Abstract
Phospholipase Cε (PLCε) has been suggested to transduce signals from small GTPases, but its biological function has not yet been clarified. Using astrocytes from PLCε-deficient mice, we demonstrate that endogenous G protein-coupled receptors (GPCRs) for lysophosphatidic acid, sphingosine 1-phosphate, and thrombin regulate phosphoinositide hydrolysis primarily through PLCε. Stimulation by lysophospholipids occurs through Gi, whereas thrombin activates PLC through Rho. Further studies reveal that PLCε is required for thrombin- but not LPA-induced sustained ERK activation and DNA synthesis, providing a novel mechanism for GPCR and Rho signaling to cell proliferation. The requirement for PLCε in this pathway can be explained by its role as a guanine nucleotide exchange factor for Rap1. Thus, PLCε serves to transduce mitogenic signals through a mechanism distinct from its role in generation of PLC-derived second messengers.
Keywords: mitogenesis, small GTPases, thrombin, lysophospholipids, guanine nucleotide exchange factor
Stimulation of a variety of cell surface receptor types, most prominently G protein-coupled receptors (GPCRs), leads to the activation of phospholipase C (PLC). Hydrolysis of phosphatidylinositol (4,5)bisphosphate (PIP2) generates the second messengers inositol (1,4,5)trisphosphate (InsP3) and diacylglycerol (DAG) (1). Thirteen mammalian PLC isozymes, divided into six families, have been identified. All contain conserved catalytic regions as well as subtype-specific domains that allow for a plethora of specific regulatory mechanisms. The best characterized of these are the PLCβ family, regulated by direct binding of the heterotrimeric G protein subunits αq and βγ, and PLCγ regulated by receptor and nonreceptor protein tyrosine kinases (2).The same second messengers are generated by all of the PLCs. Thus, regardless of the extracellular signal- and PLC isoform-activated, InsP3 will be formed and trigger Ca2+ release from intracellular stores, and DAG accumulation will lead to activation of protein kinase C (3). Undoubtedly, there are distinct spatiotemporal aspects to the signals elicited by different receptors and classes of PLC, but it remains unclear why so many PLC isoforms, capable of generating the same signal, should exist.
An emerging area in biology has been the recognition that many proteins identified on the basis of a particular activity serve additional functions. The regulators of G protein signaling (RGS) proteins are paradigmatic in this regard. These proteins contain not only canonical RGS sequences dedicated to GTP hydrolysis but also non-RGS domains and motifs with alternative functions. Thus, not only do they turn off GPCR signaling but in addition they can enhance small G protein activation, serve as effectors, and act as scaffold proteins (4). PLCε, an isoform of PLC that was discovered fewer than 6 years ago, likewise has the structure of a multifunctional signaling protein (5–7). It contains not only PLC catalytic regions but also, in its N-terminal region, a CDC25 homology domain, which acts as a guanine nucleotide exchange factor (GEF) and an activator of Ras family GTPases, specifically of Rap1 (8). The pathways for activation of PLCε are also unique. Studies using heterologous expression demonstrate that, in contrast to PLCβ, PLCε is not activated by Gαq but rather by Gβγ and Gα12 signaling (5, 9). Most remarkably, PLCε is activated by direct binding of small GTPases, including Ras, Rap1, and Rap2B (6, 7, 10–12) as well as by RhoA (13). Thus, PLCε appears to serve as a nexus for signaling, responding to inputs from a broad range of receptor activation and uniquely linking PLC to small-GTPase pathways (14, 15). The studies reported here use astrocytes derived from PLCε knockout (KO) mice and agonists for a series of endogenous GPCRs on these cells to demonstrate that PLCε is required not only to generate the expected second messengers but that it is also an obligate mediator of Rap1 and ERK activation and thus of cell proliferation in response to thrombin.
Results and Discussion
PLCε knockout mice, generated in the A.V.S. laboratory and described in previous publications (16, 17), have been shown to lack PLCε expression in a variety of tissues, including lung and heart. We used RT-PCR to confirm the presence of PLCε message in primary astrocytes from wild-type (WT) neonatal mice and the absence of PLCε mRNA in primary astrocytes prepared from the KO mice (Fig. 1).
Fig. 1.
Expression of PLCε mRNA in WT and KO astrocytes. Total cellular RNA was extracted from cultured WT and PLCε KO astrocytes, and the mRNA levels of PLCε were analyzed by RT-PCR followed by electrophoresis on 1% agarose gels and staining with ethidium bromide. The β-actin mRNA was used as a control.
Receptors for thrombin (PAR1,3,4), lysophosphatidic acid (LPA1–5 receptors), sphingosine 1-phosphate (S1P1–5 receptors), and carbachol [M1–5 muscarinic acetylcholine receptor (mAChR)] couple to G proteins that can regulate PLC activity. All four of these agonists were able to increase accumulation of inositol phosphates (InsPs) in primary mouse astrocytes, consistent with published findings (18).
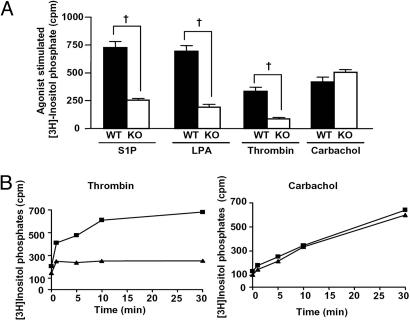
The extent to which PLCε is involved in phospholipase signaling by each of these GPCR agonists was then evaluated. InsP production in response to 0.5 unit/ml thrombin, 10 μM LPA, 5 μM S1P, and 500 μM carbachol was compared in WT and PLCε KO primary astrocytes (Fig. 2A). S1P, LPA, and thrombin responses were significantly reduced in KO cells (64 ± 2%, 73 ± 2%, and 75 ± 4% inhibition, respectively), demonstrating that PLCε is the predominant PLC mediating agonist-stimulated InsP production in response to these ligands.
Fig. 2.
S1P, LPA, and thrombin induce InsP production through PLCε. Cultured WT and PLCε KO astrocytes were serum-starved overnight in the presence of [3H]inositol. (A) Cells were treated for 30 min with vehicle (BSA), 5 μM S1P, 10 μM LPA, 0.5 unit/ml thrombin, or 500 μM carbachol in the presence of LiCl before isolation of [3H]InsPs. Data are reported as cpm of agonist-stimulated InsP production (cpm of agonists − cpm of vehicle). Cpm of vehicle were 290 ± 26 and 183 ± 10 for WT and KO cells, respectively. Data are averaged from five experiments performed in triplicate on at least two different astrocyte culture preparations and are presented as means ± SEM. †, P < 0.001, one-way ANOVA. (B) Time course of thrombin- and carbachol-induced InsP production. Cultured WT (■) and KO (▴) astrocytes were serum-starved overnight in DMEM without FBS containing [3H]inositol and challenged with 0.5 unit/ml thrombin or 500 μM carbachol in the presence of LiCl for the indicated times before isolation of [3H]InsP. Data are reported as cpm recovered per well in triplicate and are averages from two independent experiments.
The response to carbachol was notably unaffected by PLCε deletion. The differential dependence of thrombin and carbachol on PLCε was confirmed by examining a full time course of agonist-induced InsP formation in WT and PLCε KO cells (Fig. 2B). Thrombin elicited a small increase in InsP accumulation at 1 min but had no further affect in the absence of PLCε, whereas carbachol continuously increased InsP formation in both WT and KO cells. The observation that InsP generation in response to carbachol is PLCε-independent is consistent with the well known involvement of PLCβ in Gq-mediated responses to M1 and M3 mAChR (19). In addition, recent data indicate that heterologous expression of M1 receptors (20) or constitutively active Gq (5) does not activate PLCε. Taken together, these data extend previous studies using heterologous expression of PLCε (12) or receptors (20) and RNAi (21) to demonstrate that endogenous PLCε is selectively activated through a subset of GPCRs.
The effects of thrombin and carbachol on PLCε activation were further compared by real-time imaging of DAG production in living astrocytes. These studies used a recently described FRET-based reporter called DAGR (22), in which intermolecular FRET between yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) is induced by DAG binding. After expression of DAGR in WT astrocytes, the addition of 0.5 unit/ml thrombin or 500 μM carbachol induced rapid and significant changes in the YFP:CFP emission ratio, indicating formation of DAG (Fig. 3 A and B, respectively, and D). In contrast, in PLCε KO cells expressing DAGR thrombin did not elicit a FRET signal (Fig. 3 C and D). Subsequent addition of carbachol to the same cell was able to induce an increase in FRET (Fig. 3 C and D), demonstrating responsiveness of the KO cells to an alternative PLC pathway for DAG generation. These data further confirm the marked dependence of thrombin, but not carbachol, on PLCε for phospholipid hydrolysis and second-messenger generation.
Fig. 3.
Thrombin but not carbachol induces diacylglycerol production through PLCε. (A–C) DAGR was expressed in primary WT (A and B) and PLCε KO (C) astrocytes, and cells were treated with 0.5 unit/ml thrombin (A and C) or 500 μM carbachol (B and C) and imaged for changes in FRET. Data are averages from at least three experiments performed on two different cell preparations. (D) Representative image from experiments shown in A–C shows increase in DAG production (red shift of pseudocolored FRET ratio image). Thr, thrombin; Carb, carbachol.
To determine which G protein-signaling pathways might be involved in PLCε activation, we examined the effect of pertussis toxin (PTX) treatment on agonist-induced InsP formation. As shown in Fig. 4A, pretreatment with 100 ng/ml PTX revealed a nearly complete dependence on Gi/o signaling for S1P and LPA stimulation of InsP production (65 ± 8% and 74 ± 9% inhibition by PTX, respectively), whereas the response to thrombin was unaffected by PTX treatment. Similar observations were made when the PLCε-dependent component of the InsP response was calculated based on the difference in InsP production between WT and PLCε KO cells (72 ± 17% and 83 ± 12% inhibition of the S1P and LPA response by PTX, no significant inhibition of the thrombin response, data not shown). It has been reported that the α subunits of Gi do not activate PLCε (5), but PLCε can be activated by βγ (9), either directly or through activation of Ras (7). Thus, PTX sensitivity implies dependence on G protein βγ subunits for PLCε activation by S1P and LPA.
Fig. 4.
Thrombin uniquely requires Rho for PLC activation and PLCε for DNA synthesis via a Rho-dependent pathway. (A) Cultured WT astrocytes were serum-starved overnight in medium containing [3H]inositol in the absence or presence of 100 ng/ml PTX, or they were treated for 4 h before challenge with agonists with 6 μg/ml exoenzyme C3. Cells were treated for 30 min with vehicle (BSA), 5 μM S1P, 10 μM LPA, 0.5 unit/ml thrombin, or 500 μM carbachol in the presence of LiCl before isolation of [3H]InsP. (B and C) Primary culture of WT (B and C) and PLCε KO (B) astrocytes were serum-starved for 24 h in the absence (B) or presence of 100 ng/ml PTX (C). Cell-permeable exoenzyme C3 (500 ng/ml) was added 4 h (C) before treatment with vehicle (BSA), 5 μM S1P, 10 μM LPA, 0.5 unit/ml thrombin, or 500 μM carbachol for an additional 24h. (A and C) Responses in the presence of inhibitors are reported as percentage of response obtained in the absence of inhibitor. (B) Data are expressed as percentage increase of [3H]thymidine incorporation over control (BSA). Data are averaged from at least two experiments performed in triplicate on two different astrocyte culture preparations and are presented as means ± SEM. *, P < 0.05; **, P < 0.01; †, P < 0.001, one-way ANOVA.
Thrombin-mediated PLCε activation has been shown in heterologous systems to involve coupling to G12/13 and Rho (12, 20). To examine involvement of this pathway in thrombin receptor-mediated activation of endogenous PLC, WT astrocytes were treated with 6 μg/ml exoenzyme C3 to inhibit Rho function (Fig. 4A). Treatment with C3 significantly impaired production of InsPs in response to thrombin (68 ± 13% inhibition), which is consistent with evidence that RhoA binds to and can directly activate PLCε (13). In contrast, the InsP response to S1P and LPA was unaffected by C3 pretreatment (Fig. 4A). It is notable in this regard that studies using heterologous expression of PLCε indicated that S1P and LPA stimulate this enzyme through G12/13 and only partially through Gi (12, 20), whereas our findings suggest that in an endogenous system these agonists couple most efficaciously through Gi/o to PLCε activation.
Activation of GPCRs modulates various aspects of cellular growth and proliferation, processes that are primarily controlled by small Ras-related GTPases and their downstream effector, the mitogen-activated protein (MAP/ERK) kinases (23). Activation of RhoGTPases further contributes to Ras-mediated cell proliferation or transformation (24–26). Ligands such as LPA, S1P, and thrombin are uniquely poised to induce these responses because of their ability to activate RhoA as well as Ras signaling pathways. The targets through which RhoA participates in mitogenesis have not been fully identified. To establish whether PLCε could serve as a mediator of RhoA signaling to astrocyte proliferation, primary cultures of WT and PLCε KO astrocytes were treated with S1P, LPA, thrombin, or carbachol, and DNA synthesis was monitored by the incorporation of [3H]thymidine. S1P elicited a modest increase in [3H]thymidine incorporation (173 ± 43% SEM increase vs. control), whereas more robust responses were seen with thrombin (504 ± 98% SEM increase vs. control) and LPA (540 ± 71% SEM increase vs. control) (Fig. 4B). These findings confirm the previously reported ability of LPA, S1P, and thrombin to induce mitogenesis in primary astrocytes in culture (18). Thrombin-mediated [3H]thymidine incorporation was found to be markedly reduced (66 ± 2% inhibition) in PLCε KO cells. In addition, this requirement was selective because there was no effect of PLCε deletion on S1P- or LPA-mediated [3H]thymidine incorporation. Thus, although PLCε plays a significant and quantitatively similar role in the overall PLC response to all three agonists, it plays a unique and unexpectedly critical role in the mitogenic response to thrombin. The signaling pathways by which thrombin and LPA stimulate [3H]thymidine incorporation were also distinguished by their sensitivity to PTX and C3 toxin (Fig. 4C). LPA-mediated DNA synthesis was significantly inhibited by PTX (56 ± 10% inhibition) but not significantly by C3 treatment, whereas the thrombin response was PTX-independent but inhibited by C3 (64 ± 5% inhibition), indicating a unique requirement for Rho activation.
Several studies suggest that the duration of ERK activation is a key determinant of cell proliferation (27, 28). S1P, LPA, or thrombin receptor activation induce prolonged ERK1/2 phosphorylation in primary astrocytes (18, 29). Based on the findings above, we explored the possibility that PLCε was involved in the regulation of ERK1/2 and specifically critical for sustained ERK signaling in response to thrombin. The time course of thrombin- and LPA-induced ERK phosphorylation was examined over a 12-h period in both WT and KO cells. Although both LPA and thrombin induced prolonged ERK activation (Fig. 5 A and B), only the response to thrombin was decreased in the absence of PLCε (Fig. 5 B and C), which mirrors the differential dependence of the proliferative responses to the two agonists on PLCε. Also of note is that the initial ERK response (seen at 15 min) was not affected by the absence of PLCε, whereas the ability of thrombin to cause ERK1/2 activation at times between 1 and 12 h was lost. The requirement of PLCε for the sustained response to thrombin is consistent with the demonstration that PLCε can mediate sustained InsP formation and the suggestion that it may be involved in pathways that require long-term responses (21).
Fig. 5.
PLCε and its CDC25 domain are required for sustained thrombin-mediated ERK1/2 phosphorylation. (A–C) Serum-starved WT and PLCε KO astrocytes were treated with vehicle (BSA), 10 μM LPA, or 0.5 unit/ml thrombin for the times shown. Lysates were immunoblotted for P-ERK1/2 or total ERK1/2 and are representative of three independent experiments. Densitometric measurements (C) were conducted with a ChemImager and are shown graphically as means ± SEM of percentage agonist-induced increase in ERK1/2 phosphorylation over vehicle. **, P < 0.01, one-way ANOVA. (D,E) PLCε WT and KO astrocytes were infected with 200 moi of adenovirus expressing either EYFP (control), PLCε wt (PLCε) or PLCε lacking the CDC25 domain (ΔCDC25PLCε). And experiments were performed 24 h after the infection. (D) Cells were treated for 30 min with vehicle (BSA) or 0.5 unit/ml thrombin in the presence of LiCl before isolation of [3H]InsP. Data are shown as cpm of agonist-stimulated InsP production (cpm of agonists − cpm of vehicle) and are averaged from two experiments performed in triplicate on two different astrocyte culture preparations and are presented as means ± SEM. *, P < 0.05, one-way ANOVA. (E) Experiments were carried out on infected KO cells as described for A–C and are representative of two separate experiments performed on two different cell preparations.
LPA and thrombin are equally dependent on PLCε for phosphoinositide hydrolysis, but the data presented above demonstrate that only thrombin requires PLCε for ERK activation and DNA synthesis. This finding suggests that thrombin elicits ERK activation and DNA synthesis through actions of PLCε that are independent of or coincidental with the generation of the PIP2-derived second messengers, InsP3 and DAG. We hypothesized that these actions resulted from the GEF activity of the CDC25 domain, and we tested it by infecting KO cells with adenovirus encoding PLCε WT or PLCε lacking the CDC25 domain (ΔCDC25PLCε). As shown in Fig. 5D, expression of either PLCε WT or ΔCDC25PLCε rescued the loss of thrombin-stimulated InsP formation seen in the KO cells. Notably, however, only PLCε WT was able to rescue the long-term ERK1/2 phosphorylation in response to thrombin (Fig. 5E), demonstrating a requirement for the CDC25 domain on PLCε in ERK activation.
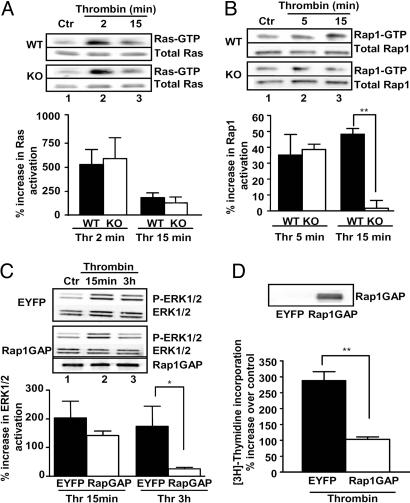
The findings presented above are consistent with a biological role for PLCε in serving as a GEF for and thereby activator of the Ras family GTPases. The role of PLCε in mediating Ras and Rap1 activation by thrombin was therefore tested. As shown in Fig. 6A, thrombin induced a rapid and transient increase in activated Ras. This response was not attenuated in cells from PLCε KO mice. In contrast, Rap1 activation induced by a 15-min treatment with thrombin was virtually abolished in cells lacking PLCε (Fig. 6B). The link between PLCε-mediated Rap activation and its role in thrombin-mediated ERK and DNA synthesis was then examined. Astrocytes were infected with an adenovirus-encoding Rap1GAP, a protein that catalyzes the hydrolysis of GTP to GDP to convert Rap1 into its inactive GDP-bound state (30). Expression of Rap1GAP markedly inhibited the ability of thrombin to elicit prolonged ERK1/2 phosphorylation (Fig. 6C) and significantly decreased thrombin-mediated thymidine incorporation (64 ± 3% inhibition; Fig. 6D) in WT astrocytes. Thus, the requirement for PLCε in thrombin-mediated ERK activation would appear to reflect the unique involvement of Rap1 in this response.
Fig. 6.
PLCε and Rap1 involvement in thrombin-induced sustained ERK activation and DNA synthesis. (A and B) Serum-starved WT and PLCε KO astrocytes were treated with vehicle (BSA) or 0.5 unit/ml thrombin (Thr) for the times shown. Active Ras and Rap1 were precipitated with GST-fused Ras-binding domain of Raf-1 (Raf-1RBD)-agarose beads or GST-fused Rap1-binding domain of RalGDS (RalGDS RBD)-agarose beads, respectively, and probed with anti-Ras or anti-Rap1 antibody (Upper); whole-cell lysates were immunoblotted for total Ras or Rap1 (Lower). Immunoblots shown are representative of three independent experiments. Data were quantitated and are shown graphically as means of percentage increase in Ras (A) or Rap1 (B) activation as means ± SEM. **, P < 0.01, one-way ANOVA. (C and D) WT astrocytes were infected with either EYFP control or Rap1GAP adenovirus (300 moi). Experiments were performed 48 h after the infection, and cells lysates were analyzed for Rap1GAP expression by Western blotting. (C) Serum-starved WT astrocytes were treated with vehicle (BSA) or thrombin for the times shown. Lysates were immunoblotted for P-ERK1/2 or total ERK1/2 and are representative of three independent experiments. Densitometric measures were conducted by using a ChemImager and are shown graphically as means ± SEM of percentage agonist-induced increase in ERK1/2 phosphorylation over vehicle, *, P < 0.05, one-way ANOVA. (D) Serum-starved WT astrocytes were treated with vehicle (BSA) or 0.5 unit/ml thrombin for 24 h, and [3H]thymidine was added for the final 4 h of agonist treatment. Data are expressed as percentage increase of [3H]thymidine incorporation over control (BSA). Data are averaged from two experiments performed in triplicate on two different astrocyte culture preparations and are presented as means ± SEM. **, P < 0.01, one-way ANOVA.
It has been shown that Rap1 can induce ERK activation through its interaction with B-Raf (8, 31, 32). Rap1 localization to endosomal or perinuclear compartments facilitates B-Raf activation (33, 34). Interestingly, PLCε has been shown to be localized to and retained at the Golgi, where it stimulates B-Raf, in response to Rap1 activation through its CDC25 domain (8). Further studies will be necessary to test the hypothesis that thrombin-induced activation of PLCε leads to its distribution to a subcellular compartment in which it is accessible for sustained signaling and mitogenesis through Rap1/B-Raf/ERK activation.
In summary, we have discovered that PLCε is a critical downstream effector for LPA-, S1P-, and thrombin-induced InsP generation by their endogenous receptors. This response occurs through the activation of a Gi pathway for the LPA- and S1P-mediated response and through a G12/13/Rho pathway for the thrombin-mediated response. Intriguingly, whereas receptors for all of these ligands depend, to a significant extent, on PLCε for phospholipid hydrolysis, thrombin also utilizes the CDC25 domain of PLCε to elicit ERK1/2 activation and to stimulate mitogenesis. These data demonstrate a unique and unexpected role for PLCε as a multifunctional signaling protein, serving to coordinate PIP2 hydrolysis with small-G protein activation and thus to mediate proliferative responses to GPCR stimulation.
Methods
Breeding and Genotyping of PLCε KO Mice.
PLCε +/− mice were bred to generate homozygous animals. PCR analysis was used to identify +/+, −/−, and +/− animals. +/+ and −/− mice were bred to generate, respectively, WT and KO pups for astrocytes isolation.
Primary Culture of Astrocytes.
Cultured astrocytes were prepared from P1–P3 postnatal mice. Cerebral cortices were isolated from adherent meninges, and tissue was dissociated and trypsinized. Dissociated cells were plated into 75-cm2 flasks by using high-glucose DMEM supplemented with 10% FBS/2 mM glutamine/100 units/ml penicillin/100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). After 11 days in culture, cells were shaken on a rotary shaker at 240 rpm for 16–22 h to remove oligodendrocytes and plated at one-third of their confluent density. Cultures were maintained at 37°C in a humidified 5% CO2-containing atmosphere. A homogeneous population of astrocytes was verified by immunofluorescence staining with glial fibrillary acidic protein. Cells were used between the first and the third passages; in all of the experiments, WT and PLCε KO cells were used at the same passage. The experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California at San Diego and followed the guidelines of the National Institute of Health (35).
Transduction of Astrocytes with Adenovirus.
Astrocytes were plated and infected the next day for 1 h in serum-free medium with either 200 multiplicity of infection (moi) of enhanced yellow fluorescent protein (EYFP), PLCε WT, or ΔCDC25PLCε or with 300 moi of Rap1GAP adenovirus. PLCε WT adenovirus expresses the PLCε gene with a FLAG tag at the C terminus and the EYFP under two separate mouse cytomegalovirus (MCMV) promoters. ΔCDC25PLCε adenovirus expresses the PLCε mutant lacking a portion of the CDC25 GEF domain, FLAG-tagged at the C terminus and the EYFP gene, under two separate MCMV promoters. PLCε WT and ΔCDC25PLCε expression was detectable at 24 h by PCR and at 48 h by PCR and Western blotting after immunoprecipitation with anti-FLAG antibody. Experiments were performed 24 h after infection, at which time the level of PLCε overexpression was comparable with the levels of endogenous PLCε expression. Rap1GAP adenovirus, driven by a cytomegalovirus promoter, was kindly provided by Patrick Casey, Duke University (36); cells were analyzed 48 h after infection to maximize RapGAP expression and Rap1 inhibition.
RT-PCR of PLCε.
Total RNA was extracted from primary astrocytes by using an RNeasy kit (Invitrogen), and first-strand cDNA was synthesized by with a SuperScript III first-strand synthesis kit for RT-PCR (Invitrogen) according to the manufacturer's instructions. Specific primers for mPLCε and β-actin have been selected on the basis of the sequences published previously (37). The PCR-mediated amplification of cDNA used Taq polymerase, in optimized conditions.
[3H]InsP Production.
Subconfluent cells were serum-starved for 18 h in DMEM containing 0.1% BSA and 1 μCi/ml [3H]inositol (PerkinElmer, Waltham, MA). Cells were placed in medium supplemented with 10 mM LiCl for 15 min before challenge with agonists, and they were then washed with ice-cold PBS and rinsed with 10% trichloroacetic acid. Scraped cells were centrifuged and trichloroacetic acid was extracted with water-saturated ether. [3H]InsPs were isolated by ion exchange chromatography, and radioactivity was then measured by liquid scintillation counting.
[3H]Thymidine Incorporation Assay.
Subconfluent cells were serum-starved overnight in DMEM containing 0.1% BSA. Medium was then replaced, and cells were stimulated with the agonists for 24 h and incubated with [3H]thymidine (PerkinElmer) for the last 4 h at a final concentration of 1 μCi/ml. Cells were washed with ice-cold PBS, and acid-soluble radioactivity was removed by treatment with 10% trichloroacetic acid. The acid-insoluble portion was recovered by digestion with 2% Na2CO3 in 0.1 M NaOH. Radioactivity was then measured by liquid scintillation counting.
ERK1/2 Phosphorylation.
Subconfluent cells were serum-starved overnight in DMEM containing 0.1% BSA, and stimulations at 37°C were terminated by the addition of ice-cold lysis buffer (20 mM Tris·HCl, pH 7.6/250 mM NaCl/3 mM EGTA/3 mM EDTA/20 mM glycerophosphate/1 mM phenylmethylsulfonyl fluoride/1 mM DTT/0.1 mM Na3VO4/10 μg/ml leupeptin/10 μg/ml aprotinin/0.5% Nonidet P-40). The protein content was measured and compensated for before SDS/PAGE. Cell lysates were solubilized by boiling at 95°C for 5 min in Laemmli buffer and subjected to electrophoresis and immunoblotting by using an anti-phospho-ERK1/2 antibody. The immunoreactive proteins were visualized by chemiluminescence with a ChemImager (Alpha Innotech Corp., San Leandro, CA). Membranes were stripped and reincubated with anti-ERK1/2 antibody.
Ras and Rap1 Pull-Down Assay.
Ras and Rap1 activation assay was performed following the affinity precipitation protocol provided by the manufacturer (Ras and Rap1 pull-down assay kit; Upstate Biotechnology, Lake Placid, NY). Briefly, cells were serum-starved overnight, treated with appropriate stimuli, and then lysed. Lysates were incubated with 5 μg of Raf-1 RBD or 26 μg of RalGDS RBD for 45 min at 4°C and then centrifuged to pellet the agarose beads. Agarose beads were washed with lysate buffer, and the pellets were resuspended in 2× Laemmli sample buffer containing DTT, boiled for 5 min, and finally centrifuged. The supernatant was collected, and cellular proteins were resolved by SDS/PAGE and analyzed by Western blotting (see above).
Cell Imaging.
Astrocytes were plated onto glass coverslips in 35-mm dishes, and at 60% confluence cells were transfected with FuGENE 6 (Roche Applied Sciences, Indianapolis, IN). Twenty-four hours later, cells were washed with PBS containing 1 mM Ca2+ and imaged in the dark at room temperature. DAGR images were acquired as described previously (22) on a Zeiss Axiovert microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) with a cooled charge-couple device camera (Photometric) controlled by MetaFluor software (Universal Imaging Corp., Downingtown, PA). Measurements were restricted to cells with similar DAGR expression.
Statistical Analysis.
Statistical differences between pairwise comparisons were evaluated by using Tukey's multicomparison analysis after one-way ANOVA with Prism software (GraphPad, San Diego, CA).
Acknowledgments
This work was supported by National Institutes of Health Grants GM 36927 (to J.H.B.) and GM 53536 (to A.V.S.) and Oral Cellular and Molecular Training Grant T32 DE07202 (to E.A.O.).
Abbreviations
- CFP
cyan fluorescent protein
- DAG
diacylglycerol
- EYFP
enhanced yellow fluorescent protein
- GEF
guanine nucleotide exchange factor
- GPCR
G protein-coupled receptor
- InsP
inositol phosphate
- InsP3
inositol (1,4,5)trisphosphate
- KO
knockout
- LPA
lysophosphatidic acid
- mAChR
muscarinic acetylcholine receptor
- moi
multiplicity of infection
- PIP2
phosphatidylinositol (4,5)bisphosphate
- PLC
phospholipase C
- PTX
pertussis toxin
- S1P
sphingosine 1-phosphate
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Berridge MJ, Irvine RF. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- 2.Rhee SG. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berridge MJ. Annu Rev Biochem. 1987;56:159–193. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- 4.Zhong H, Neubig RR. J Pharmacol Exp Ther. 2001;297:837–845. [PubMed] [Google Scholar]
- 5.Lopez I, Mak EC, Ding J, Hamm HE, Lomasney JW. J Biol Chem. 2001;276:2758–2765. doi: 10.1074/jbc.M008119200. [DOI] [PubMed] [Google Scholar]
- 6.Song C, Hu CD, Masago M, Kariyai K, Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T, Kataoka T. J Biol Chem. 2001;276:2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- 7.Kelley GG, Reks SE, Ondrako JM, Smrcka AV. EMBO J. 2001;20:743–754. doi: 10.1093/emboj/20.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin TG, Satoh T, Liao Y, Song C, Gao X, Kariya K, Hu CD, Kataoka T. J Biol Chem. 2001;276:30301–30307. doi: 10.1074/jbc.M103530200. [DOI] [PubMed] [Google Scholar]
- 9.Wing MR, Houston D, Kelley GG, Der CJ, Siderovski DP, Harden TK. J Biol Chem. 2001;276:48257–48261. doi: 10.1074/jbc.C100574200. [DOI] [PubMed] [Google Scholar]
- 10.Song C, Satoh T, Edamatsu H, Wu D, Tadano M, Gao X, Kataoka T. Oncogene. 2002;21:8105–8113. doi: 10.1038/sj.onc.1206003. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, Evellin S, Weernink PA, von Dorp F, Rehmann H, Lomasney JW, Jakobs KH. Nat Cell Biol. 2001;3:1020–1024. doi: 10.1038/ncb1101-1020. [DOI] [PubMed] [Google Scholar]
- 12.Kelley GG, Reks SE, Smrcka AV. Biochem J. 2004;378:129–139. doi: 10.1042/BJ20031370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wing MR, Snyder JT, Sondek J, Harden TK. J Biol Chem. 2003;278:41253–41258. doi: 10.1074/jbc.M306904200. [DOI] [PubMed] [Google Scholar]
- 14.Wing MR, Bourdon DM, Harden TK. Mol Interv. 2003;3:273–280. doi: 10.1124/mi.3.5.273. [DOI] [PubMed] [Google Scholar]
- 15.Bunney TD, Katan M. Trends Cell Biol. 2006;16:640–648. doi: 10.1016/j.tcb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Oestreich EA, Maekawa N, Bullard TA, Vikstrom KL, Dirksen RT, Kelley GG, Blaxall BC, Smrcka AV. Circ Res. 2005;97:1305–1313. doi: 10.1161/01.RES.0000196578.15385.bb. [DOI] [PubMed] [Google Scholar]
- 17.Oestreich EA, Wang H, Malik S, Kaproth-Joslin KA, Blaxall BC, Kelley GG, Dirksen RT, Smrcka AV. J Biol Chem. 2007;282:5488–5495. doi: 10.1074/jbc.M608495200. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen SD, Nicole O, Peavy RD, Montoya LM, Lee CJ, Murphy TJ, Traynelis SF, Hepler JR. Mol Pharmacol. 2003;64:1199–1209. doi: 10.1124/mol.64.5.1199. [DOI] [PubMed] [Google Scholar]
- 19.Berstein G, Blank JL, Smrcka AV, Higashijima T, Sternweis PC, Exton JH, Ross EM. J Biol Chem. 1992;267:8081–8088. [PubMed] [Google Scholar]
- 20.Hains MD, Wing MR, Maddileti S, Siderovski DP, Harden TK. Mol Pharmacol. 2006;69:2068–2075. doi: 10.1124/mol.105.017921. [DOI] [PubMed] [Google Scholar]
- 21.Kelley GG, Kaproth-Joslin KA, Reks SE, Smrcka AV, Wojcikiewicz RJ. J Biol Chem. 2006;281:2639–2648. doi: 10.1074/jbc.M507681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutkind JS. Oncogene. 1998;17:1331–1342. doi: 10.1038/sj.onc.1202186. [DOI] [PubMed] [Google Scholar]
- 24.Olson MF, Ashworth A, Hall A. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 25.Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu RG, Chen J, McCormick F, Symons M. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottazzi ME, Zhu X, Bohmer RM, Assoian RK. J Cell Biol. 1999;146:1255–1264. doi: 10.1083/jcb.146.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roovers K, Davey G, Zhu X, Bottazzi ME, Assoian RK. Mol Biol Cell. 1999;10:3197–3204. doi: 10.1091/mbc.10.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicole O, Goldshmidt A, Hamill CE, Sorensen SD, Sastre A, Lyuboslavsky P, Hepler JR, McKeon RJ, Traynelis SF. J Neurosci. 2005;25:4319–4329. doi: 10.1523/JNEUROSCI.5200-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, Barkett M, Ram KT, Quintanilla A, Hariharan IK. Proc Natl Acad Sci USA. 1997;94:12485–12490. doi: 10.1073/pnas.94.23.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattori M, Minato N. J Biochem (Tokyo) 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- 32.York RD, Yao H, Dillon T, Ellig CL, Eckert SP, McCleskey EW, Stork PJ. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 33.York RD, Molliver DC, Grewal SS, Stenberg PE, McCleskey EW, Stork PJ. Mol Cell Biol. 2000;20:8069–8083. doi: 10.1128/mcb.20.21.8069-8083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y, Satoh T, Gao X, Jin TG, Hu CD, Kataoka T. J Biol Chem. 2001;276:28478–28483. doi: 10.1074/jbc.M101737200. [DOI] [PubMed] [Google Scholar]
- 35.Committee on Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. Bethesda: Natl Inst Health; DHHS Publ No (NIH) 85-23. [Google Scholar]
- 36.Wittchen ES, Worthylake RA, Kelly P, Casey PJ, Quilliam LA, Burridge K. J Biol Chem. 2005;280:11675–11682. doi: 10.1074/jbc.M412595200. [DOI] [PubMed] [Google Scholar]
- 37.Wu D, Tadano M, Edamatsu H, Masago-Toda M, Yamawaki-Kataoka Y, Terashima T, Mizoguchi A, Minami Y, Satoh T, Kataoka T. Eur J Neurosci. 2003;17:1571–1580. doi: 10.1046/j.1460-9568.2003.02591.x. [DOI] [PubMed] [Google Scholar]