in the TN promoter were generated by PCR using the 4.2-kb TN genomic fragment (15) as template, and fragments were inserted in the promoterless vector pBasic (Promega). Full-length cDNAs for Krox20, Krox24, Brn2, Brn4, and SCIP were generated by reverse transcription–PCR and inserted into the EcoRI–HindIII sites of a modified pcDNA3 expression vector (Invitrogen) containing an N-terminal myc tag. Proteins of the correct size were produced from these gene constructs using an in vitro transcription/translation system (Promega).
Mouse NIH 3T3 cells were cultured in DMEM (Life Technologies, Gaithersburg, MD) containing 10% calf serum. N2A, C6, and COS-1 cells were maintained in DMEM containing 10% fetal bovine serum. Cells were transfected with constructs using lipofectamine (Life Technologies) as described (15). For cotransfection experiments, POU-HD effector plasmids (1.6 μg) were cotransfected in N2A or C6 cells with 0.4 μg of either TN11 or TN12 promoter construct and 0.4 μg of the CMV-βgal plasmid as described (15). Chloramphenicol acetyltransferase assays were performed as described (15) and quantitated using a PhosphorImager (Molecular Dynamics). The chloramphenicol acetyltransferase activities presented in Tables 1 and 2 are the average of six independent experiments, performed in duplicate.
Table 1.
Relative activity of TN promoter constructs
| Construct | Element(s) examined | Relative activity
|
||
|---|---|---|---|---|
| NIH 3T3 | C6 | N2A | ||
| TN9 | Krox | 28.2 | 30.0 | 11.4 |
| TN7 | TCE | 28.2 | 20.8 | 23.0 |
| TN10 | dT | 22.0 | 18.4 | 8.0 |
| TN11 | octamer | 16.6 | 16.3 | 7.4 |
| TN12 | NF-1 | 16.1 | 20.6 | 6.1 |
| TN12M | NF-1 mut | 4.5 | 3.6 | 2.8 |
| TN13 | dA; Ant-HD | 9.8 | 6.6 | 2.4 |
| TN8 | TATA | 7.1 | 5.7 | 5.2 |
| pBasic | N/A | 1.0 | 1.0 | 1.0 |
N/A, Not applicable.
Table 2.
Regulation of TN promoter activity by Brn2
| Construct | Activity
|
|||
|---|---|---|---|---|
| N2A
|
C6
|
|||
| pcDNA3 | Brn2 | pcDNA3 | Brn2 | |
| pBasic | 1.3 | 1.1 | 0.6 | 0.7 |
| TN11 | 15.2 | 34.6 | 4.8 | 4.1 |
| TN12 | 11.4 | 7.8 | 4.8 | 3.9 |
For binding experiments, nuclear extracts were prepared as described from NIH 3T3 cells (21), and C6, N2A, and COS-1 cells (22). DNase I footprinting analysis was performed as described (21) using a probe generated by PCR with an end-labeled 5′ primer (bases −287 to −266), a 3′ primer complementary to bases +15 to −7, and the 4.2-kb fragment of the TN gene as template (15). Oligonucleotides corresponding to the Krox, TCE, NF-1, and octamer elements were annealed, and probes were purified on a 6% polyacrylamide gel. For electrophoretic mobility-shift assays (EMSAs), 5 pmol of probes was end-labeled using the Klenow fragment of DNA polymerase I (Boehringer Mannheim). Between 2 and 7.5 μg of nuclear extract was preincubated in a buffer containing 12 mM Hepes (pH 7.9), 4% Ficoll type 400, 0.1% Nonidet P-40, 0.5 mM DTT, 100–200 mM KCl, 10 μg of BSA, and 1 μg of dI/dC for 10 min at room temperature. Approximately 20,000 cpm (15–20 fmol) of probe was then added, and binding reactions were incubated for an additional 30 min. Competitor DNAs were added simultaneously with probe. For supershift analyses, either a polyclonal antibody to Krox24 (Santa Cruz Biotechnology) or a monoclonal antibody to the N-terminal myc tag (Invitrogen) were preincubated with nuclear extract for 10 min before addition of probe. Complexes were resolved on 6%/0.25× Tris-borate polyacrylamide gels. For the Southwestern blot analyses, NIH 3T3 extracts (64 μg) were separated on a 4.5–15% polyacrylamide gradient gel (Bio-Rad) and electroblotted to nitrocellulose. Transferred proteins were denatured in 6 M guanidine hydrochloride, renatured, and hybridized with a probe containing 6 tandem copies of the TCE as described (23). For immunoblot analyses of Brn2, 30 μg of cell extracts was heated at 100°C, separated on a polyacrylamide gradient gel, and electroblotted to nitrocellulose. The blot was blocked in 0.1% Triton X-100 in phosphate-buffered saline with 1.5% bovine serum albumin. Brn2 proteins were detected using a monoclonal anti-myc primary antibody, a secondary rabbit anti-mouse IgG (H and L) polyclonal antibody (Zymed), and 125I-protein A.
RESULTS
Activity of TN Promoter Elements.
Several regulatory elements are conserved in the promoters from the chicken, mouse, and human TN genes (14, 15, 19). These sequences (Fig. 1) include a Krox element, an octamer motif, a binding site for NF-1, homopolymeric tracts of dA/dT, and a binding site for Antennapedia class HD proteins. To determine how these and other elements contribute to TN promoter activity, we prepared a series of chloramphenicol acetyltransferase reporter constructs in which elements were successively deleted from the 5′ end of the promoter (Fig. 1). These constructs were transfected into mouse NIH 3T3 fibroblasts, rat C6 glioma cells, and mouse N2A neuroblastoma cells, all of which show expression of TN mRNA by reverse transcription–PCR (data not shown).
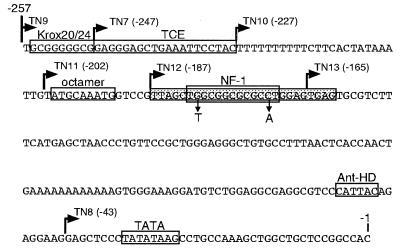
Figure 1.
Sequence of the proximal promoter of the mouse TN gene. The 5′ terminus for each deletion construct is indicated with a right arrow, and regulatory elements are indicated with boxes. The promoter region protected in footprinting experiments is indicated by a shaded box. The NF-1 site within this region is indicated, and the two mutations made in the NF-1 site are shown under the small vertical arrows.
As shown in Table 1, deletion of each element from the TN promoter resulted in activity profiles that were cell-type-dependent. Deletion of the Krox element (compare TN9 to TN7) did not affect TN promoter activity in NIH 3T3 cells, resulted in a 30% reduction of activity in C6 cells, and increased activity 2-fold in N2A cells. Deletion of an additional 20 bp (the TCE) from −247 (TN7) to −227 (TN10) resulted in a 25% reduction in NIH 3T3 cells, a negligible reduction in C6 cells, and a 70% reduction of promoter activity in N2A cells. Deletion of the poly(dT) tract (TN11) reduced promoter activity 20% in NIH 3T3 cells but showed negligible effects on activity in both C6 and N2A cells. Deletion of the octamer motif (TN12) resulted in a 25% increase in chloramphenicol acetyltransferase activity in C6 cells but had no effect in the other two cell lines. Deletion of the NF-1 site (TN13) resulted in a significant reduction of promoter activity in all three cell lines: 60% in NIH 3T3, 70% in C6, and 40% in N2A cells (Table 1). Moreover, the TN12M construct containing a two-base substitution in the NF-1 site (see Fig. 1), which has been shown to abolish binding of NF-1 (24), reduced TN promoter activity between 55% and 80% relative to TN12 in all three cell lines (Table 1). Deletion of the remaining 122 bp, including a poly(dA) tract and an Antennapedia HD binding site (TN8), had little effect on TN promoter activity in NIH 3T3 and C6 cells, but resulted in a 2-fold increase in activity in N2A cells. These data indicate that the NF-1 site is essential for the positive activity of the TN proximal promoter in all three cell lines. Other elements such as binding sites for Krox proteins, the region between −247 and −227 (hereafter designated the TCE), and the poly(dA)/Antennapedia HD region have different cell-specific effects on TN promoter activity.
Binding and Activity.
To determine whether the four TN promoter elements bind nuclear proteins and to relate the formation of DNA–protein complexes to the positive or negative activities of these elements in each cell type, a series of binding experiments was carried out; these experiments involved the regulatory elements and nuclear extracts isolated from the different cell lines. The Krox element, which showed both positive and negative effects on TN promoter activity, formed different complexes in each of these cell lines. The TCE and NF-1 site, each of which contributed to positive regulation of the TN promoter, each showed similar patterns of bound complexes in EMSA experiments in all three cell lines. The octamer motif had no effect in deletion experiments but in cotransfection experiments was found to be required for the induction of TN promoter activity by Brn2. We consider below the detailed data for each of the elements, Krox, TCE, NF-1, and octamer, in order.
Binding Complexes Formed with the Krox Element Are Correlated with Its Positive and Negative Effects on TN Promoter Activity.
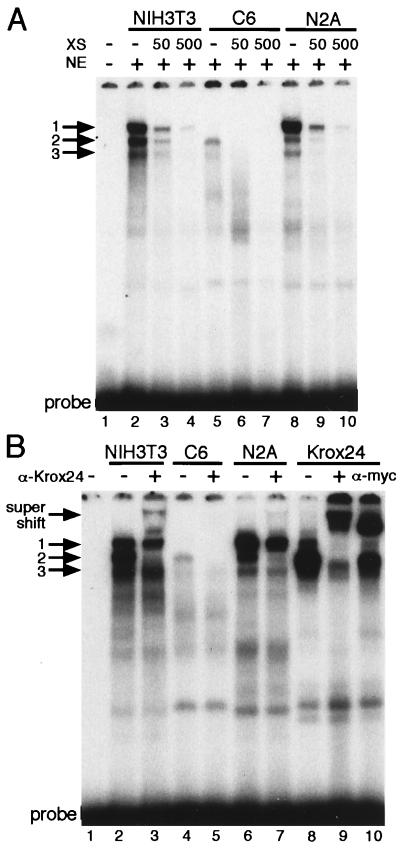
Deletion of the Krox element had no effect in NIH 3T3 cells, a negative effect in C6 cells, and a positive effect in N2A cells (Table 1). In binding experiments using the Krox element with nuclear extracts from the three cell lines, three different DNA–protein complexes (complexes 1–3; see Fig. 2A) were observed, and formation of all complexes was competed with an excess of unlabeled Krox element. Although complex 3 was observed in all three cell lines, only complexes 1 and 2 were correlated with differences in TN promoter activity. In NIH 3T3 cells, in which the Krox element did not affect TN promoter activity, the intensities of complexes 1 and 2 were approximately equal (Fig. 2A, lane 2). In C6 cells, in which the Krox element enhanced TN promoter activity, only complex 2 was observed (Fig. 2A, lane 5). In N2A cells, in which the Krox element reduced TN promoter activity, complex 1 was significantly more abundant than complex 2 (Fig. 2A, lane 8). Thus, complex 2 is correlated with the positive activity, and complex 1 is correlated with the negative activity of the Krox element.
Figure 2.
Binding of nuclear proteins to the TN promoter Krox element. (A) NIH 3T3, C6, and N2A extracts formed three complexes (complexes 1–3), which are indicated by arrows. (B) Complexes 1 and 2 contain Krox24 and are supershifted (see band labeled supershift) with antibodies to either the N-terminal myc tag or Krox24. Abbreviations for binding components: CD, competitor DNA (μg); NE, nucleus extract (μg); XS, molar excess.
Krox20 and Krox24 are proteins that bind to the Krox element and thus are candidates for regulation of the TN promoter. To examine whether complexes 1 and 2 contained these proteins, we prepared expression vectors for Krox20 and Krox24, transfected them into COS-1 cells, and tested cell extracts for binding to the TN promoter Krox element. Krox20 did not comigrate with any of the DNA–protein complexes in NIH 3T3, C6, and N2A cells (data not shown). However, in all three cell types, Krox24 formed a major band that comigrated with complex 2 and a minor band that comigrated with complex 1 (Fig. 2B, compare lanes 2, 4, and 6 to lane 8). These data indicate that Krox24 is likely to be a component of complexes 1 and 2. To show this directly, Krox24 containing the myc tag was produced in COS-1 cells. We then examined whether a polyclonal antibody to Krox24 or an antibody to the myc tag could supershift complexes 1 and 2. This Krox24 fusion protein formed complexes 1 and 2 which were both disrupted and supershifted when antibodies to Krox24 were included in the binding reaction (see Fig. 2B and compare lanes 9 and 8). Addition of an antibody to the myc tag also supershifted these complexes (Fig. 2B, compare lanes 10 and 8).
Complexes 1 and 2, which were formed between nuclear extracts from NIH 3T3, C6, and N2A cells and the Krox element, were also disrupted and supershifted with an antibody to Krox24. In NIH 3T3 cells, Krox24 antibody eliminated complex 2, decreased the amount of complex 1, and produced a supershifted Krox24 complex (Fig. 2B, compare lane 3 to lanes 2 and 9). In C6 cells, the Krox24 antibody disrupted complex 2 completely (Fig. 2B, compare lanes 5 and 4). In N2A cells, the Krox24 antibody prevented the formation of complex 2, decreased the amount of complex 1, and produced a very faint supershifted band (Fig. 2B, compare lane 7 to lanes 6 and 9). These data demonstrate that Krox24 is a component of complexes 1 and 2. Combined with the correlations discussed earlier, they reinforce the conclusion that Krox24 can behave as both a positive and negative regulator of TN expression in different cells; when present in complex 2, Krox24 behaves as an activator, and when present in complex 1, it behaves as a repressor.
Binding Complexes Formed with the TCE and the NF-1 Site.
The TCE did not resemble a binding site for any known transcription factor in the transcription factor database (Genetics Computer Group, tfsites7.2 database). This element formed two binding complexes in all three cell types, and these complexes were competed with an excess of unlabeled TCE (Fig. 3A). To identify proteins that bind to this element, we applied a Southwestern blot procedure using a concatamer probe containing six copies of the TCE (23). As shown in Fig. 3B, this probe bound to two polypeptides with approximate molecular masses of 70 and 130 kDa.
Figure 3.
The TCE binds to nuclear proteins in NIH 3T3, C6 and N2A cells. (A) EMSA of the TCE. An arrow and bracket define the upper and lower complexes, respectively. CD, Competitor DNA; XS, -fold molar excess; NE, nuclear extract. (B) Southwestern blot analysis of proteins that bind to the TCE. NIH 3T3 nuclear extract (64 μg) was separated on a gradient polyacrylamide gel, transferred to nitrocellulose, and hybridized with a probe consisting of six tandem copies of the TCE. The mobility of markers are indicated in kilodaltons (kDa). Arrows mark two proteins with approximate molecular masses of 70 and 130 kDa, respectively.
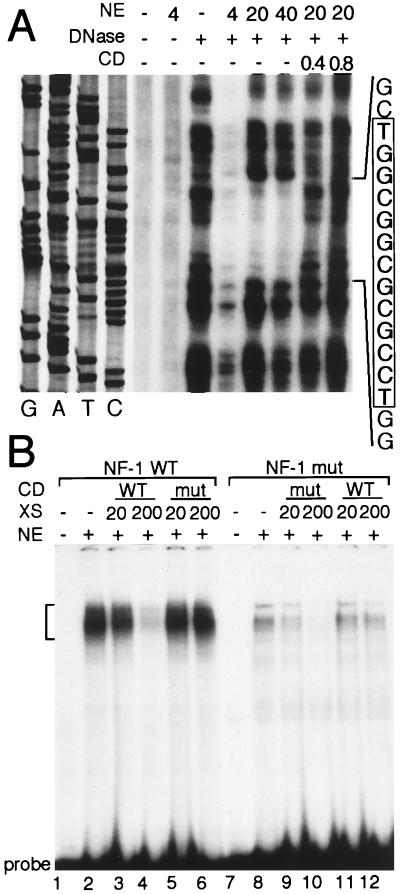
In a DNase I footprinting analysis using nuclear extracts from NIH 3T3 cells, the region between −191 and −166, including the NF-1 site, was protected from nuclease digestion, and a double-stranded competitor containing the NF-1 site eliminated this footprint (Fig. 4A). In an EMSA, the NF-1 site formed similar DNA–protein complexes with nuclear proteins from all three cell lines (Fig. 4B and data not shown). These NF-1 complexes were competed by a 200-fold excess of unlabeled NF-1 site but not by a 200-fold excess of the mutated NF-1 site. In direct binding experiments, a 32P-labeled mutant NF-1 site showed little binding to the NIH 3T3 extracts (Fig. 4B). Moreover, the complexes formed with the mutant NF-1 site did not appear to contain NF-1 inasmuch as addition of unmutated NF-1 competitor DNA did not interfere with formation of these complexes.
Figure 4.
The NF-1 site binds to nuclear proteins of NIH 3T3 cells. (A) DNase I footprint analysis of a portion of the mouse TN proximal promoter. The DNA sequence shown was generated using the 3′ PCR primer. The amount of nuclear extract (NE) and and competitor DNA (CD) is indicated above each lane. The final concentration of DNase I was 0.5 μg/ml in lane 3 and 5 μg/ml in lanes 4–8. The sequence at the right corresponds to the complement of the region protected from DNase I digestion. The NF-1 site within the protected region is boxed and shaded. (B) EMSA of the NF-1 region of the TN promoter. Binding was performed with either the wild-type (WT) or mutant (mut) NF-1 probes. Brackets mark the positions of the shifted and competed complexes. The addition of 7.5 μg of NIH 3T3 nuclear extract (NE) is indicated above each lane. Competitor DNA (CD) and the molar excess (XS) of each are also indicated above each panel.
The Octamer Motif Mediates Activation of the TN Promoter by the POU-HD Protein, Brn2.
The octamer motif, which binds to members of the POU-HD family of transcription factors (25), is conserved among TN promoters from three vertebrate species (1). To investigate whether this element controls the induction of TN promoter activity by POU-HD proteins, we tested the activity of two promoter constructs that either contain or lack the octamer motif (TN11 and TN12, respectively) in cotransfection experiments with expression constructs for three class III POU-HD proteins, Brn2, Brn4, and SCIP. These proteins are all expressed during early neural development (26). Only Brn2 activated the TN promoter in an octamer-dependent manner, and this induction occurred only in N2A cells, not in C6 or NIH 3T3 cells (Table 2 and data not shown).
The Brn2 gene produces three proteins by differential translation initiation. All three forms of Brn2 contain the DNA-binding domain and in DNA-binding experiments can be discriminated as three different complexes, N-Oct3, N-Oct5A, and N-Oct5B (27). N-Oct3 is the full-length translation product of Brn2, whereas N-Oct5A and N-Oct5B are generated by internal translation initiation at two AUG codons that are located downstream of the activation domain. Consequently, N-Oct3 is the only transcriptionally active form of Brn2. N-Oct5A and N-Oct5B lack the N terminus and activation domain of N-Oct3, and although they are capable of binding to the octamer motif, they cannot activate transcription (27).
To examine whether differences in the DNA–protein complexes formed in Brn2-transfected N2A or C6 cells were correlated with the preferential induction of TN promoter activity in N2A cells, we performed a series of binding experiments. A full-length N-Oct3 translation product containing an N-terminal myc tag was synthesized from the Brn2 gene template in an in vitro translation system. This N-Oct3–myc tag fusion protein formed a single complex with the TN promoter octamer motif (Fig. 5A, lane 3) which was supershifted when an antibody to the myc tag was included in the binding reaction (Fig. 5A, lane 4). The positions of these native and supershifted N-Oct3 complexes provided reference standards for comparison of Brn2 complexes formed in N2A and C6 cells.
Figure 5.
Brn2 complexes are assembled on the octamer motif from the TN promoter. (A) Brn2 protein synthesized by in vitro translation or extracts prepared from N2A or C6 cells transfected with a plasmid expressing Brn2 containing an N-terminal myc tag were assayed for binding to a 32P-labeled octamer probe. The three distinct Brn2 complexes N-Oct3, Oct5A, and Oct5B are indicated by arrows. The presence or absence of antibody to the N-terminal myc tag is indicated above each lane. The position of the [35S]methionine-labeled Brn2 synthesized in vitro is indicated with a bracket. (B) Immunoblot analysis of Brn2 expressed in C6 and N2A cells using an antibody to the myc tag. The sizes of molecular mass standards are indicated in kilodaltons.
As shown in Fig. 5A, in N2A cells transfected with the Brn2 expression vector, the octamer motif formed a major complex that comigrated with N-Oct3, and this complex was supershifted with the myc tag antibody (Fig. 5A, lanes 7 and 8). In addition to N-Oct3, minor amounts of the two lower molecular weight complexes N-Oct5A and N-Oct5B were formed (Fig. 5A, lane 7). However, extracts from C6 cells transfected with Brn2 formed complexes lacking N-Oct3 but containing N-Oct5A and N-Oct5B exclusively (Fig. 5A, lane 5). These complexes did not produce a supershifted band when the myc tag antibody was added to the binding reaction, a finding consistent with the lack of an N terminus in the mature Brn2 product N-Oct3 (Fig. 5A, lane 6). Thus, N2A cells expressed a full-length, transcriptionally active form of Brn-2, whereas C6 cells expressed truncated, transcriptionally inactive forms of the protein.
To confirm this conclusion with an independent technique, we performed immunoblot analyses of nuclear extracts from Brn2-transfected N2A and C6 cells using the myc tag antibody to determine whether Brn2 produced in these cells contained the mature N terminus. As shown in Fig. 5B (lane 1), Brn2 expressed in N2A cells contained the myc tag and was detected as a single band of 55 kDa. However, Brn2 expressed in C6 cells was not detected with the antibody, indicating that it did not contain the mature N terminus (Fig. 5B, lane 2). Together, these results suggest that N-Oct3, the transcriptionally active form of Brn2, is produced in N2A cells, whereas N-Oct5A and N-Oct5B, transcriptionally inactive forms of Brn2, are produced in C6 cells. These differences in the Brn2 proteins found in N2A and C6 cells are correlated with the cell-specific and octamer-dependent activation of the TN promoter.
DISCUSSION
TN is expressed in dynamic and place-specific patterns during vertebrate morphogenesis and is reinduced in the adult during tumorigenesis, wound healing, and peripheral nerve regeneration. These dynamic characteristics make it a particular challenge to identify the different regulatory elements and transcription factors that determine the various contexts of TN expression. Collectively, the data that the four TN promoter elements described here are used differentially to produce various patterns of TN gene expression.
In previous studies, we isolated the promoters for the chicken and mouse TN genes (14, 15) so that we could compare their promoter elements and select those that were likely to function in vivo. We found that both TN promoters contain binding sites for HD proteins of the Antennapedia family and are activated by them (16). In this study, we identified various DNA elements in the proximal promoter that are involved in the regulation of TN gene expression. In the three cell lines that we analyzed, at least four different cis regulatory elements and trans factors were found to contribute to TN gene expression. We consider here the separate and coordinate roles of each of these elements and the factors to which they bind.
Krox.
The expression patterns of Krox24 and TN overlap at several embryonic sites, including the brain, bone, cartilage, and various sites of epithelial–mesenchymal transformation. Krox24 has been implicated in cell proliferation and differentiation, and it is rapidly induced by mitogens, tissue or radiation injury, and neuronal excitation (28). It is induced by growth factors, including platelet-derived growth factor, basic fibroblast growth factor, epidermal growth factor, and BMP-4, a member of the transforming growth factor type β superfamily (28, 29). Likewise, TN has been shown to be induced by several growth factors, including members of the transforming growth factor type β and basic fibroblast growth factor families (1, 9).
The differential effects on TN promoter activity by the Krox element are correlated in this work with two distinct DNA–protein complexes that contained the protein Krox24. Complex 2 was correlated with activation, whereas complex 1 was correlated with repression of TN promoter activity. Krox24 is an immediate-early gene that encodes a zinc-finger protein known to act as both a transcriptional activator and a repressor. The protein is phosphorylated, but the functional significance of this modification is unclear (28). Phosphorylation might be responsible for the difference in the size between the Krox24 complexes that we observed in binding experiments. Alternatively, since Krox24 contains an inhibitory domain of 34 amino acids termed R1 (30) that has been shown to interact with several corepressors (31, 32), the larger size of complex 1, which is correlated with the repressor function of Krox24, may represent a ternary complex that includes a corepressor.
TCE.
The TCE appears to be a novel element. It formed two prominent complexes that were similar in the three cell lines examined but contributed significantly to TN promoter activity only in the N2A cell line. Favorable interactions between proteins bound at the TCE and those bound at other sites in the TN promoter, as well as to components of the basic transcription machinery, may be necessary for this element to activate the TN promoter. Using a Southwestern blot analysis, the TCE was shown to bind two proteins of 70 and 130 kDa, which are likely to correspond to the two complexes observed in the EMSA. The identity of these proteins is currently being determined.
NF-1.
NF-1 proteins are a large family of proteins that regulate both constitutive and glial-specific transcription (33, 34) and bind to DNA as homo- and heterodimers (35). A form of NF-1 called NF1-A1 is enriched in the cerebellum (34) and has been shown to transactivate glial-specific promoters (36). In the present study, it was found that the NF-1 site was required for high levels of TN promoter activity in NIH 3T3, C6, and N2A cells, but deletion of this element had the most dramatic effect on TN promoter activity in C6 glioma cells. TN is highly expressed by Bergmann glia during development of the cerebellum and in glial tumors in the adult (4, 7, 13). Recent work in our laboratory has indicated that anibodies to NF1-A1 supershift TN promoter NF-1 complexes in C6 and NIH 3T3 cells (unpublished observations). Thus NF1-A1 is a likely candidate for regulation of TN in glia and possibly in other non-neural cells.
NF-1 has also been found to mediate the response to growth factors of genes for extracellular matrix proteins. For example, an NF-1 site in the α2(I) collagen promoter controls induction of this gene by transforming growth factor type β1 (37). Given these correlations, it will be revealing to compare the roles of the Krox and NF-1 elements in the induction or repression of TN expression by different growth factors.
Octamer.
The octamer motif is recognized by POU-HD proteins. Such factors, particularly those of the POU-III family, which include Brn2, Brn4, and SCIP, play roles in the differentiation of neuronal and glial cell populations. For example, gene knockout of Brn2 (38) disrupts the differentiation of neurons in the hypothalamus and pituitary gland, and knockout of SCIP (39, 40) prevents differentiation of Schwann cells. In our cotransfection experiments that examined the regulation of the TN promoter by Brn2, Brn4, and SCIP, only Brn2 activated the TN promoter in an octamer-dependent manner. Although uncovering this activity in cellular transfection experiments is not sufficient to establish that this POU-HD protein is a natural regulator of TN gene expression, many POU-HD proteins are expressed in the developing nervous system in patterns that correlate with the spatiotemporal expression of TN in neuroepithelial precursors and glial cell lineages (26). Moreover, in the adult, POU-HD proteins are expressed in neuroblastomas and glioblastomas, in which TN expression is prominent.
Extracts from N2A cells transfected with Brn2 formed a high molecular weight complex with the octamer motif containing N-Oct3, a transcriptionally active form of Brn2, whereas extracts from C6 cells formed only lower molecular weight complexes containing N-Oct5A and 5B, both of which are transcriptionally inactive forms of Brn2. N-Oct5A and N-Oct5B arise from internal translation initiation within the Brn2 mRNA at two AUG codons located downstream of the transcriptional activation domain (27). The N2A and C6 cell lines together provide a useful model system to analyze the cis RNA sequences and trans factors that differentially control translation initiation of Brn2 and possibly other POU-HD proteins.
The present studies reveal some of the rich complexity of the interactions of the promoter elements of TN. To define how the patterns of TN gene expression are actually determined during development, a combinatorial analysis of mutated and unmutated promoter elements will have to carried out in transgenic mice. The results of the in vitro experiments described here provide a sound basis for such an analysis.
Acknowledgments
We are grateful to Michael Metzler and Madhu Katragadda for excellent technical assistance and Stephen Jenkinson for help with DNA binding studies. We thank Drs. Bruce Cunningham and George Miklos for critical reading of the manuscript. This work was supported by U.S. Public Health Service Grants HD33576 (to G.M.E.) and NS34493 (to F.S.J.) and a grant from the G. Harold and Leila Mathers Charitable Trust (to G.M.E.). G.M.E. is a consultant to Becton Dickinson and Co.
ABBREVIATIONS
- TN
tenascin
- TCE
TN control element
- HD
homeodomain
- EMSA
electrophoretic mobility-shift assay
References
- 1.Jones F S, Copertino D W. In: Tenascin and Counteradhesive Molecules of the ECM. Crossin K L, editor. Amsterdam: Harwood; 1996. pp. 1–22. [Google Scholar]
- 2.Bourdon M A, Ruoslahti E. J Cell Biol. 1989;108:1149–1155. doi: 10.1083/jcb.108.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prieto A L, Edelman G M, Crossin K L. Proc Natl Acad Sci USA. 1993;90:10154–10158. doi: 10.1073/pnas.90.21.10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossin K L, Hoffman S, Grumet M, Thiery J-P, Edelman G M. J Cell Biol. 1986;102:1917–1930. doi: 10.1083/jcb.102.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossin K L, Prieto A L, Mauro V P. In: Tenascin and Counteradhesive Molecules of the ECM. Crossin K L, editor. Amsterdam: Harwood; 1996. pp. 23–46. [Google Scholar]
- 6.Chuong C-M, Crossin K L, Edelman G M. J Cell Biol. 1987;104:331–342. doi: 10.1083/jcb.104.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prieto A L, Jones F S, Cunningham B A, Crossin K L, Edelman G M. J Cell Biol. 1990;111:685–698. doi: 10.1083/jcb.111.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson H P, Bourdon M A. Annu Rev Cell Biol. 1989;5:71–92. doi: 10.1146/annurev.cb.05.110189.000443. [DOI] [PubMed] [Google Scholar]
- 9.Pearson C A, Pearson D, Shibahara S, Hofsteenge J, Chiquet-Ehrismann R. EMBO J. 1988;7:2977–2982. doi: 10.1002/j.1460-2075.1988.tb03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiners S, Marone M, Rittenhouse J L, Geller H M. Dev Biol. 1993;160:480–493. doi: 10.1006/dbio.1993.1322. [DOI] [PubMed] [Google Scholar]
- 11.Rettig W J, Triche T J, Garin-Chesa P. Brain Res. 1989;487:171–177. doi: 10.1016/0006-8993(89)90954-2. [DOI] [PubMed] [Google Scholar]
- 12.Carnemolla B, Siri A, Borsi L, Zardi L. In: Tenascin and Counteradhesive Molecules of the ECM. Crossin K L, editor. Amsterdam: Harwood; 1996. pp. 89–108. [Google Scholar]
- 13.Faissner A. In: Tenascin and Counteradhesive Molecules of the ECM. Crossin K L, editor. Amsterdam: Harwood; 1996. pp. 47–88. [Google Scholar]
- 14.Jones F S, Crossin K L, Cunningham B A, Edelman G M. Proc Natl Acad Sci USA. 1990;87:6497–6501. doi: 10.1073/pnas.87.17.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Copertino D W, Jenkinson S, Jones F S, Edelman G M. Proc Natl Acad Sci USA. 1995;92:2131–2135. doi: 10.1073/pnas.92.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones F S, Chalepakis G, Gruss P, Edelman G M. Proc Natl Acad Sci USA. 1992;89:2091–2095. doi: 10.1073/pnas.89.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumgartner S, Martin D, Hagios C, Chiquet-Ehrismann R. EMBO J. 1994;13:3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R. Cell. 1994;77:587–598. doi: 10.1016/0092-8674(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 19.Gherzi R, Carnemolla B, Siri A, Ponassi M, Balza E, Zardi L. J Biol Chem. 1995;7:1–6. doi: 10.1074/jbc.270.7.3429. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman G, editors. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1990. [Google Scholar]
- 22.Schreiber E, Matthias P, Müller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419–6423. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 24.Vries E, van Driel W, van den Heuvel S J L, van der Vliet P C. EMBO J. 1987;6:161–168. doi: 10.1002/j.1460-2075.1987.tb04734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schöler H R. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- 26.Wegner M, Drolet D W, Rosenfeld M G. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 27.Scheiber E, Tobler A, Malipiero U, Schaffner W, Fontana A. Nucleic Acids Res. 1993;21:253–258. doi: 10.1093/nar/21.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gashler A, Sukhatme V P. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 29.Vainio S, Karavanova I, Jowett A, Thesleff I. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 30.Russo M W, Matheny C, Milbrandt J. Mol Cell Biol. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo M W, Sevetson B R, Milbrandt J. Proc Natl Acad Sci USA. 1995;92:6873–6977. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svaren J, Sevetson B R, Apel E D, Zimonjic D B, Popescu N C, Milbrandt J. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santoro C, Mermod N, Andrews P C, Tjian R. Nature (London) 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 34.Inoue T, Tamura T-a, Furuichi T, Mikoshiba K. J Biol Chem. 1990;265:19065–19070. [PubMed] [Google Scholar]
- 35.Kruse U, Sippel A E. FEBS Lett. 1994;348:46–50. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- 36.Krebs C J, Dey B, Kumar G. J Neurochem. 1996;66:1354–1361. doi: 10.1046/j.1471-4159.1996.66041354.x. [DOI] [PubMed] [Google Scholar]
- 37.Rossi P, Karsenty G, Roberts A B, Roche N S, Sporn M B, deCombrugghe B. Cell. 1988;52:405–414. doi: 10.1016/s0092-8674(88)80033-3. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama J, Fukuda M N, Fredette B, Ranscht B, Fukuda M. Proc Natl Acad Sci USA. 1995;92:7031–7035. doi: 10.1073/pnas.92.15.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bermingham J B, Jr, Scherer S S, O’Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Genes Dev. 1996;10:1751–1761. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 40.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]