Abstract
It is possible to evolve RNA enzymes in a continuous manner by employing a simple serial-transfer procedure. This method was previously applied only to descendants of one unusually fast-reacting RNA enzyme with RNA ligase activity. The present study establishes a second continuously evolving RNA enzyme, also with RNA ligase activity, but with a completely independent evolutionary origin. Critical to achieving the fast catalytic rate necessary for continuous evolution, development of this enzyme entailed the addition and evolutionary maturation of a 35-nucleotide accessory domain and the application of highly stringent selection pressure, with reaction times as short as 15 ms. Once established, continuous evolution was carried out for 80 successive transfers, maintaining the population against an overall dilution of 10207-fold. The resulting RNA enzymes exhibited ≈105-fold improvement in catalytic efficiency, compared with the starting molecules, and became dependent on a structural feature of the substrate that previously conferred no selective advantage. This adaptation was eliminated by deleting the substrate feature and then carrying out 20 additional transfers of continuous evolution, which resulted in molecules with even greater catalytic activity. Now that two distinct species of continuously evolving enzymes have been established, it is possible to conduct molecular ecology experiments in which the two are made to compete for limited resources within a common environment.
Keywords: in vitro evolution, RNA catalysis, RNA ligase, selection, serial transfer
Darwinian evolution has been applied to populations of RNA molecules in the laboratory to generate RNA enzymes (ribozymes) that catalyze a variety of reactions (1–3). These experiments involve repeated rounds of selective amplification and mutagenesis to drive the evolving population toward desired catalytic behaviors. Of the many ribozymes that have been obtained by in vitro evolution, six different classes catalyze the RNA-templated joining of an oligonucleotide 3′-hydroxyl and an oligonucleotide 5′-triphosphate, forming a 3′,5′-phosphodiester and releasing inorganic pyrophosphate (4–9). This reaction is similar to that carried out by protein polymerases that catalyze the synthesis of biological RNAs.
All six of the known 3′,5′-ligase ribozymes achieve rate enhancements of several orders of magnitude, compared with the uncatalyzed rate of templated RNA ligation, which is ≈10−7 min−1 (10). Notably, only derivatives of the class I ligase, first described by Bartel, Szostak, and colleagues (4, 11), have been able to achieve a catalytic rate of >1 min−1. Because of the superior catalytic rate of this ribozyme, it has been chosen as the starting point for several subsequent in vitro evolution experiments. One series of studies led to the development of a sequence-general, RNA-dependent RNA polymerase ribozyme that can add as many as 20 NTPs (12, 13). This effort involved several structural modifications of the class I ligase, including the addition of a large random-sequence accessory domain, followed by 24 rounds of stepwise evolution and extensive screening of cloned individuals.
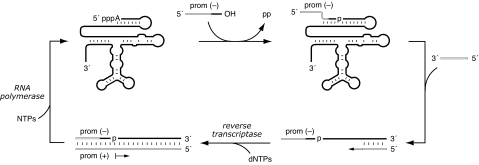
Another line of investigation beginning with the class I ligase resulted in the establishment of a system for the continuous in vitro evolution of ribozymes (14). This system operates at a constant temperature within a common reaction vessel. It employs a promoter-containing oligonucleotide substrate and two proteins, reverse transcriptase and RNA polymerase, to bring about the selective amplification of reactive ribozymes (Fig. 1). The RNA population expands exponentially and can be diluted serially to allow sustained exponential growth. The serial transfers also provide an opportunity to change the selection pressures applied to the evolving population, for example, lowering the Mg2+ concentration (15), raising or lowering the pH (16), or introducing a ribozyme-cleaving DNA enzyme (17).
Fig. 1.
Scheme for continuous evolution of ligase ribozymes. RNA and DNA are depicted as solid and open lines, respectively. Ligation of a chimeric DNA–RNA substrate by the ribozyme results in attachment of the T7 promoter sequence (prom) to the 5′ end of the ribozyme. Reverse transcription of reacted, but not unreacted, ribozymes results in the formation of a cDNA that serves as a template for transcription of multiple progeny RNAs.
Continuous evolution is the most efficient and powerful method for evolving functional molecules in vitro, but thus far has only been applied to descendants of the class I ligase. It has been hypothesized that continuous evolution could be used to evolve other ligase ribozymes and perhaps RNA molecules that catalyze other bond-forming reactions (18). However, no other ligase ribozyme has a catalytic rate as fast as that of the class I ligase. A fast catalytic rate is essential for continuous evolution because the ribozyme must react before it becomes reverse transcribed. Only if the ribozyme reacts before it is reverse transcribed will it acquire a promoter sequence that allows the resulting cDNA to give rise to progeny ribozymes.
The goal of the present study was to develop a second ribozyme that can evolve in a continuous manner. This goal is a challenge for RNA catalysis and an opportunity to develop a distinct species of continuously evolving ribozymes that might be made to operate in competition or cooperation with the class I ligase. The DSL ligase ribozyme, described previously by Inoue and colleagues (8), was chosen as the starting material for this effort. The DSL ligase was obtained by using a combination of rational design and in vitro selection. The framework for this ribozyme is a structural scaffold based on a naturally occurring RNA motif, to which was added a duplex region for templated ligation and a potential catalytic domain of 30 random-sequence nucleotides. A large pool of these RNA molecules was subjected to 10 rounds of in vitro selection, resulting in a variety of ligase ribozymes. The most active catalyst identified is the cis-DSL-1S ribozyme, which contains 140 nucleotides and has a kcat of 0.066 min−1 and a Km of 940 nM, measured in the presence of 50 mM MgCl2 and 200 mM KCl at pH 7.5 and 37°C (8, 19).
The catalytic activity of the DSL ribozyme is noteworthy for a ligase obtained after only 10 rounds of selective amplification and derived from only 30 random-sequence nucleotides. The small size and modular construction of the DSL ligase were regarded as advantageous features for the development of a second continuously evolving ribozyme. In the present study, the DSL ribozyme was structurally modified to accommodate the promoter-containing substrate needed for continuous evolution, and an accessory domain of 35 random-sequence nucleotides was added to provide the opportunity for substantial improvement in catalytic rate. A population of these modified ribozymes was subjected to vigorous stepwise evolution, with reaction times as short as 15 ms, to drive the improvement of catalytic function. This stringent selection pressure ultimately resulted in a population of DSL-derived ribozymes that were capable of undergoing continuous evolution. Once established, continuous evolution can be continued indefinitely, resulting in further improvement in the catalytic activity of the ribozymes. This work demonstrates that continuous evolution can be applied to ribozymes in addition to the class I ligase, setting the stage for molecular ecology studies in which two or more species evolve within a common environment.
Results
Stepwise Evolution.
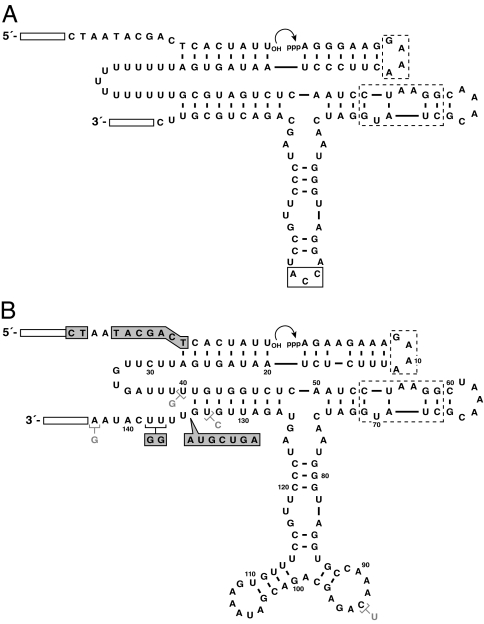
The DSL ribozyme was modified in several ways to render it compatible with the continuous evolution system (Fig. 2A). Continuous evolution requires the RNA-catalyzed ligation of a chimeric DNA–RNA substrate that has the sequence of an RNA polymerase promoter. This substrate is more AU-rich at its 3′ end, compared with the preferred substrate for the DSL ribozyme, making it necessary to extend the template region of the ribozyme so that it could bind the substrate with sufficient affinity. Extension of the template region necessitated removal of a triplex structural element, which was replaced by a flexible oligouridylate linker (Fig. 2A). Additionally, the primer binding site at the 3′ end of the ribozyme was replaced by nucleotides of a different sequence that were known to be compatible with continuous evolution.
Fig. 2.
Sequence and secondary structure of the starting and evolved DSL ribozymes. Open rectangles indicate primer binding sites, which are immutable during the course of evolution. Dashed boxes indicate the tetraloop and tetraloop receptor. (A) Modified DSL ligase that was used as the basis for constructing the starting pool of ribozymes. Boxed residues were replaced by 35 random-sequence nucleotides. (B) T80-1 ligase, which was obtained after transfer 80 of continuous evolution. Ribozyme residues in shaded boxes indicate changes present in all clones obtained after transfer 100. These changes confer complementarity to substrate residues shown in shaded boxes. Additional mutations present in the T100-1 ligase are shown in gray.
The modified DSL ligase had substantially diminished activity with the promoter-containing substrate, exhibiting a kcat of 1.76 ± 0.02 × 10−2 min−1 and a Km of 4.0 ± 1.0 μM when the reaction was carried out in the presence of 25 mM MgCl2 at pH 8.5 and 37°C and almost no detectible activity (kobs < 5 × 10−4 min−1) when the reaction was carried out under the conditions necessary for continuous evolution. The continuous evolution mixture contains 8.8 mM NTPs and dNTPs, which reduce the concentration of free Mg2+ to 16.2 mM, accounting for the reduction in activity under these conditions. An improvement in catalytic rate of >10,000-fold under continuous evolution conditions would be needed for the modified DSL ribozyme to have a rate comparable to that of the class I ligase.
A starting pool of ≈1014 variants was prepared based on partial randomization of the modified form of the DSL ligase ribozyme. Random mutations were introduced at a frequency of 9% per nucleotide position at all positions except the tetraloop, tetraloop receptor, and 3′ primer binding site (Fig. 2A). This pool was subjected to eight rounds of stepwise evolution, in which the molecules were required to react in progressively shorter times, eventually reaching 30 s. The ligated RNAs, representing catalytically competent molecules, were isolated by gel electrophoresis and then amplified to generate progeny RNAs to begin the next round of evolution. During the first three rounds, the reaction mixture contained only ribozymes, substrate, 25 mM MgCl2, and 50 mM EPPS (pH 8.5). In subsequent rounds, the reaction mixture more closely mimicked that used in continuous evolution, also containing the cDNA primer, NTPs, dNTPs, spermidine, and DTT, but not the polymerase proteins. Diversity was maintained within the population by performing standard mutagenic PCR after round seven, which introduces mutations as a frequency of 0.7% per nucleotide position (20).
The activity of the evolving population showed steady improvement through the eighth round before reaching a maximum. Individuals were cloned from the population after the eighth round and sequenced. A representative clone, designated as R8-11, which conformed to the consensus sequence of all of the analyzed clones, was subjected to kinetic analysis. It exhibited a kcat of 0.14 ± 0.01 min−1 and a Km of 1.3 ± 0.2 μM in the presence of 25 mM MgCl2, and a kcat of 5.5 ± 0.4 × 10−3 min−1 and a Km of 1.2 ± 0.3 μM under continuous evolution conditions. Despite the >10-fold improvement in catalytic rate under the desired reaction conditions, further selection, with greater emphasis on rate enhancement, would be required for the ligase to be capable of undergoing continuous evolution.
A second phase of stepwise evolution was initiated beginning with a pool of randomized variants of the R8-11 ribozyme. As with the first pool, random mutations were introduced at a frequency of 9% per nucleotide position, sparing the tetraloop and tetraloop receptor. Additionally, the primer binding sites at the 5′ end of the substrate and the 3′ end of the ribozyme were changed to prevent cross-contamination with previously amplified molecules. Finally, an accessory domain of 35 random-sequence nucleotides was added to the distal portion of the central stem-loop within the catalytic domain (Fig. 2A). This second pool was subjected to 13 rounds of rigorous selective amplification, with progressively decreasing reaction times ranging from 60 min to 15 ms. The shortest reaction time that reliably could be achieved by manual pipetting was 15 s. Reaction times as short as 15 ms were achieved by using a quench-flow apparatus. Such reaction times are the fastest ever reported for an in vitro selection experiment.
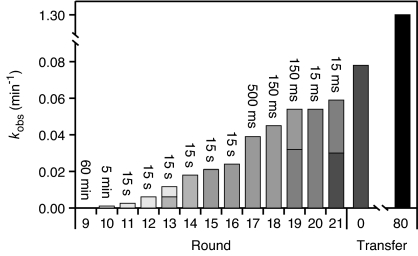
Mutagenic PCR was carried out after rounds 12, 14, 18, and 21 to maintain diversity within the population. The reaction rate of the population was monitored at every round and showed steady improvement throughout rounds 9–21 of the evolution process (Fig. 3). At several points during this procedure, the population was assayed for its ability to initiate continuous evolution. RNAs isolated after round 21, which demonstrated a kobs of 0.080 ± 0.002 min−1 under continuous evolution conditions, proved capable of evolving continuously. It is likely that only the fastest individuals within this heterogeneous population were competent in continuous evolution, and thus poised to dominate the population upon further evolution.
Fig. 3.
Increase in catalytic activity of the population over successive rounds of evolution. Reaction times for each round of stepwise evolution are given above each bar. The activity of the populations used to initiate continuous evolution (transfer 0) and obtained after transfer 80 are shown at the right. Reactions were carried out under continuous evolution conditions, but excluding the protein enzymes. Increasingly darker shading during the stepwise rounds indicates successive applications of mutagenic PCR, with split bars indicating the activity of both the mutagenized and nonmutagenized populations.
Continuous Evolution.
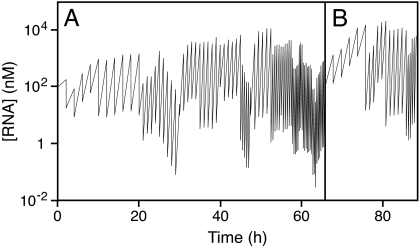
A seed of 100 nM (2 pmol) RNA from the population obtained after round 21 was used to initiate continuous evolution. After a 2-h incubation, a small aliquot was taken from the continuous evolution mixture and transferred to a new reaction vessel that contained a fresh supply of substrate and other reagents. This process was continued for 80 successive transfers (Fig. 4A), with error-prone PCR performed after transfers 10, 20, 30, 40, and 50. The concentration of RNA was measured before and after each transfer to monitor the course of evolution. The dilution factor between transfers was increased as tolerated, and the time between transfers was reduced as tolerated. Over the course of 80 transfers, the evolving population was maintained against an overall dilution of ≈10207 (the product of 80 dilutions of 10- to 1,000-fold each). By the end of this process, the population was capable of 1,000-fold selective amplification in a 20-min interval. This rate of growth is comparable to that of the continuously evolving class I ligase ribozyme (14).
Fig. 4.
Time course of continuous evolution. The concentration of RNA was measured before and after each transfer (zigzag line). The time between transfers was decreased as tolerated. (A) Eighty transfers were carried out over 65 h, with a dilution of 10- to 1,000-fold between transfers. (B) Twenty additional transfers were carried out over 23 h by using the 5′-truncated substrate.
Individuals were cloned from the population at the outset of continuous evolution and after transfers 15, 50, and 80. Sequence analysis of these individuals was used to monitor the genetic composition of the population, and ligation assays were carried out to assess the evolution of catalytic function. Among the ribozymes isolated at the outset of continuous evolution, two distinct sequence classes within the 35-nucleotide accessory domain were identified (type I and II). The sequences are listed in supporting information (SI) Tables 1 and 2.
After 15 transfers of continuous evolution, both sequence classes were still well represented in the population. The consensus of 12 type I sequences contained 14 point mutations, two deletions, and two insertions, compared with the type I consensus sequence at the outset of continuous evolution. In contrast, the consensus of nine type II sequences obtained after 15 transfers contained only one point mutation. A representative clone from each sequence class was chosen for further study. The type I ribozyme exhibited a kobs of 0.39 ± 0.02 min−1, whereas the type II ribozyme had a kobs of 0.28 ± 0.01 min−1. Both ribozymes were tested for their ability to undergo exponential amplification in the continuous evolution system and exhibited nearly superimposable growth profiles (SI Fig. 6).
After 50 transfers of continuous evolution, only type I ribozymes remained among the clones that were sequenced (SI Table 1). These individuals had only a few scattered mutations within the 35-nucleotide accessory domain, including one highly conserved point mutation, compared with the consensus type I sequence identified after transfer 15. In addition, two clones isolated after transfer 50 exhibited a seven- or eight-nucleotide deletion near the 5′ end of the accessory domain. This deletion became fixed within the population by transfer 80. Two clones contained a point mutation at the 3′ end of the accessory domain that also became fixed after transfer 80.
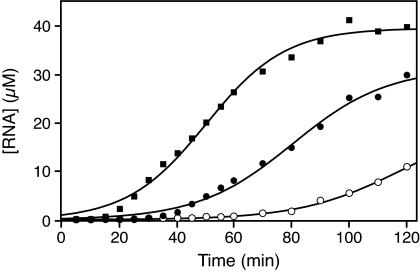
There was little sequence variation among 10 clones that were isolated after the 80th transfer (SI Tables 1 and 2). A typical clone, designated as T80-1, whose sequence conforms to the consensus sequence, was chosen for further study. The amplification profile of the T80-1 ribozyme is consistent with a logistic growth curve, indicative of exponential growth subject to a limiting constraint (Fig. 5). Here the constraint is the finite amount of substrate and other resources present in the continuous evolution mixture.
Fig. 5.
Amplification profiles of individual ribozymes in the continuous evolution mixture. Filled and open circles, T80-1 ligase using either the full-length or truncated substrate, respectively; filled square, T100-1 ligase using the truncated substrate. The concentration of RNA was determined at various times, and the data were fit to the logistic equation: [RNA] = a/(1 + be−ct), where a is the maximum extent of amplification and c is the exponential growth rate. Curvilinear regression coefficients were in the range of 0.996–0.999.
Kinetic and Secondary Structure Analyses.
The catalytic activity of the T80-1 ligase ribozyme was examined under continuous evolution conditions (excluding the protein enzymes), exhibiting a kcat of 1.39 ± 0.06 min−1 and a Km of 0.14 ± 0.04 μM (SI Fig. 7). This result corresponds to a catalytic efficiency, kcat/Km, of 1 × 107 M−1·min−1, which is a 2,000-fold improvement compared with the behavior of the R8-11 ligase and a >80,000-fold improvement compared with the starting DSL ligase ribozyme. The parent cis-DSL-1S ribozyme has a catalytic efficiency of 7 × 104 M−1·min−1 under its preferred reaction conditions (19).
To verify that the continuously evolving ribozyme retained the ability to form a 3′,5′-phosphodiester, the ligation junction was probed by using RNase T2, which cleaves 3′,5′- but not 2′,5′-phosphodiester linkages of RNA and does not cleave DNA. An unlabeled ribozyme was allowed to react with a 5′-32P-labeled substrate that contained a single ribonucleotide at its 3′ end. The ligated products were isolated by polyacrylamide gel electrophoresis and then digested with RNase T2 (SI Fig. 8). Because the substrate only contained a single 3′-terminal ribonucleotide, the labeled product of RNase T2 digestion would be the same length as the product of complete alkaline digestion if a 3′,5′-phosphodiester had formed, which was indeed the case.
Enzymatic and chemical probing studies were carried out to analyze the secondary structure of the T80-1 ligase, especially the newly evolved accessory domain. 5′-32P-labeled ligated products were partially digested with either RNase A or RNase T1 under either native conditions (mimicking the conditions of continuous evolution) or denaturing conditions (SI Fig. 9A). Unlabeled ligated products were subjected to chemical modification, employing either dimethyl sulfate or kethoxal under either native or denaturing conditions, followed by reverse transcription using a 5′-32P-labeled DNA primer (SI Fig. 9B). These two methods were used in combination to confirm that the overall secondary structure of the DSL ribozyme had not changed during the 80 transfers of continuous evolution. The data also were used to establish a secondary structural model for the newly evolved accessory domain, which appears to form two short stem-loop elements that join to the remainder of the ribozyme by a three-way junction (Fig. 2B).
Substrate Dependency of the Evolved Ribozyme.
The T80-1 ligase was challenged to ligate a 5′-truncated substrate that contains the entire RNA polymerase promoter sequence, but lacks the upstream primer binding site that was used for PCR amplification and cloning. The ribozyme reacted poorly with this truncated substrate, with a kobs of only 0.032 ± 0.001 min−1. This finding corresponds to a rate reduction of ≈50-fold compared with the reaction with the full-length substrate. The T80-1 ribozyme was tested for its ability to undergo exponential amplification by using the truncated substrate and demonstrated much slower exponential growth compared with its behavior with the full-length substrate (Fig. 5).
The population of ribozymes obtained after transfer 80 was subjected to mutagenic PCR and then used to initiate 20 additional transfers of continuous evolution employing the truncated substrate (Fig. 4B). The evolving population quickly regained the ability to undergo rapid exponential growth. After transfer 100, individuals were cloned from the population and sequenced. There was little sequence variation among 10 clones that were analyzed (SI Tables 1 and 2), and all contained a seven-nucleotide insertion (5′-AGUCGUA-3′) and two U → G changes located near the 3′ end of the ribozyme (Fig. 2B). These mutations confer complementarity between this region of the ribozyme and 11 nucleotides at the 5′ end of the truncated substrate.
One of the cloned individuals, designated as T100-1, whose sequence conforms to the consensus of all sequenced clones, was found to undergo rapid exponential growth in the presence of the truncated substrate, even exceeding that of the T80-1 ribozyme with the full-length substrate (Fig. 5). The kobs of the T100-1 ribozyme with the truncated substrate is 2.9 ± 0.3 min−1, which is about twice as fast as the rate of the T80-1 ribozyme with the full-length substrate.
Mutations were introduced within the seven-nucleotide insertion of the T100-1 ribozyme, replacing the sequence 5′-AGUCGUA-3′ by 5′-ACAGCAA-3′ (mutated residues in italics). The mutant ribozyme was unable to ligate the truncated substrate (SI Fig. 10). Full ligation activity was restored, however, by employing a different substrate that contained five compensatory mutations. This result indicates that the T100-1 ribozyme binds the truncated substrate through a newly evolved Watson–Crick pairing interaction that augments the preexisting substrate-binding domain (Fig. 2B). To exclude the possibility that the two pairings engage two different substrate molecules, the mutant ribozyme was tested with an equimolar mixture of the standard truncated substrate and an all-DNA version of the substrate bearing the compensatory mutations. The latter cannot undergo ligation, but in principle could satisfy the requirement for a second pairing interaction. There was no detectable activity under these conditions, consistent with the hypothesis that the same substrate molecule is involved in both pairing interactions.
Discussion
The Need for Speed.
Continuous evolution is the most powerful method currently available for the in vitro evolution of catalytic function. However, since the development of this method a decade ago (14), its application has been limited to variants of the class I ligase ribozyme. With the present study, the DSL ribozyme becomes the second continuously evolving functional macromolecule. It is desirable to have multiple classes of ribozymes that are capable of undergoing continuous evolution, not only to facilitate the evolution of each catalyst individually, but also to enable more complex experiments in which two or more molecular species evolve within a common environment.
The most significant obstacle to achieving the continuous evolution of a ribozyme is the need to attain a sufficiently fast reaction rate. The ribozyme must catalyze ligation of the promoter-containing substrate before becoming inactivated by reverse transcription. The minimum catalytic rate needed depends on the sequence and structure of the ribozyme and various aspects of the reaction environment. For the class I and DSL ribozymes under the conditions of continuous evolution, the kobs for reverse transcription was determined to be 4.1 ± 0.3 min−1 and 2.4 ± 0.2 min−1, respectively. Thus, it appears that a catalytic rate for the ribozyme of ≈1 min−1, in addition to other context-dependent adaptive features, is required for it to cross the threshold to continuous evolution. The class I ligase was preadapted for continuous evolution because its catalytic rate was already >10 min−1 under the reaction conditions for which it was originally optimized (4, 11). Yet many rounds of stepwise evolution were required to adapt the ribozyme to the promoter-containing substrate and to the reaction conditions necessary for continuous evolution (14). In contrast, the slow-reacting DSL ligase required substantial improvement in its catalytic rate. This goal was achieved by using a quench-flow apparatus to select molecules that could react in times as short as 15 ms. This technique could be applied more broadly, apart from the goal of continuous evolution, to obtain other fast-reacting catalysts.
A critical step in the development of the continuously evolving DSL ribozyme was the addition and evolutionary maturation of the 35-nucleotide accessory domain, which greatly augments the behavior of the catalytic core. The added nucleotides provided an opportunity to evolve peripheral elements that result in enhanced catalytic performance. A similar situation exists for various biological ribozymes, where the catalytic core alone provides a modest level of activity and peripheral elements bring about a substantial increase in the catalytic rate. The hammerhead and Neurospora Varkud satellite ribozymes, for example, both catalyze a self-cleavage reaction that yields products bearing a 2′,3′-cyclic phosphate and 5′-hydroxyl. Minimized forms of these ribozymes that contain only the phylogenetically conserved residues necessary for catalysis have markedly reduced activity, compared with the full-length motifs that occur in nature (21, 22). In both cases, the additional peripheral elements allow formation of tertiary contacts that stabilize the active conformation of the ribozyme and enhance its catalytic rate.
Adaptation to Substrate Features.
The substrate molecule that is used in continuous evolution contains the sequence of an RNA polymerase promoter. For convenience, additional nucleotides often are placed upstream of the promoter sequence to serve as a primer binding site for PCR amplification and subsequent cloning. In the present study, 18 nucleotides were placed upstream of the promoter, having a sequence that was irrelevant to the substrate-recognition properties of the ribozyme. During 80 transfers of continuous evolution, however, the ribozyme became dependent on the added nucleotides. The activity of the T80-1 ribozyme is reduced ≈50-fold when provided a substrate that lacks the added nucleotides, and its rate of exponential amplification in the continuous evolution system decreases accordingly (Fig. 5). Thus, what began as an irrelevant appendage was coopted by evolution to confer enhanced selective advantage to the ribozyme.
It is not uncommon in biological evolution for traits to arise that initially have no selective advantage, but subsequently become coopted in a way that enhances fitness (23). Evolutionary biologists are challenged to distinguish between a primary adaptation and a neutral change that subsequently was “exapted” (24) for selective advantage. Laboratory evolution, in contrast, generates a complete historical record in the form of the molecules that are archived at each step of the evolution process. One can return to any point along an evolutionary pathway and determine which molecular traits are present and whether they provide a selective advantage.
The added substrate nucleotides, which initially had no effect on catalysis, led to an exaptation, seen in the strong dependence of the activity of the T80-1 ribozyme on this feature of the substrate. When the basis for this exaptation was removed, the ribozymes exhibited a substantial decrease in fitness. After 20 additional transfers of continuous evolution, however, the population adapted in a new way. The ribozymes obtained after the 100th transfer had acquired a seven-nucleotide insertion and a two-nucleotide substitution that improved their ability to bind the truncated substrate (Fig. 2B). These changes enhanced the catalytic activity of the ribozymes and their corresponding rate of amplification in the continuous evolution mixture (Fig. 5), an example of how continuous evolution can be used as a tool to address questions pertaining to the biochemical basis for evolutionary adaptation. By developing a second ribozyme that is capable of continuous evolution, the scope of these experiments can be broadened considerably.
Materials and Methods
Stepwise Evolution.
DNA templates for the starting pool were generated by a 10-cycle PCR employing two synthetic oligonucleotides having the sequence 5′-GAAGTAATACGACTCACTATTAGGGAAGGAAACTTCCCTAATAGTGATTTT T T T T T TTTTTTGCGTAGTCTCAATCCTAAGGCAAACGCTATG-3′ and 5′-CGTGACCGAGCCACTAGGAACGCAGTCTG C T A G GGAACGGATGGTCCTACCCATTGATCCATAGCG T T T G CCTTAG-3′ (T7 RNA polymerase promoter sequence underlined; nucleotides randomized at 9% degeneracy in italics). The PCR products, consisting of ≈1014 different sequences, were used as templates for in vitro transcription. The resulting RNAs were purified by denaturing PAGE. A starting population of ≈20 copies each of 1014 different RNA molecules (3.3 nmol) were challenged to ligate a chimeric DNA-RNA substrate having the sequence 5′-CTTGACGTCAGCCTGGACTAATACGACTCACUAUU-3′ (T7 promoter sequence underlined; RNA residues in bold). During rounds 1–3 of in vitro evolution, the ligation reaction was carried out in the presence of 1 μM ribozyme, 2.5 μM substrate, 25 mM MgCl2, and 50 mM EPPS (pH 8.5) at 37°C. During rounds 4–8, the reaction mixture also contained 2.5 μM cDNA primer having the sequence 5′-CGTGACCGAGCCACTAG-3′, 2 mM each NTP, 0.2 mM each dNTP, 50 mM KCl, 2 mM spermidine, and 4 mM DTT. The ligated products were purified by PAGE, eluted from the gel, reverse transcribed, PCR amplified, and then forward transcribed to produce progeny RNAs, which again were gel purified.
DNA templates for the second pool (after round 8) were generated by a single-cycle extension reaction employing two oligonucleotides having the sequence 5′-GAAGTAATACGACTCACTATTAGAGAAGGAAACTTCCCTAATAGTGATTTTTGTGGTTTTTTGCGCAGTCTCAATCCTAAGGCAAACGCTATGG-3′ and 5′-GGATGGCACGGAGTCAGGGAGCGCAGTCTGCTAGGGAACGGA-N35-TCCTACCCATTGATCCATAGCGTTTGCCTTA-3′ (T7 promoter sequence underlined; nucleotides randomized at 9% degeneracy in italics; random nucleotides denoted as N). The resulting double-stranded DNAs, consisting of ≈1014 different sequences, were transcribed to produce RNAs, which were gel purified. A starting population of ≈20 copies each of 1014 different RNA molecules (3.0 nmol) were challenged to ligate a substrate having the sequence 5′-CATCGTGCCTTGCTGCTCTAATACGACTCACUAUU-3′. The reaction conditions for rounds 9–16 were as in rounds 4–8, except that a different cDNA primer was used, having the sequence 5′-GGATGGCACGGAGTCAG-3′.
Rounds 17–21 were carried out by using a KinTek model RQF-3 quench-flow apparatus to achieve progressively shorter reaction times, ranging from 500 to 15 ms. Separate syringes of the quench-flow apparatus were used to deliver 15 μl each of a solution of ribozyme and substrate, each also containing all of the other reaction components and maintained at 37°C. A third syringe was used to deliver 87 μl of a quench solution containing 40 mM Na2EDTA after a precisely specified interval. The drive solution used to propel the other solutions through the reaction loop contained 25 mM MgCl2, 50 mM KCl, 50 mM EPPS (8.5), 2 mM spermidine, and 4 mM DTT. Five sequential 30-μL scale reactions were carried out and pooled before the reacted RNAs were selectively amplified.
Continuous Evolution.
After the last round of stepwise evolution (round 21), ligated RNAs were isolated by PAGE, eluted from the gel, reverse transcribed, PCR amplified, forward transcribed, and gel purified as described earlier. Two pmol of the purified RNAs were used to initiate continuous evolution, which was carried out as described previously (14). The reaction mixture contained 5 μM substrate and 2.5 μM cDNA primer (having the same sequence as in rounds 9–21 of stepwise evolution), 2 mM each NTP, 0.2 mM each dNTP, 25 mM MgCl2, 50 mM KCl, 50 mM EPPS (8.5), 2 mM spermidine, 4 mM DTT, 10 units/μl SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), 2.5 units/μl T7 RNA polymerase, and 0.001 units/μl inorganic pyrophosphatase. Successive reaction mixtures were incubated for times decreasing from 2 h to 20 min, always at 37°C. A small aliquot of each completed reaction mixture was transferred to the next reaction vessel, with the dilution increasing from 10- to 1,000-fold over the course of 80 transfers. The same procedure was used for transfers 81–100 except that the substrate had the sequence 5′-CTAATACGACTCACUAUU-3′.
During the course of continuous evolution, error-prone PCR was performed periodically on aliquots taken from the population, employing either standard mutagenic PCR with an error rate of 0.7% per nucleotide position (20), which was done after transfers 10, 20, and 80, or hypermutagenic PCR with an error rate of ≈10% per nucleotide position (25), which was done after transfers 30, 40, and 50. In each case, the PCR products were transcribed, the resulting RNAs were purified by PAGE, and the purified RNAs were used to reinitiate continuous evolution.
Kinetic Analysis.
Ligation reactions were carried out under substrate-excess conditions, employing [α32-P]ATP-labeled ribozyme and 10 different concentrations of substrate ranging from 50 nM to 10 μM. In all cases, the concentration of substrate was in at least 10-fold excess over the concentration of ribozyme. The other components of the reaction mixture were identical to those used in continuous evolution except the protein enzymes were omitted. The reaction products were separated by PAGE and quantitated by using a PhosophorImager (Molecular Dynamics, Sunnyvale, CA). Values for kobs were determined for each concentration of substrate based on at least six data points, which were fit to the equation y = a (1− e−kobst · t), where y is the fraction reacted and a is the maximum extent as determined from long time points. Values for kcat and Km were obtained from a Michaelis–Menten plot of kobs versus substrate concentration.
RNase T2 digestion was performed to confirm the 3′,5′-regiospecificity of the reaction. Ligation was carried out by using 5′-32P-labeled 5′-CATCGTGCCTTGCTGCTCTAATACGACTCACTATU-3′. The ligated products were purified by PAGE and then subjected to complete RNase T2 digestion in a mixture containing 10 nM ligated products, 1 unit/μl RNase T2, 2 mM Na2EDTA, and 50 mM NaOAc (pH 5.2), which was incubated at 37°C for 30 or 60 min. The digested products were analyzed by PAGE, comparing them to materials resulting from alkaline digestion employing 100 mM sodium carbonate (pH 9.0) at 95°C for 60 min.
The observed rate of reverse transcription was measured under continuous evolution conditions, excluding substrate and T7 RNA polymerase and employing 0.5 μM ribozyme and 2.5 μM fluorescently labeled cDNA primer. After incubation for 0.25, 0.5, 1, 4, or 10 min, the reaction products were separated by PAGE, and the fraction of extended primer was determined. These data were fit to the equation y = a (1 − e−kobst · t), where y is the fraction extended and a is the fraction extended at long time points.
Secondary Structure Analysis.
Partial RNase digestions were carried out by using 5′-32P-labeled ligated products, which were incubated under either continuous evolution conditions (but excluding protein enzymes, NTPs, dNTPs, and DTT) or denaturing conditions, employing 6 M urea, 25 mM sodium citrate (pH 5.0), and 1 mM Na2EDTA at 55°C. Carrier tRNA at 0.17 μg/μl was included in the mixture to ensure uniform enzymatic digestion. Chemical modification assays employing the ligated products and either dimethyl sulfate or kethoxal were carried out as previously described (26). The reaction conditions were as before except that the cDNA primer was omitted.
Supplementary Material
Acknowledgments
We thank Sarah Hamilton for assistance in cloning molecules obtained at the start and after transfer 15 of continuous evolution. This work was supported by National Aeronautics and Space Administration Grant NNX07AJ23G and National Science Foundation Grant MCB-0614614.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707490104/DC1.
References
- 1.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Joyce GF. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Li N, Ellington AD. Chem Biodiversity. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- 4.Ekland EH, Szostak JW, Bartel DP. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 5.Robertson MP, Ellington AD. Nat Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger L, Wright MC, Joyce GF. Proc Natl Acad Sci USA. 1999;96:14712–14717. doi: 10.1073/pnas.96.26.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers J, Joyce GF. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikawa Y, Tsuda K, Matsumura S, Inoue T. Proc Natl Acad Sci USA. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshioka W, Ikawa Y, Jaeger L, Shiraishi H, Inoue T. RNA. 2004;10:1900–1906. doi: 10.1261/rna.7170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohatgi R, Bartel DP, Szostak JW. J Am Chem Soc. 1996;118:3332–3339. doi: 10.1021/ja953712b. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP, Szostak JW. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 12.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 13.Zaher HS, Unrau PJ. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright MC, Joyce GF. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt T, Lehman N. Chem Biol. 1999;6:857–869. doi: 10.1016/s1074-5521(00)80005-8. [DOI] [PubMed] [Google Scholar]
- 16.Kühne H, Joyce GF. J Mol Evol. 2003;57:292–298. doi: 10.1007/s00239-003-2480-z. [DOI] [PubMed] [Google Scholar]
- 17.Ordoukhanian P, Joyce GF. Chem Biol. 1999;6:881–889. doi: 10.1016/s1074-5521(00)80007-1. [DOI] [PubMed] [Google Scholar]
- 18.Johns GC, Joyce GF. J Mol Evol. 2005;61:253–263. doi: 10.1007/s00239-004-0307-1. [DOI] [PubMed] [Google Scholar]
- 19.Horie S, Ikawa Y, Inoue T. Biochem Biophys Res Comm. 2006;339:115–121. doi: 10.1016/j.bbrc.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Cadwell RC, Joyce GF. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 21.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 22.Zamel R, Poon A, Jaikaran D, Anderson A, Olive J, De Abreu D, Collins RA. Proc Natl Acad Sci USA. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace AR. J Linnaean Soc. 1859;3:53–62. [Google Scholar]
- 24.Gould SJ, Vrba ES. Paleobiology. 1982;8:4–15. [Google Scholar]
- 25.Vartanian J-P, Henry M, Wain-Hobson S. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern S, Moazed D, Noller HF. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.