Abstract
Dictyostelium discoideum is a useful model for studying mechanisms of cisplatin drug sensitivity. Our previous findings, that mutations in sphingolipid metabolism genes confer cisplatin resistance in D. discoideum and in human cells, raised interest in the resistance mechanisms and their implications for cisplatin chemotherapy. Here we used expression microarrays to monitor physiological changes and to identify pathways that are affected by cisplatin treatment of D. discoideum. We found >400 genes whose regulation was altered by cisplatin treatment of wild-type cells, including groups of genes that participate in cell proliferation and in nucleotide and protein metabolism, showing that the cisplatin response is orderly and multifaceted. Transcriptional profiling of two isogenic cisplatin-resistant mutants, impaired in different sphingolipid metabolism steps, showed that the effect of cisplatin treatment was greater than the effect of the mutations, indicating that cisplatin resistance in the mutants is due to specific abilities to overcome the drug effects rather than to general drug insensitivity. Nevertheless, the mutants exhibited significantly different responses to cisplatin compared with the parent, and >200 genes accounted for that difference. Mutations in five cisplatin response genes (sgkB, csbA, acbA, smlA, and atg8) resulted in altered drug sensitivity, implicating novel pathways in cisplatin response. Our data illustrate how modeling complex cellular responses to drugs in genetically stable and tractable systems can uncover new targets with the potential for improving chemotherapy.
Keywords: drug resistance, sphingolipids, sphingosine kinase, sphingosine-1-phosphate lyase, transcriptional profiling
Dictyostelium discoideum is well known for studies of cell and developmental biology and is gaining interest as a model for studying human disease, including cancer (1). A substantial body of work has accumulated using D. discoideum to identify genes and proteins associated with resistance to the anti-cancer drug cisplatin (2).
Cisplatin [cis-diamminedichloroplatinum(II)] is used to treat a variety of solid tumors (3), but efficacy is often limited by drug resistance (4). Studies in human tumor cells and cell lines have implicated a number of resistance mechanisms and demonstrated that resistance is multifaceted (5).
D. discoideum provides a genetically tractable system to the problem of cisplatin resistance. It is a eukaryote with cellular properties similar to those of human cells (6). Its genome is sequenced, and many genes are homologous to human genes (7). Using insertional mutagenesis screens (8), we isolated mutants with single gene disruptions that have decreased sensitivity to cisplatin (9). One of these strains was null for the sphingosine-1-phosphate (S-1-P) lyase gene sglA, establishing the first link between sphingolipid metabolism and cisplatin sensitivity and defining S-1-P lyase as a potential target for controlling cisplatin sensitivity.
S-1-P synthesis is catalyzed by sphingosine kinase from sphingosine and ATP, and the S-1-P lyase degrades S-1-P to phosphoethanolamine and hexadecanal (10). S-1-P is a signaling molecule that promotes cell growth and survival whereas ceramide, another bioactive sphingolipid in the same pathway, promotes cell death. The two lipids work as a rheostat (11, 12) in which their relative levels promote proliferation (high S-1-P) or death (high ceramide). It was proposed that modulating the activity of S-1-P synthesis or degradation enzymes would affect cisplatin sensitivity.
Further genetic, biochemical, and pharmacological studies in D. discoideum established a clear role for the S-1-P lyase and the sphingosine kinases in controlling cisplatin sensitivity. Cells overexpressing sphingosine kinase or null for S-1-P lyase are less sensitive to cisplatin, and cells null for sphingosine kinase or overexpressing S-1-P lyase are more sensitive to cisplatin. The change in sensitivity is specific to cisplatin and carboplatin (9, 13, 14) and is associated with the activation of MAP kinases and signal transduction pathways in human cells (11, 12).
We have now extended these studies by transcriptional profiling of the cellular response to cisplatin in D. discoideum. Transcriptional profiling with cDNA microarrays can identify groups of genes whose expression is altered by experimental treatments. It is also used as a surrogate measure of cell physiology and to detect differences between mutant strains (15–20). The availability of two isogenic cisplatin-resistant strains with alterations in sphingolipid metabolism allowed us to compare the mutants' transcriptional responses to that of wild-type cells after cisplatin exposure and to understand how the mutations affected the response. We show that the transcription of a select number of genes is reproducibly altered after cisplatin treatment and that mutating some of the cisplatin response genes confers altered cisplatin sensitivity, defining new drug response pathways. The analysis of the responding genes in the mutants identified differential expression of cell proliferation genes, implicating sphingolipid metabolism in regulating cell proliferation after cisplatin treatment. Our results demonstrate the utility of genomic methods in D. discoideum to identify molecular pathways for studying drug resistance in cancer.
Results
Cisplatin Treatment Induces a Reproducible Transcriptional Response.
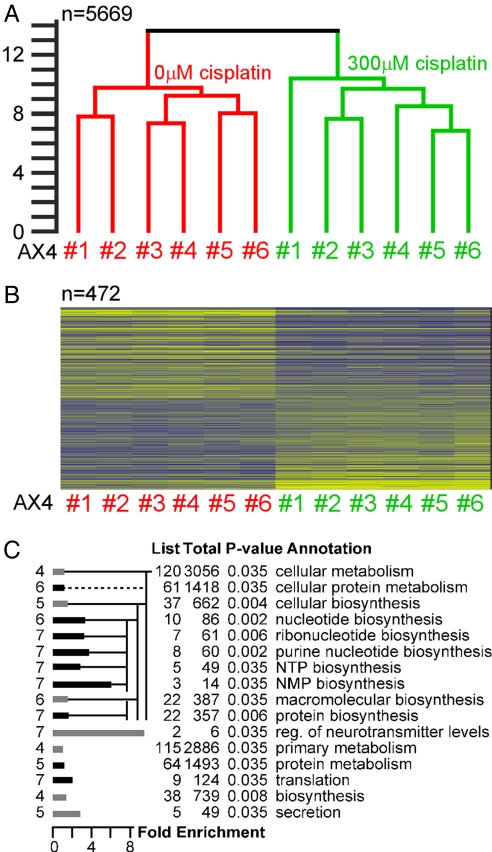
Cisplatin causes DNA damage and apoptotic cell death, but some reports show that cisplatin can also cause chaotic, necrotic cell death (21, 22). Studies in D. discoideum have shown that specific mutations can protect or sensitize cells to the cytotoxic effects of cisplatin, but not to other genotoxic agents (2, 9). These findings suggest that the cellular response to cisplatin is regulated and reproducible. Transcriptional analysis with microarrays is an established indicator of cell physiology in D. discoideum (18, 19). Thus, we tested whether cisplatin causes orderly physiological changes by examining the transcriptional response of cells to cisplatin treatment. RNA samples from cells treated with or without cisplatin were collected (six biological replicates) and analyzed on a microarray (five technical replicates) containing most of the D. discoideum-expressed genes (19). If the transcriptional response to cisplatin were robust and reproducible, one would expect to observe larger differences between the treated and untreated samples and smaller differences among the biological replicates. We calculated the Euclidean distances between the 12 RNA samples (six treated and six untreated) and found that all of the untreated samples were similar to each other and different from all of the treated samples (Fig. 1A). The variation among the treated samples was not significantly different from the variation among the untreated samples. These results show that cisplatin induces a robust and reproducible transcriptional response, confirming the idea that cisplatin induces a regulated physiological response.
Fig. 1.
The transcriptional response to cisplatin treatment. Expression data were collected from wild-type cells treated with cisplatin for 3 h and compared with data collected from untreated cells. Both treatments were repeated six independent times (#1 to #6). (A) The similarity between each sample and every other sample was determined by calculating the by-gene Euclidean distances between the samples, and the results are shown in a dendrogram that represents the distances between the replications. All of the microarray genes were used to calculate the distances (n = 5,669). The vertical distance between leaves and joins represents the dissimilarity between them (scale bar on the left, arbitrary units). Red, untreated samples (0 μM cisplatin); green, treated samples (300 μM cisplatin). Confidence levels, calculated by bootstrap analysis, were ≥0.98 for each join. (B) The heat map represents a group of 472 genes that were expressed differently between the untreated and treated cells (ANOVA, P < 0.05, followed by contrast analysis). Each column represents a sample, either untreated (red) or treated (green) with cisplatin. Each row represents a gene. The rows are ranked according to the outcome of the contrast analysis to illustrate the differences caused by the treatment. Yellow, higher-than-average expression; blue, lower-than-average expression. (C) The 472 genes from B were GO-annotated, and the “biological process” annotation of significantly enriched groups is shown. The GO tree levels are shown as numbers to the left of the bars (ranging from 2 to 9). The table indicates the number of genes in each group (List), genes with that annotation on the entire array (Total), the P value, and the annotation. Bar lengths represent the fold enrichment (scale: x axis). Annotation subgroups are indicated by the branching pattern. Black bars highlight nucleotide- and protein-related processes.
To identify which genes were differentially affected by the treatment, we performed an ANOVA, considering treatment, biological replication, and technical replication as factors. We also performed contrast analysis to find genes that had opposite expression patterns in the presence or absence of cisplatin. We then identified the overlap between the results of the two methods and found 472 genes whose expression was variable because of the cisplatin treatment (Fig. 1B). The degree of confidence in this finding is high because of the large number of replicas (P < 0.05). Twenty-one of these genes were tested by RT-PCR, and 13 differed significantly between the treated and untreated samples (P < 0.05) [supporting information (SI) Tables 2 and 3].
D. discoideum genes can be grouped into 17 expression modes based on their developmental expression patterns (15). We examined the developmental regulation of 467 of the 472 genes in our published microarray data (19) using these modes. We found that 252 of the 467 cisplatin response genes are expressed in vegetative cells and in early stages of development and are down-regulated during later stages of development (modes 1–6), and 131 genes were developmentally up-regulated (modes 8–16) (SI Table 4). The vegetative modes 1 and 3 were significantly overrepresented among the cisplatin-responsive genes (χ2 test, P = 1.48 × 10−7 and P = 0.006, respectively) (SI Table 4).
Annotation Analysis.
The correlation between an individual gene's expression and gene function is rather poor, but it is quite good for groups of genes (15, 16, 20, 23). We therefore used GOAT, a computational tool for identifying groups of gene annotations that are overrepresented in expression profiles (23), to analyze the 472 genes. We found overrepresentation of the cisplatin response genes in two major processes: nucleotide metabolism, including ribonucleotide, purine, NTP, and NMP metabolism (e.g., purB, C/E, D, and H), and protein metabolism, including translation and protein biosynthesis (e.g., many of the ribosomal protein large and small subunits; rpl and rps, respectively) (Fig. 1C and SI Table 5). These results agree with functional findings from genetic screens in yeast (24, 25), underscoring the confidence in both analyses.
Decreased Sensitivity to Cisplatin Caused by Perturbation of Specific Pathways Does Not Abolish the Overall Physiological Response to the Drug.
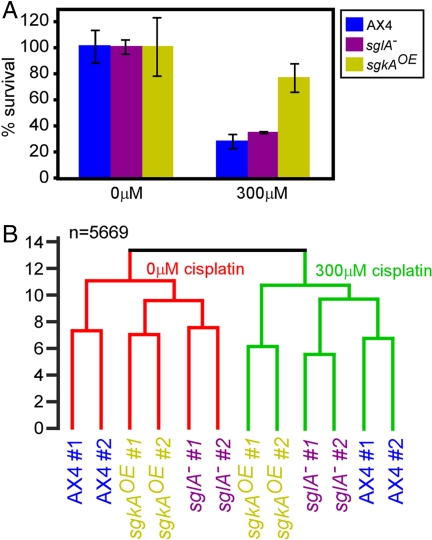
We tested whether cisplatin-resistant mutants would mount the stereotypical physiological drug response observed in wild-type cells. We have previously shown that cisplatin sensitivity in D. discoideum and in human cells is reduced by overexpression of the sphingosine kinase gene sgkA (sgkAOE) or by deletion of the S-1-P lyase gene sglA (sglA−) (13, 14, 26). Current models propose that sphingolipid metabolism modulates the cisplatin response through second-messenger signaling (2). Sphingolipid metabolism might also affect the permeability of various membranes, although previous studies indicate that cisplatin uptake in the mutants is indistinguishable from the wild-type uptake (27, 28). If the sgkAOE and sglA− cells were resistant to cisplatin because they became insensitive to the drug, one would expect the mutant cell physiology to be unperturbed by the drug. We therefore compared the transcriptional profiles of the two mutants to those of the parental cells with and without cisplatin treatment (Fig. 2). The survival rates were similar to those reported previously (9, 13, 14): no significant difference between the survival rates of the untreated strains (0 μM) (Fig. 2A), a small but significant increase in the sglA− cell survival, and a large and significant increase in the sgkAOE cell survival compared with the parental cells after cisplatin treatment (300 μM) (Fig. 2A).
Fig. 2.
The transcriptional response to cisplatin treatment in wild-type and mutant cells. Wild-type cells and two cisplatin-resistant mutants were treated with cisplatin for 3 h. Survival and gene expression data were collected from each sample, each experiment was repeated twice independently, and measurements were repeated several times as indicated below. (A) Cell survival of wild-type (AX4) and two cisplatin-resistant mutants (sglA− and sgkAOE) cells incubated without (0 μM) or with (300 μM) cisplatin for 3 h. Results are averages and SDs of three measurements of each of the two independent replications. (B) The similarity between each sample and every other sample was determined, and the data are displayed as in Fig. 1A. The different strains are indicated by different color text, and the two independent replications are indicated (#1 and #2). Confidence levels from bootstrap analysis were ≥0.98 for each join.
Transcriptional profile analysis resulted in several observations (Fig. 2B). First, the effect of cisplatin treatment was more significant than the effect of the mutations. All of the cisplatin-treated samples (green clade, Fig. 2B) clustered separately from the untreated samples (red clade, Fig. 2B), regardless of the genotypes. This finding indicates that cisplatin treatment induces similar physiological perturbations in the wild type and in the S-1-P metabolism mutants, supporting the notion that the reduced cisplatin sensitivity is caused by alteration of a specific signaling pathway rather than by global drug insensitivity. Second, in the untreated cells, the transcriptional profiles of the two mutants were more similar to each other than to the profile of the wild-type cells (red clade, Fig. 2B). This finding is consistent with the metabolic function of the mutated genes. Finally, we observed a high similarity between the transcriptional profiles of the wild-type and the sglA− cells in the cisplatin-treated group, whereas the sgkAOE profile was significantly different (green clade, Fig. 2B). These findings are consistent with the survival data (300 μM) (Fig. 2A) and suggest that mutations in sgkA and in sglA have distinct effects on the cell. Importantly, the differences between the branches were always larger than the technical and biological differences between the replicate experiments, as indicated by the pairing of samples 1 and 2 under each condition.
Differential Gene Expression in Cisplatin-Resistant Cells.
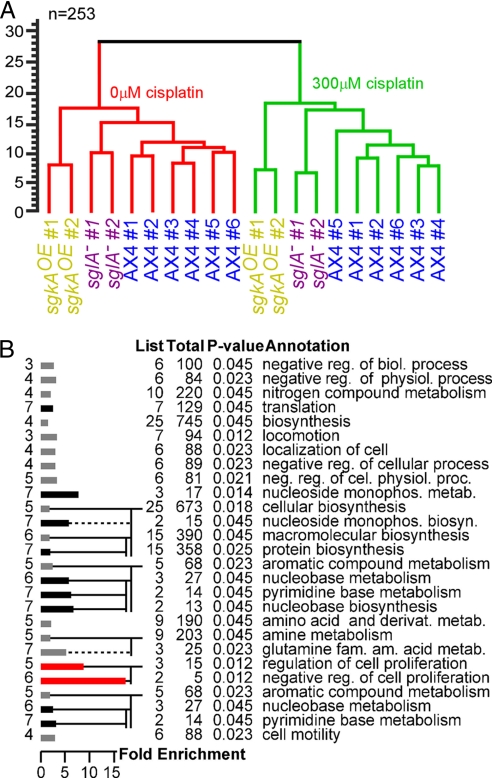
Wild type and cisplatin-resistant mutants exhibited similar transcriptional profiles (Fig. 2B), but the cells were differentially resistant to the drug. Thus, we searched for transcriptional responses that could distinguish between the treated strains. We used the transcriptional information from Figs. 1 and 2 and performed an ANOVA using genotype (wild type, sglA−, and sgkAOE) and treatment (0 μM and 300 μM cisplatin) as factors. We found 253 genes that distinguished the samples most significantly (P < 0.05), tested eight of the genes by RT-PCR, and confirmed the microarray results in six (SI Table 3).
Using the 253 genes to calculate the distances between the samples revealed that the most dominant factor in the difference between the samples was cisplatin treatment (Fig. 3A). The largest distance in the dendrogram was between the untreated samples (red clade, Fig. 3A) and the cisplatin-treated samples (green clade, Fig. 3A). All of the wild-type samples clustered together and the sglA− samples were more similar to the wild-type samples than to the sgkAOE samples. These results are similar to the ones found by using information from all of the genes (Fig. 2), supporting the above conclusions. The main difference between the dendrogram in Fig. 3A and the one in Fig. 2B was the clustering of the sgkAOE samples away from the other samples in the untreated conditions (red clades, Figs. 2A and 3A). These results support the notion that sgkA and sglA may have distinct roles in the regulation of S-1-P levels.
Fig. 3.
Differential transcriptional responses to cisplatin between wild-type and mutant cells. Gene expression data from Figs. 1 and 2 were tested by ANOVA to identify genes whose expression varied as a function of genotype and treatment, yielding 253 significant genes (P < 0.05). (A) The similarity between each sample and every other sample was determined by calculating the by-gene Euclidean distances between the samples, and the results are shown in a dendrogram that represents the distances between the different replications. Only the 253 genes found by ANOVA were used. The vertical distance between leaves and joins represents the dissimilarity between them (scale bar on the left, arbitrary units). Red, untreated samples (0 μM cisplatin); green, treated samples (300 μM cisplatin). The different strains are indicated by different color text, and the independent replications are indicated (#1 to #6). Confidence levels, calculated by bootstrap analysis, were ≥0.98 for each join. (B) The 253 genes were GO-annotated, and the biological process annotation of significantly enriched groups is shown as described in Fig. 1C. Black bars, nucleotide- and protein-related processes; red bars, cell proliferation.
We analyzed the annotations of the 253 genes using GOAT as described in Fig. 1D. Again we found genes involved in protein biosynthesis and nucleotide metabolism, but we also found genes involved in amino acid metabolism and in cell proliferation (e.g., the cytokinesis gene spkA_1 and the cell proliferation regulator aprA) (Fig. 3B and SI Table 6). These results are consistent with results from genetic screens in yeast (24, 25, 29) and from transcriptional profiling of human cells (30–32), increasing the confidence in our findings.
Comparing the Gene Ontology (GO) annotation analyses (Figs. 1C and 3B) reveals that nucleotide metabolism and protein biosynthesis are common to both analyses whereas cell proliferation genes were enriched only after considering mutations in sphingosine metabolism genes (Fig. 3B). This suggests that transcriptional regulation of nucleotide metabolism and protein synthesis genes is part of the general drug response, whereas regulation of cell proliferation is unique to the mutants and may be part of the response that allows the mutants to resist cisplatin treatment. The latter notion is consistent with the known effects of sphingosine metabolism on cell proliferation (13).
Identification of New Regulators of Cisplatin Sensitivity.
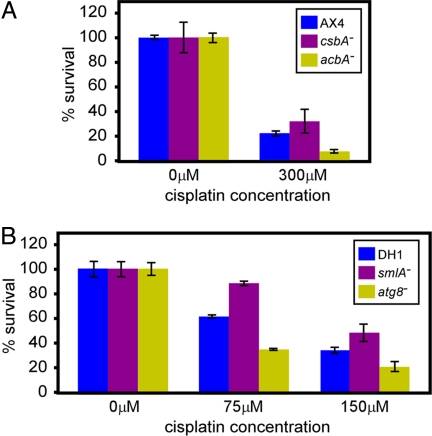
Our analysis revealed several hundred cisplatin response genes (SI Tables 7 and 8). One of the genes was sgkB, which has a known effect on the cellular response to cisplatin (14), supporting the notion that the genes we found have causative roles in the cisplatin response. We selected four additional genes to test that possibility. We chose the genes only on the basis of mutant strain availability, so they can be considered as randomly chosen. The csbA− (33), acbA− (34), and atg8− (35) strains were selected from the list of 472 genes (Fig. 1), and the smlA− (36) strain was from the list of 253 genes (Fig. 3). csbA− and acbA− cells (AX4 background) and smlA− and atg8− cells (DH1 background) were tested for survival under various cisplatin concentrations in comparison with the respective parental strains. In the AX4 background, the most significant results were found at 300 μM cisplatin. Inactivation of csbA reduced drug sensitivity, but the statistical significance was marginal (t test, P = 0.064) (Fig. 4A). Inactivation of acbA significantly increased the cisplatin sensitivity (t test, P < 0.05) (Fig. 4A). In the DH1 background the most significant results were found at 75 μM and 150 μM cisplatin. Inactivation of smlA reduced drug sensitivity, and inactivation of atg8 increased drug sensitivity (t test, P < 0.05 for both) (Fig. 4B). Therefore, three of the four genes found by microarray profiling have effects on cisplatin sensitivity, lending significant credibility to the microarray study.
Fig. 4.
Survival of new mutants in the presence of cisplatin. Mutant strains were grown in shaking suspension in the presence of cisplatin at the indicated concentrations (μM). Cells were counted and plated clonally, and survival (percentage of control) was plotted as the average ± SD of three independent replications. (A) Survival of csbA− and acbA− cells compared with the parental AX4 cells. (B) Survival of smlA− and atg8− cells compared with the parental DH1 cells.
Discussion
The completion of the D. discoideum genome sequence enabled the global analyses of gene function using insertional mutagenesis and transcriptional profiling with microarrays (37, 38). The latter is a surrogate measure of cell physiology and an indicator of differences between mutant strains (15–20). Recently, the organism has been used as a model for drug target discovery using genetics and genomic approaches (1, 2). The small genome, the precisely controlled growth conditions, and the fact that isogenic mutant and wild-type strains differ by single mutations make this organism ideal for microarray analyses. These advantages are confirmed by the reproducibly of our microarray data and the ultimate discovery of functional genes.
Our study shows that the response to cisplatin is programmed and tightly regulates a specific set of genes. These findings are significant because previous work in mammalian cells has indicated that cisplatin can cause both apoptotic and necrotic cell death and that the latter is associated with chaotic gene expression (21, 39).
Comparison of the mutant and parental strains indicates that the mutations did not confer total immunity to the drug, because the mutants retained most of the physiological responses of the wild type. These data support the idea that the mutations affect specific signaling pathways and suggest new pathways that are involved in the cisplatin response and can be targeted to improve efficacy.
Comparing the response of the sgkAOE and sglA− cells to each other reveals that the mutants have distinct transcriptional responses to cisplatin despite the fact that both genotypes regulate S-1-P levels (2, 40). These data are consistent with the biochemical properties of the pathway. The sgkAOE strain generates S-1-P constitutively, thus depleting the ceramide pool, whereas the sglA− mutation blocks S-1-P degradation and the resulting consumption of sphingosine and ceramide (2, 40). Therefore, the different transcriptional profiles reflect the biochemical differences between the mutants. This type of analysis could be extended to other mutants known to effect cisplatin sensitivity such as the cAMP phosphodiesterase regA (9).
The identification of >600 cisplatin response genes confirms the idea that the cellular response to cisplatin is multifaceted, and it is consistent with the many genes and proteins that underlie cisplatin resistance in human tumors (5). Even a 2-fold increase in resistance can be fatal in humans undergoing chemotherapy because cisplatin is used near its toxic threshold. Thus, the identification of new targets that can increase cisplatin sensitivity is of clinical interest. The genes discovered in our study fall into several functional groups including nucleotide metabolism, protein metabolism, and regulation of cell proliferation. These functions have also been implicated by genetic screens for cisplatin resistance in yeast (24, 29, 41) and by transcriptional profiling of human cells (30–32). These correlated findings, made in organisms from three different taxa (7), suggest that some of the cisplatin response mechanisms are conserved between D. discoideum and humans.
One of the distinct advantages of working with D. discoideum is the public availability of mutants (http://dictybase.org). We found five mutations in cisplatin response genes in the mutant collection. sgkB is a sphingosine kinase gene that we have already described as having a role in cisplatin resistance (14), and we now find that three other genes (acbA, atg8, and smlA) also had altered cisplatin sensitivity. Interestingly, acbA encodes an acyl-CoA binding protein, which is involved in sphingolipid metabolism and in developmental intercellular signaling, including the regulation of long-chain base sphingolipid by the yeast homolog Acb1p (34, 42–44). atg8 participates in the regulation of autophagy, a process in which cells use internal organelles and macromolecules as a source of nutrients and energy and in which conjugation to phosphatidylethanolamine plays a key role (35, 45, 46). smlA participate in aggregate size regulation (47, 48). Therefore, acbA, atg8, and sgkB are linked to sphingolipid metabolism, which is involved in regulating sensitivity to cisplatin in both D. discoideum and human cells (2).
This study continues to establish D. discoideum as a model system for understanding the biochemistry underlying the sensitivity to cisplatin and other chemotherapeutic drugs and for the discovery of novel drug targets that could improve the efficacy of existing drugs. Some of the genes and pathways we found have been associated with cisplatin resistance before, and others represent targets that should be examined in human cells. Specifically, the identification of altered cell proliferation genes in the mutant strains improves our understanding of the mechanisms by which sphingolipids regulate cisplatin sensitivity. Tumor cells and cancer cell lines are notoriously genetically complex and unstable. D. discoideum allows us to discover new drug targets and cisplatin resistance pathways and to test them in a genetically defined system. The similarity between the cisplatin responses of D. discoideum and human cells promises that findings in this system would be translatable to human health.
Materials and Methods
Strains and Cell Growth.
The strains used are described in Table 1. Cells were grown in shaking suspension in HL5 medium (49) with the appropriate supplements for auxotrophy or drug selection.
Table 1.
Strains used in this study
| Strain | Parental strain | Genotype | Reference |
|---|---|---|---|
| AX4 | AX3-K | axeA1,*axeB1,*axeC1* (axeA1, B1, C1) | 55 |
| TL72 (csbA) | HL330 | axeA1, B1, C1, pyr5-6−† [pΔPYR-BglII], ura+, csbA−‡, csbB−‡, csbC−‡ [csb-KO] | 39 |
| TL177 (acbA) | AX4 | axeA1, B1, C1, acbA− [acbA-bsr], bsR§ | 40 |
| DH1 | AX3 | axeA1, B1, C1, pyr5-6− [pRG24], ura− | 56 |
| smlA | DH1 | axeA1, B1, C1, pyr5-6− [pRG24], ura+, smlA−¶ [pSmlA-KO] | 57 |
| atg8 | DH1 | axeA1, B1, C1, pyr5-6− [pRG24], ura−, atg8−‖ [atg8-KO], bsR | 41 |
| SA550 | AX4 | axeA1, B1, C1, sglA−** [sglA-KO], bsR | 13 |
| SA604 | AX3-ORF | axeA1, B1, C1, sgkAOE†† [sgkA-actin 15], G418R‡‡ | 14 |
*Mutations required for axenic growth.
†Uridine synthase.
‡Contact site B.
§Blasticidin S resistance.
¶Small A, aggregate size.
‖Autophagy regulation.
**S-1-P lyase.
††Sphingosine kinase A.
‡‡G418 resistance.
Drug Treatment, Viability, and RNA Collection.
Cisplatin (Sigma–Aldrich, St. Louis, MO) was dissolved in PT buffer (3 mM NaCl/1 mM sodium phosphate, pH 7.4) immediately before use and protected from light. Concentration was adjusted by spectrophotometry at 220 nm by using an extinction coefficient of 1.957 mM−1·cm−1 (50).
Exponentially growing cells (2 × 106 cells per milliliter) were divided into 12 50-ml aliquots in 500-ml flasks. The cells were shaken for 30 min, 300 μM cisplatin was added to six cultures (PT buffer was added to the six controls), and the cells were shaken for 3 h at 22°C in the dark. Samples were assayed for viability (51), the remaining cells were washed with PBS and resuspended in 1.0 ml of TRIzol reagent (Life Technologies, Gaithersburg, MD), and RNA was extracted.
Microarray Experiments, Normalization, and Multiarray Scaling.
Microarray experiments were performed as described (19). A two-color assay was performed: total RNA was reverse-transcribed with a fluorescent oligo(dT) primer, and a reference sample (total RNA pooled from several developmental time points of wild-type cells) was labeled with a second fluorophore. The samples were cohybridized to a microarray of 5,669 unique targets (Gene Expression Omnibus accession no. GPL5382) (19). Bioconductor software was used to normalize each microarray (52). Multiarray scaling was performed as described (16, 18, 19).
We performed three kinds of replication for each sample. Each hybridization target was printed twice on the array for single-chip normalization. For technical variation, RNA samples from each experiment were hybridized to at least five arrays (technical replication). For biological variation, each treatment (with or without cisplatin) was repeated twice for each mutant and six times for the wild type (biological replication). The data are available in the Gene Expression Omnibus (accession no. GSE8287).
Dissimilarities Between RNA Samples.
To calculate dissimilarity, we averaged the technical replications and calculated the distances between the samples (18). Pairwise Euclidean distances were calculated by converting the 5,669 row by n samples matrix to a 5,669 × n vector. Distance calculation, followed by average-linkage clustering, was performed by using the software package R. The results were visualized as dendrograms, and the confidence was calculated by bootstrapping (53).
Detection of Differently Expressed Genes.
In the case of wild-type cells, ANOVA was performed by using biological replication (P > 0.05), technical replication (P > 0.05), and treatment (P < 0.05) as factors. In addition, contrast analysis was used to find opposite patterns in the treated and untreated samples (19). Two gene lists were obtained, and the overlap between them was determined. Analysis of GO annotations of the gene lists was carried out by using GOAT (23).
To find the differential transcriptional responses to cisplatin between wild type and mutant cells we performed ANOVA with genotype (P < 0.05), treatment (P < 0.05), and technical replication (P > 0.05) as factors. Hierarchical clustering on the resulting gene list and GO analysis were done as described above.
The gene expression differences were confirmed in at least two biological replications by quantitative RT-PCR and repeated three times as described (54).
Cell Survival.
Cells were incubated in shaking suspension in 2 ml of HL-5 medium in triplicates with 0, 75, 150, and 300 μM cisplatin for 24 h. Cell viability was tested by clonal plating in 24-well plates (51). The cell cultures were serially diluted in 10 mM NaCl, 10 mM KCl, and 2.7 mM CaCl2, and 15-μl aliquots were mixed with stationary-phase Klebsiella aerogenes plated on 1 ml of SM agar in each of the 24 wells. The plates were incubated at 22°C, and the plaques on the bacterial lawn (viable cells) were counted.
Supplementary Material
Acknowledgments
We thank E. Huang for assistance with statistical data analysis. This work was supported by the National Institutes of Health (Grant HD39691 to A.K. and G.S. and Grants GM53929 and CA95872 to S.A.).
Abbreviations
- S-1-P
sphingosine-1-phosphate
- GO
Gene Ontology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8287).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705996104/DC1.
References
- 1.Williams RS, Boeckeler K, Graf R, Muller-Taubenberger A, Li Z, Isberg RR, Wessels D, Soll DR, Alexander H, Alexander S. Trends Mol Med. 2006;12:415–424. doi: 10.1016/j.molmed.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Alexander S, Min J, Alexander H. Biochim Biophys Acta. 2006;1760:301–309. doi: 10.1016/j.bbagen.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Kelland LR, Farrell PN. Platinum-Based Drugs in Cancer Therapy. Totowa, NJ: Humana; 2000. [Google Scholar]
- 4.Perez RP. Eur J Cancer. 1998;34:1535–1542. doi: 10.1016/s0959-8049(98)00227-5. [DOI] [PubMed] [Google Scholar]
- 5.Siddik ZH. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 6.Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge, UK: Cambridge Univ Press; 2001. [Google Scholar]
- 7.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuspa A, Loomis WF. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Alexander H, Schneider N, Alexander S. Microbiology. 2000;146:2219–2227. doi: 10.1099/00221287-146-9-2219. [DOI] [PubMed] [Google Scholar]
- 10.van Veldhoven PP, Mannaerts GP. Adv Lipid Res. 1993;26:69–98. [PubMed] [Google Scholar]
- 11.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 12.Spiegel S. J Leukocyte Biol. 1999;65:341–344. doi: 10.1002/jlb.65.3.341. [DOI] [PubMed] [Google Scholar]
- 13.Min J, Stegner AL, Alexander H, Alexander S. Eukaryotic Cell. 2004;3:795–805. doi: 10.1128/EC.3.3.795-805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min J, Traynor D, Stegner AL, Zhang L, Hanigan MH, Alexander H, Alexander S. Eukaryotic Cell. 2005;4:178–189. doi: 10.1128/EC.4.1.178-189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth E, Van Driessche N, Zhuchenko O, Kuspa A, Shaulsky G. Bioinformatics. 2005;21:4371–4377. doi: 10.1093/bioinformatics/bti726. [DOI] [PubMed] [Google Scholar]
- 16.Katoh M, Shaw C, Xu Q, Van Driessche N, Morio T, Kuwayama H, Obara S, Urushihara H, Tanaka Y, Shaulsky G. Proc Natl Acad Sci USA. 2004;101:7005–7010. doi: 10.1073/pnas.0306983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza MC, Booth E, Shaulsky G, Firtel RA. Mol Cell Biol. 2007;27:3817–3827. doi: 10.1128/MCB.02194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Driessche N, Demsar J, Booth E, Hill P, Juvan P, Zupan B, Kuspa A, Shaulsky G. Nat Genet. 2005;37:471–477. doi: 10.1038/ng1545. [DOI] [PubMed] [Google Scholar]
- 19.Van Driessche N, Shaw C, Katoh M, Morio T, Sucgang R, Ibarra M, Kuwayama H, Saito T, Urushihara H, Maeda M, et al. Development (Cambridge, UK) 2002;129:1543–1552. doi: 10.1242/dev.129.7.1543. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, Ibarra M, Mahadeo D, Shaw C, Huang E, Kuspa A, Cotter D, Shaulsky G. Eukaryotic Cell. 2004;3:1101–1110. doi: 10.1128/EC.3.5.1101-1110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fietta P. Riv Biol. 2006;99:69–83. [PubMed] [Google Scholar]
- 22.Syntichaki P, Tavernarakis N. EMBO Rep. 2002;3:604–609. doi: 10.1093/embo-reports/kvf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q, Shaulsky G. Appl Bioinformatics. 2005;4:281–283. doi: 10.2165/00822942-200504040-00008. [DOI] [PubMed] [Google Scholar]
- 24.Huang RY, Eddy M, Vujcic M, Kowalski D. Cancer Res. 2005;65:5890–5897. doi: 10.1158/0008-5472.CAN-04-4093. [DOI] [PubMed] [Google Scholar]
- 25.Liao C, Hu B, Arno MJ, Panaretou B. Mol Pharmacol. 2007;71:416–425. doi: 10.1124/mol.106.030494. [DOI] [PubMed] [Google Scholar]
- 26.Min J, Van Veldhoven PP, Zhang L, Hanigan MH, Alexander H, Alexander S. Mol Cancer Res. 2005;3:287–296. doi: 10.1158/1541-7786.MCR-04-0197. [DOI] [PubMed] [Google Scholar]
- 27.Hassoun SM, Lancel S, Petillot P, Decoster B, Favory R, Marchetti P, Neviere R. Mitochondrion. 2006;6:149–154. doi: 10.1016/j.mito.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Contreras FX, Sot J, Alonso A, Goni FM. Biophys J. 2006;90:4085–4092. doi: 10.1529/biophysj.105.076471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM. Cancer Res. 2004;64:3940–3948. doi: 10.1158/0008-5472.CAN-03-3113. [DOI] [PubMed] [Google Scholar]
- 30.Voland C, Bord A, Peleraux A, Penarier G, Carriere D, Galiegue S, Cvitkovic E, Jbilo O, Casellas P. Mol Cancer Ther. 2006;5:2149–2157. doi: 10.1158/1535-7163.MCT-05-0212. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Chuang L, Zhang X, Colton S, Dombkowski A, Reiners J, Diakiw A, Xu XS. Nucleic Acids Res. 2004;32:2231–2240. doi: 10.1093/nar/gkh541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Dombkowski A, Chuang L, Xu X. Cell Res. 2004:14303–314. doi: 10.1038/sj.cr.7290375. [DOI] [PubMed] [Google Scholar]
- 33.Xu XS, Kuspa A, Fuller D, Loomis WF, Knecht DA. Dev Biol. 1996;175:218–226. doi: 10.1006/dbio.1996.0109. [DOI] [PubMed] [Google Scholar]
- 34.Anjard C, Loomis WF. Proc Natl Acad Sci USA. 2005;102:7607–7611. doi: 10.1073/pnas.0501820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. J Biol Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 36.Brock DA, Buczynski G, Spann TP, Wood SA, Cardelli J, Gomer RH. Development (Cambridge, UK) 1996;122:2569–2578. doi: 10.1242/dev.122.9.2569. [DOI] [PubMed] [Google Scholar]
- 37.Kuspa A, Loomis WF. Methods Mol Biol. 2006;346:15–30. doi: 10.1385/1-59745-144-4:15. [DOI] [PubMed] [Google Scholar]
- 38.Shaulsky G, Loomis WF. Protist. 2002;153:93–98. doi: 10.1078/1434-4610-00087. [DOI] [PubMed] [Google Scholar]
- 39.Tan B, Piwnica-Worms D, Ratner L. Curr Opin Oncol. 2000;12:450–458. doi: 10.1097/00001622-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Merrill AH., Jr J Biol Chem. 2002;277:25843–25846. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- 41.Burger H, Capello A, Schenk PW, Stoter G, Brouwer J, Nooter K. Biochem Biophys Res Commun. 2000;269:767–774. doi: 10.1006/bbrc.2000.2361. [DOI] [PubMed] [Google Scholar]
- 42.Cabral M, Anjard C, Loomis WF, Kuspa A. Eukaryotic Cell. 2006;5:2024–2032. doi: 10.1128/EC.00287-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anjard C, Loomis WF. Development (Cambridge, UK) 2006;133:2253–2261. doi: 10.1242/dev.02399. [DOI] [PubMed] [Google Scholar]
- 44.Faergeman NJ, Wadum M, Feddersen S, Burton M, Kragelund BB, Knudsen J. Mol Cell Biochem. 2007;299:55–65. doi: 10.1007/s11010-005-9040-3. [DOI] [PubMed] [Google Scholar]
- 45.Tekinay T, Wu MY, Otto GP, Anderson OR, Kessin RH. Eukaryotic Cell. 2006;5:1797–1806. doi: 10.1128/EC.00342-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. J Biol Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 47.Tang L, Ammann R, Gao T, Gomer RH. J Biol Chem. 2001;276:27663–27669. doi: 10.1074/jbc.M102205200. [DOI] [PubMed] [Google Scholar]
- 48.Brock DA, Gomer RH. Genes Dev. 1999;13:1960–1969. doi: 10.1101/gad.13.15.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sussman M. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 50.Li GC, Foote C, Alexander S, Alexander H. Development (Cambridge, UK) 2001;128:3473–3483. doi: 10.1242/dev.128.18.3473. [DOI] [PubMed] [Google Scholar]
- 51.Alexander H, Vomund AN, Alexander S. BioTechniques. 2003;35:464–470. doi: 10.2144/03353bm05. [DOI] [PubMed] [Google Scholar]
- 52.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efron B, Halloran E, Holmes S. Proc Natl Acad Sci USA. 1996;93:13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang E, Blagg SL, Keller T, Katoh M, Shaulsky G, Thompson CR. Development (Cambridge, UK) 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.