Abstract
MicroRNAs (miRNAs) are a class of 19- to 23-nt, small, noncoding RNAs, which bind the 3′ UTR of target mRNAs to mediate translational repression in animals. miRNAs have been shown to regulate developmental processes, such as self-renewal of stem cells, neuronal differentiation, myogenesis, and cancer. A functional role of miRNAs in the regulation of neurotransmitter synthesis has yet to be ascribed. We used mesenchymal stem cells (MSCs) as a model to study miRNA-mediated neurotransmitter regulation in developing neuronal cells. MSCs are mesoderm-derived cells, primarily resident in adult bone marrow, which can generate functional neuronal cells. We have previously shown that human MSC-derived neuronal cells express the neurotransmitter gene, Tac1, but do not synthesize the gene's encoded peptide, the neurotransmitter substance P (SP), unless stimulated with the inflammatory mediator IL-1α. These findings suggested a potential role for miRNAs in the regulation of SP synthesis. Here, we report on the miRNA profile of undifferentiated human MSCs and MSC-derived neuronal cells by using miRNA-specific bioarrays. miRNAs that were increased in the neuronal cells and decreased after IL-1α stimulation were analyzed by the miRanda algorithm to predict Tac1 mRNA targets. Putative miR-130a, miR-206, and miR-302a binding sites were predicted within the 3′ UTR of Tac1. Target validation using a luciferase reporter system confirmed the miR-130a and miR-206 sites. Specific inhibition of miR-130a and miR-206 in the neuronal cells resulted in SP synthesis and release. The studies provide a different approach in ascribing a new regulatory role for miRNAs in regulating neurotransmitter synthesis.
Keywords: neuron, Tac1, tachykinin, transdifferentiation, IL-1α
MicroRNAs (miRNAs) are a novel class of 19- to 23-nt, small, noncoding RNA molecules encoded in the genomes of plants and animals (1). The functions of miRNAs are different in plants and animals. In both, miRNAs bind to the 3′ UTR of target mRNAs (1). However, miRNAs mediate mRNA degradation in plants and translational repression in animals (1). Hundreds of miRNAs have been described, with many sequences exhibiting homology among species, thus ascribing to their evolutionary conservation (2). Most miRNAs are found in noncoding regions of the genome (3).
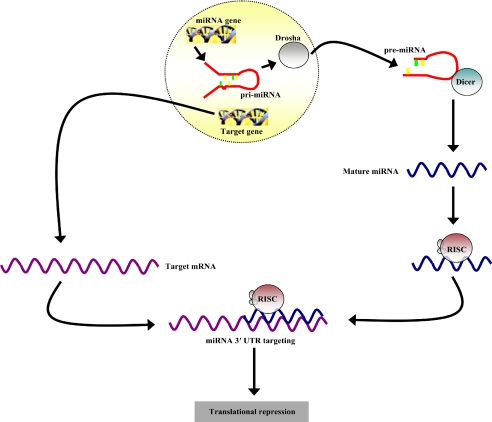
Primary miRNA molecules are transcribed by RNA polymerase II (Fig. 1) and then digested by the ribonuclease Drosha to generate precursor miRNAs (pre-miRNAs) of ≈70 nt (4–6). The pre-miRNAs are then exported to the cytoplasm for processing by Dicer to generate mature miRNAs, which associate with the RNA-induced silencing complex for loading onto 3′ UTR of targeted mRNAs (5–7). Animal miRNAs typically have imperfect homology with their target mRNAs (8). This inexact complementarity causes translational repression, as opposed to siRNAs, which mediate mRNA degradation through near-perfect base-pairing. The regulatory effects of miRNAs are typically seen to increase when more than one miRNA binds its target (9).
Fig. 1.
Cartoon depicting the mechanism of miRNA transcription, processing, and regulatory activity. miRNA genes are transcribed by RNA polymerase II to form primary miRNA (pri-miRNA) molecules. The ribonuclease, Drosha, then cleaves the pri-miRNA to release the pre-miRNA for cytoplasmic export and processing by Dicer. The mature miRNA product associates with the RNA-induced silencing complex for loading onto the 3′ UTR of target mRNAs to mediate translational repression.
Because miRNAs exhibit imperfect complementarity with their target mRNA, previous efforts to computationally predict and functionally validate miRNA targets have been difficult (10). More recently, new target prediction algorithms have estimated that thousands of human gene products are regulated by miRNAs (11). Current functional studies have shown that miRNAs are key regulators of developmental processes, such as self-renewal of stem cells, myogenesis, embryogenesis, cellular differentiation, and brain development (12–16). miRNAs have also been implicated in many cancers and viral infection (17, 18).
Mesenchymal stem cells (MSCs) are adult stem cells mostly found in the adult bone marrow (BM), surrounding the blood vessels and trabecula (19). Physiologically, MSCs show lineage-specific differentiation along osteogenic, chrondrogenic, and adipogenic paths (20). MSCs have also been reported to transdifferentiate into functional neuronal cells (21, 22). The utilization of neuronal cells generated from human MSCs underscores their potential as a source of autologous stem cells for neural tissue repair.
We have reported the ability of MSC-derived neurons to express the neurotransmitter gene, Tac1, at the level of transcription, but without translation to its encoded neurotransmitter substance P (SP) (21, 23). However, stimulation with the proinflammatory mediator, IL-1α, resulted in Tac1 mRNA translation (21, 23). IL-1α therefore served as a model to recapitulate the in vivo effects of an inflammatory microenvironment, as would be expected at regions of tissue insults. We hypothesize that IL-1α alleviates translational repression of Tac1 mRNA through negative effects on miRNAs. Here, we explore the role of miRNAs in neurotransmitter regulation in developing neuronal cells by using an approach involving microarray, computational, and functional cellular analyses.
Results
miRNA Profile of Uninduced MSCs.
The miRNA profile of MSCs has not been described, thus we investigated miRNA expression in uninduced MSCs. miRNAs were studied with mirVana miRNA Bioarrays, which consisted of 385 miRNA-specific oligonucleotides identified in human, mouse, or rat. The analyses were done with total RNA, from three different human BM donors. There were 32 different miRNAs identified in all three donors (hsa_let_7i, hsa_miR_191, hsa_miR_222, hsa_miR_199a, hsa_miR_125b, hsa_miR_31, hsa_miR_23b, hsa_miR_22, hsa_miR_221, hsa_miR_125a, hsa_miR_214, hsa_lrt_7b, hsa_miR_30a_5p, hsa_miR_29a, hsa_miR_27a, hsa_miR_152, hsa_miR_16, hsa_let_7f, hsa_let_7a, hsa_miR_320, hsa_let_7c, hsa_miR_100, hsa_miR_199a_AS, hsa_miR_24, hsa_miR_145, hsa_miR_23a, hsa_let_7d, hsa_miR_27b, hsa_miR_143, hsa_let_7e, hsa_miR_26a, and hsa_miR_21).
Profile of Up-Regulated miRNAs in Day 12 Neuronal Induction (D12) MSCs.
MSCs provide an alternate source of neuronal cells for tissue repair, with potential for autologous transplantation. We have used the transdifferentiation of MSCs as a model to recapitulate the in vivo effects of an inflammatory microenvironment on developing neuronal cells. Because MSC-derived neuronal cells express Tac1, but do not produce the neurotransmitter SP unless stimulated with the inflammatory microenvironmental factor IL-1α, we explored the role of miRNAs in the observed results (21, 23). The miRNA profile of MSCs incubated in neuronal induction media for 12 days was studied. There were 16 different miRNAs found in all three donor samples that were up-regulated (Table 1) compared with uninduced MSCs. To identify miRNAs potentially involved in Tac1 regulation, the miRNA profile of D12 cells stimulated for 16 h with IL-1α was explored (Table 1). There were nine different miRNAs that were decreased in response to IL-1α stimulation (Table 1). These identified miRNAs were further investigated by computational and functional analyses.
Table 1.
miRNAs in MSC-derived neurons (D12) in the presence or absence of IL-1α
| miRNA | Unstimulated | IL-1α |
|---|---|---|
| mmu_miR_140_AS | + | − |
| hsa_miR_130a | + | − |
| hsa_miR_30d | + | − |
| hsa_miR_206 | + | − |
| hsa_miR_30e_5p | + | − |
| hsa_miR_15b | + | − |
| hsa_miR_342 | + | − |
| hsa_miR_302a | + | − |
| hsa_miR_99b | + | − |
| hsa_miR_210 | + | + |
| hsa_miR_34a | + | + |
| hsa_miR_99a | + | + |
| hsa_miR_103 | + | + |
| hsa_miR_107 | + | + |
| hsa_let_7 g | + | + |
| hsa_miR_181a | + | + |
miRNA from D12 MSC-derived neurons, unstimulated or stimulated with IL-1α, was analyzed with mirVana miRNA Bioarrays. +, miRNA detection; −, undetectable miRNA.
Identification and Quantification of miRNAs with Predicted Tac1 Targets.
We next determined whether miRNAs decreased by IL-1α stimulation (Table 1) have putative binding sites within Tac1 3′ UTR. Because miRNAs exhibit imperfect complementarity with their mRNA target, the miRNA-specific target detection algorithm, miRanda, was used. Table 2 lists the names and sequences of the three miRNAs predicted to have one or more binding sites within Tac1 3′ UTR.
Table 2.
Prediction of miRNAs with putative Tac1 mRNA 3′ UTR targets
| miRNA | Sequence (5′ → 3′) | Tac1 target | Known targets |
|---|---|---|---|
| hsa_miR_130a | cag ugc aau guu aaa agg gca u | Yes | MAFB (24) |
| hsa_miR_206 | ugg aau gua agg aag ugu gug g | Yes | hERα, connexin 43 (25, 26) |
| hsa_miR_302a | uaa gug cuu cca ugu uuu ggu ga | Yes | ESC-specific, unknown (27) |
miRNA sequences of those increased in D12 MSCs and decreased by IL-1α (Table 1), and their known and predicted targets to Tac1 mRNA, as indicated by miRanda algorithm. Listed miRNAs are predicted to target one or more sites within the 3′ UTR of Tac1.
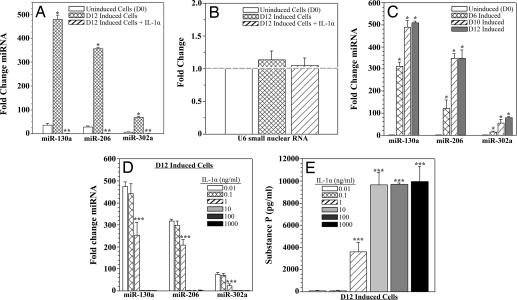
To quantify the observed miRNA bioarray results, candidate miRNAs (Table 2) were examined by real-time RT-PCR. D12-induced cells showed significantly (P < 0.05) elevated levels of miR-130a, miR-206, and miR-302a (Fig. 2A, hatched bars) compared with uninduced cells (Fig. 2A, open bars). IL-1α stimulation of D12 cells significantly (P < 0.05) decreased all miRNAs to undetectable levels (Fig. 2A, diagonal bars). U6 small nuclear RNA levels were not affected by neuronal induction or IL-1α stimulation, and thus served as a small RNA negative control (Fig. 2B). The results observed in Fig. 2A are consistent with developmental up-regulation of the candidate miRNAs, because increasing levels were found at day 6 neuronal induction (D6) and day 10 neuronal induction (D10) (Fig. 2C, hatched and diagonal bars). Varying the concentration of IL-1α stimulation produced a significant (P < 0.05) decrease in candidate miRNA levels (Fig. 2D) and an increase in SP production (Fig. 2E) at 1 ng/ml (Fig. 2E, diagonal bars), with maximal effect seen at 10 ng/ml (Fig. 2E, light gray bars). In summary, quantification of the endogenous candidate miRNAs in uninduced and induced MSCs verified the findings of the miRNA bioarrays.
Fig. 2.
miRNA and SP levels in D0 and D12 cells with or without IL-1α stimulation. (A and B) Fold change in candidate miRNAs (miR-130a, miR-206, and miR-302a) (A) and U6 small nuclear RNA in D0 and D12 cells (B) with or without IL-1α stimulation. (C and D) Candidate miRNAs were re-examined at D6 and D10 (C) and in D12 cells stimulated with various concentrations of IL-1α (D). All RNA levels were determined by real-time RT-PCR. Results are presented as mean fold change ± SD; n = 5. Normalizations with 5S rRNA were arbitrarily assigned values of 1. (E) D12 cells were stimulated for 16 h with various concentrations of IL-1α, and SP release was quantified by ELISA. Results are presented as mean ± SD; n = 5. *, P < 0.05 vs. D0; **, P < 0.05 vs. unstimulated D12 cells; ***, P < 0.05 vs. D12 cells stimulated with 0.01 ng/ml IL-1α.
Functional Target Validation of Candidate miRNAs.
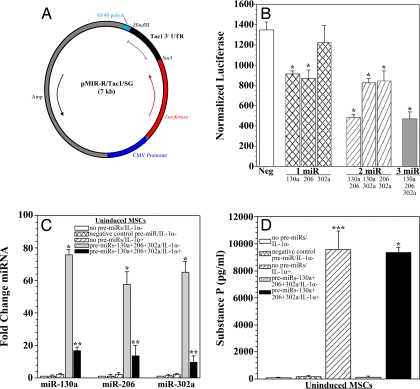
We assessed whether the candidate miRNAs can target Tac1 3′ UTR. We performed this investigation by cloning the Tac1 3′ UTR into the pMIR-REPORT miRNA luciferase reporter vector (pMIR-R/Tac1/SG) (Fig. 3A). Uninduced MSCs were cotransfected with pMIR-R/Tac1/SG and miR-130a, miR-206, and/or miR-302a pre-miRs (Fig. 3B). In parallel studies, cells were transfected with negative control pre-miRs. Transfectants with pre-miR-130a or pre-miR-206 alone (Fig. 3B, left and middle hatched bars), two pre-miRs (Fig. 3B, diagonal bars) or three pre-miRs (Fig. 3B, gray bar) showed significantly (P < 0.05) decreased reporter activity compared with negative control. The most significant (P < 0.05) decreases in reporter activity were observed in cells cotransfected with pre-miR-130a and pre-miR-206 (Fig. 3B, left diagonal bar) or all three pre-miRs (Fig. 3B, gray bar). Only transfectants with pre-miR-302a alone did not display a significant (P > 0.05) difference in reporter activity. Results in uninduced MSCs are only shown, because the assay measures the relative effect of exogenously applied miRNAs on the 3′ UTR reporter, regardless of cell-specific endogenous miRNAs. Similar results were observed for D12 cells (data not shown). In summary, the findings demonstrate that both miR-130a and miR-206 can target Tac1 3′ UTR.
Fig. 3.
Target validation of candidate miRNAs and effects of IL-1α on transfected pre-miRs. (A) The Tac1 3′ UTR was cloned into the pMIR-REPORT miRNA luciferase reporter system (pMIR-R/Tac1/SG). (B) D0 cells were cotransfected with pMIR-R/Tac1/SG and candidate pre-miRs (miR-130a, miR-206, and/or miR-302a), and luciferase and β-gal activities were measured. In parallel studies, uninduced cells were transfected with pre-miR negative control. Results are presented as the mean ± SD of normalized luciferase; n = 5. Normalizations were performed with luciferase/β-gal activities in cells transfected with pMIR-R/Tac1/SG alone, arbitrarily assigning a value of 1,500. (C) D0 cells were transfected with candidate or negative control pre-miRs or left untransfected. Cells were then stimulated for 16 h with IL-1α or unstimulated, and candidate miRNA levels were determined by real-time RT-PCR. Results are presented as mean fold change ± SD; n = 5. Normalizations with 5S rRNA were arbitrarily assigned values of 1. (D) D0 cells were prepared as in C, and SP levels were quantified by ELISA. Results are presented as mean ± SD; n = 5. *, P < 0.05 vs. negative control cells; **, P < 0.05 vs. untransfected cells stimulated with IL-1α; ***, P < 0.05 vs. untransfected, unstimulated cells.
Pre-miRNA Degradation by IL-1α.
We investigated whether down-regulation of candidate miRNAs by IL-1α (Fig. 2 A and D) is caused by miRNA degradation. We examined the effects of IL-1α on pre-miRNAs transfected in uninduced MSCs by real-time RT-PCR (Fig. 3C). Uninduced MSCs were chosen, because they endogenously express minimal levels of candidate miRNAs (Fig. 2A). MSCs cotransfected with all three candidate pre-miRNAs and stimulated with IL-1α (Fig. 3C, black bars) showed significantly (P < 0.05) decreased miRNA levels compared with unstimulated cells (Fig. 3C, gray bars). This decrease in candidate miRNAs was reflected at the protein level, because transfectants stimulated with IL-1α produced high levels of SP (Fig. 3D, black bar). In summary, IL-1α stimulation promotes the degradation of miR-130a, miR-206, and miR302a.
Inhibition of Endogenous miRNAs.
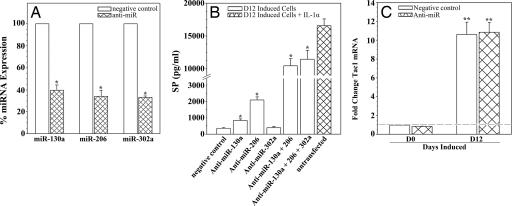
To determine whether the findings of the target validation reporter assay recapitulate the cell-specific effects of endogenous miRNAs, we used specific miRNA inhibitors (anti-miRs). Uninduced MSCs (D0) and induced MSCs (D12) were transfected with anti-miR-130a, anti-miR-206, or anti-miR-302a, and percent endogenous miRNA expression was measured by real-time RT-PCR (Fig. 4A). In parallel studies, cells were tranfected with negative control anti-miRs (Fig. 4A, open bars). The results showed a significant (P < 0.05) decrease in percent miRNA expression with each respective anti-miR (Fig. 4A, hatched bars). The data from D0 and D12 cells were pooled, because similar inhibitions were observed in both cell types.
Fig. 4.
Synergistic translational repression of Tac1 mRNA by miR-130a and miR-206. (A) D0 and D12 cells were transfected with anti-miR-130a, anti-miR-206, anti-miR-302a, or negative control. Endogenous miRNA levels were determined by real-time RT-PCR. Results are presented as mean ± SD percent expression; n = 5. Normalizations were performed with 5S rRNA in negative control cells, arbitrarily assigning a value of 100%. (B) D12 cells were transfected with anti-miR-130a, anti-miR-206, and/or anti-miR-302a or negative control, and SP release was quantified by ELISA. D12 cells stimulated for 16 h with IL-1α served as positive control. Results are presented as mean ± SD; n = 5. (C) D0 and D12 cells were transfected with anti-miR-130a, anti-miR-206, and anti-miR-302a or negative control, and Tac1 mRNA expression was determined by real-time RT-PCR. Results are presented as mean ± SD fold change; n = 5. Normalizations were performed with 5S rRNA in negative control cells, arbitrarily assigning a value of 1. *, P < 0.05 vs. negative control cells; **, P < 0.05 vs. D0 cells.
Effect of miRNA Inhibition on SP Production.
We next assessed whether specific inhibition of miR-130a, miR-206, and miR-302a had an effect on Tac1 translation. Detection of SP in the growth media served as readout for this investigation. D12 MSCs were transfected with anti-miR-130a, anti-miR-206, and/or anti-miR-302a, and growth media were collected for SP quantification by ELISA (Fig. 4B, open bars). In parallel studies, cells were transfected with negative control anti-miRs. D12 cells stimulated for 16 h with 10 ng/ml IL-1α served as positive control (Fig. 4B, hatched bar). The findings recapitulate the results of the target validation reporter assay, because significant (P < 0.05) SP levels were found only in cells transfected with anti-miR-130a or anti-miR-206 alone (Fig. 4B, second and third open bars from left), together (Fig. 4B, third open bar from right), or with all three inhibitors (Fig. 4B, second open bar from right). Significant levels of SP were not found (P > 0.05) in similarly transfected, uninduced MSCs (data not shown). In summary, the results demonstrate that miR-130a and miR-206 synergistically mediate translational repression of the neurotransmitter SP in MSC-derived neuronal cells.
Effect of miRNA Inhibition on Tac1 Transcription.
To determine whether miRNA inhibition influences Tac1 at the transcriptional level, D0 and D12 MSCs were cotransfected with anti-miR-130a, anti-miR-206, and anti-miR-302a, and Tac1 mRNA was measured by real-time RT-PCR (Fig. 4C). In parallel studies, cells were transfected with negative control anti-miRs. All candidate anti-miRs were cotransfected, because significant (P < 0.05) SP production was observed with these inhibitors (Fig. 4B, second bar from right). As expected from previous reports, Tac1 mRNA was significantly (P < 0.05) elevated in D12 cells transfected with negative control (Fig. 4C, right open bar), as compared with uninduced MSCs (Fig. 4C, left open bar) (21, 23). However, no significant (P > 0.05) difference in Tac1 levels was observed in D12 cells transfected with negative control as compared with anti-miRs (Fig. 4C, right hatched bar).
Discussion
IL-1α can recapitulate the response of an injured microenvironment where stem cells and/or the cells that they generate are implanted. Here, we report IL-1α alleviating miRNA-mediated repression of SP synthesis, which represents a unique finding, in particular to neuronal development. Specifically, IL-1α promoted miRNA degradation in D12 MSCs, although inhibitory effects on miRNA transcription cannot be ruled out (Fig. 3C). Effects of IL-1α on miRNA levels were seen at concentrations as low as 1 ng/ml, which closely mimics the in vivo environment (Fig. 2D). The physiological significance of this finding with regard to tissue repair is the proinflammatory property of SP (28). Excessive production of SP by implanted D12 MSCs might lead to immune cell infiltration and transplant rejection. Cellular transplantation alone generates a local immune response in the host tissue and exposure of donor cells to an inflammatory milieu. In places such as the brain or spinal cord, an exacerbated immune response could be extremely detrimental to the transplant recipient.
Roles for miRNA in the regulation of neurotransmitter synthesis hold physiological significance in the synaptic plasticity of neurons. miRNAs have been observed bound to mRNA transcripts and packaged with polyribosomes in dendrites (29). Such findings would implicate local protein synthesis in neurites, with the potential for rapid neurotransmitter production at synaptic termini. In this model, synapses could locally respond to an external stimulus by translating different prepackaged neurotransmitter transcripts to mediate synaptic plasticity.
We have combined computational and functional approaches to verify the specific effects of miRNAs to study translational regulation of Tac1 mRNA. Three candidate miRNAs were investigated (Table 2). Although miR-302a was predicted, functional analyses showed no evidence of interaction with the 3′ UTR of Tac1 mRNA (Figs. 3B and 4B). This conclusion was supported by the observation that miR-130a and miR-206 were just as effective in repressing luciferase translation as miR-130a, miR-206, and miR-302a together (Fig. 3B).
Inhibition of endogenous miR-130a and miR-206 in D12 cells caused significantly (P < 0.05) increased Tac1 mRNA translation, as indicated by high levels of its encoded peptide, SP (Fig. 4B). However, the levels of SP were significantly (P < 0.05) less than untransfected D12 neuronal cells, stimulated with IL-1α. An explanation for this disparity is incomplete inhibition of endogenous miR-130a and miR-206 by the anti-miRs (Fig. 4A).
Inhibition of miR-130a and miR-206 alone caused considerably less translation of Tac1 than in studies where both were inhibited (Fig. 4B). This synergism between miR-130a and miR-206 is consistent with other reports that show efficient inhibition by more than one miRNA per target (9).
Our studies focused on three miRNAs predicted to regulate Tac1. The results from the miRNA bioarrays showed many other miRNAs that were up-regulated in uninduced and induced MSCs. Several of the miRNAs expressed in the uninduced MSCs, namely miR-222, miR-221, miR-214, and miR-143, have functions linked to angiogenesis, myogenesis, adipogenesis, and hematopoietic regulation (30–33). These miRNAs are consistent with known functions of MSCs. Interestingly, miR-125a, miR-125b, and miR-23b are linked to neuronal differentiation (34, 35). These findings imply that undifferentiated MSCs possess intrinsic mechanisms to guide neuronal differentiation, when exposed to the proper environment.
Whether the profile of miRNAs up-regulated in D12 cells is similar to primary neurons is currently unknown. The type of primary neuron most similar to those derived from MSCs is still being investigated, thus parallel comparison is currently difficult. We have validated many of the miRNAs present in IL-1α-stimulated D12 cells in retinoic acid-treated neuroblastoma SY5Y cells (data not shown). Extensions to the present investigation with MSC-derived neurons are needed with other neuronal cells that produce SP, such as those in the hippocampus. Tac1 is induced in nonneuronal cells, such as BM stromal cells, although induction correlates with transcription and translation (36). However, it would be interesting to study whether similar roles are operative by miRNAs in stroma (36).
In summary, the present studies undertook an interdisciplinary methodology that traversed from global array analyses to computational target prediction to functional analyses by cellular and molecular approaches. The findings present a different role for miRNAs in the regulation of neurotransmitters.
Materials and Methods
Reagents.
DMEM with high glucose, DMEM/F12, l-glutamine, and B-27 supplement were purchased from Gibco (Carlsbad, CA). FCS, all-trans retinoic acid, and phospatase substrate were from Sigma (St. Louis, MO), and defined FCS was from Atlanta Biologicals (Lawrenceville, GA). IL-1α was obtained from Roche (Nutley, NJ), biotinylated SP was from Arnell Products (New York, NY), rabbit anti-SP was from Biogenesis (Kingston, NH), and alkaline phosphatase-conjugated goat anti-rabbit IgG was from Kirkegaard & Perry Laboratories (Gaithersburg, MD).
Culture of Human MSCs.
Human MSCs were cultured from BM aspirates as described (37). The use of human BM aspirates followed a protocol approved by the Institutional Review Board of The University of Medicine and Dentistry of New Jersey-Newark campus. Briefly, unfractionated BM aspirates were cultured in DMEM with 10% FCS on plasma-treated surfaces. At day 3, red blood cells and granulocytes were removed by Ficoll Hypaque density gradient separation. The cultures were incubated until 80% confluence. After four cell passages, the adherent cells were symmetric: CD14−, CD29+, CD44+, CD34−, CD45−, SH2+, prolyl-4-hydroxylase−. All experiments using MSCs were repeated with different BM donors.
Induction of MSCs.
At 20% confluence, MSCs subcultured in DMEM with 10% FCS were induced with neuronal induction media, which were comprised of Ham's DMEM/F12, 2% FCS (Sigma), B27 supplement, 20 μM retinoic acid, and 12.5 ng/ml basic fibroblast growth factor, and cells were cultured as described (22). In parallel studies, MSCs were cultured in media alone with vehicle used for reconstitution of retinoic acid and growth factors (D0). The data in all experiments were similar for D0 and those cultured in expansion media (DMEM with 10% FCS), with the latter referred to as undifferentiated MSCs. All experimental endpoints were performed with a maximal confluence of 70% to control for contact inhibition. Endpoints for induced MSCs at 6 and 12 days induction have been shown to correspond to partially differentiated and fully differentiated neuronal phenotype (21, 22).
miRNA Bioarray Analyses.
Total RNA (10 μg) from three different human BM donors was extracted from uninduced and IL-1α stimulated or unstimulated D12 MSCs. Samples were processed by Asuragen (Austin, TX) for miRNA profiling studies. The miRNA enriched fraction was obtained by passing total RNA through a flashPAGE Fractionator apparatus (Ambion, Austin, TX). The 3′ ends of the RNA molecules were tailed and then labeled with the mirVana miRNA Labeling Kit (Ambion). Amine-modified nucleotides were incorporated during the poly (A) polymerase-mediated tailing reaction, and Cy3 succinimide esters (Amersham Biosciences-GE Healthcare, Piscataway, NJ) were conjugated to the amine moieties on the miRNAs, and then hybridized to the mirVana miRNA Bioarrays (Ambion). The Cy3 fluorescence on the arrays was scanned at an excitation wavelength of 532 nm with a GenePix 4200AL scanner (Molecular Devices, Union City, CA). The fluorescent signal associated with the probes and local background was extracted by using GenePix Pro (version 6.0; Molecular Devices).
Thresholding and signal scaling were generated by using algorithms selected by Asuragen. The background adjusted fluorescent values generated by GenePix Pro were normalized for each miRNA using a variation stabilization transformation method followed by hypothesis testing with one-way ANOVA or t test depending on the number of groupings in the experimental design. For multiple group comparisons, the one-way ANOVA model was used to test the null hypothesis and filter out genes that had the same expression level across all groups. Pairwise comparisons were carried out on differentially expressed genes identified by ANOVA to see how they differed from each other. For each pair of treatments, a two-sample t test was carried out for every gene and multiplicity correction was followed to control the false discovery rate by using a step-up approach of 5%.
Real-Time RT-PCR.
miRNAs were analyzed for binding sites within Tac1 3′ UTR by the miRanda algorithm (www.microrna.org). Candidate miRNAs with putative Tac1 binding sites were analyzed by real-time RT-PCR to determine relative gene expression. For quantification of miR-130a, miR-206, and miR-302a, 2 μg total RNA was reverse-transcribed and amplified by using the mirVana qRT-PCR miRNA Detection Kit (Ambion), according to the manufacturer's specified guidelines. Primers for the RT-PCRs to quantitate miR-130a (GenBank accession no. MI0000448), miR-206 (GenBank accession no. MI0000490), and miR-302a (GenBank accession no. MI0000738) were purchased from Ambion. PCRs were normalized with the same test samples and 5S rRNA-specific primers. The normalization reactions were arbitrarily assigned values of 1 or 100%. The primers for 5S rRNA (GenBank accession no. V00589) and U6 small nuclear RNA (GenBank accession no. NR_002752) were purchased from Ambion.
Real-time PCR for Tac1 mRNA was initiated with 2 μg of total RNA in reverse transcription reactions. Two hundred micrograms of cDNA was used in amplification reactions with a Platinum SYBR Green qPCR SuperMix-UDG Kit (Invitrogen, Carlsbad, CA). The primers for Tac1 span + 60/+328 (GenBank accession no. NM_003182) with the following sequences: (forward) 5′-act gtc cgt cgc aaa atc-3′ and (reverse) 5′-ggg cca ctt gtt ttt caa-3′. PCRs were normalized by amplifying the same sample of cDNA with primers specific for β-actin and arbitrarily assigning a value of 1. The primers for β-actin span +842/+1037 (GenBank accession no. NM_001101), with the following sequence: (forward) 5′-tgc cct gag gca ctc ttc-3′ and (reverse) 5′-gtg cca ggg cag tga tct-3′. Real-time PCRs were performed with a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). The cycling profile for miRNA quantification (40 cycles) and 5S rRNA (40 cycles) was 95°C for 15 s and 60°C for 30 s. The cycling profile for Tac1 quantification (40 cycles) was 95°C for 15 s and 60°C for 45 s. Gene expression analysis was performed with 7500 System SDS Software (Applied Biosystems).
Cloning of 3′ UTR of Tac1 mRNA.
cDNA (200 ng) from D12 induced MSCs served as template to amplify Tac1 3′ UTR. Tac1 3′ UTR primers spanned +521/1054 (GenBank accession no. NM_003182): (forward) 5′-tcg aag ctt (HindIII site) gaa aga aga cgt taa-3′ and (reverse) 5′-atc gag ctc (SacI site) gct atc aca aca cat-3′. The underlined nucleotides indicate the respective restriction sites, shown in parenthesis. The amplified PCR product was gel-purified and subcloned into pCR2.1 (Invitrogen). The insert was excised with FastDigest HindIII and SacI restriction enzymes (Fermentas, Hanover, MD), gel-purified, and ligated into the pMIR-REPORT miRNA luciferase reporter vector (Ambion), referred to as pMIR-R/Tac1/SG. The reporter vector expresses a luciferase gene under the control of a CMV promoter and is therefore constitutively expressed. Because the multiple cloning regions are located downstream of the luciferase gene, the cloned fragment was therefore ligated 3′ of the luciferase gene.
Transient Transfection and Reporter Gene Assay.
At 50% confluence, D0 or D12 MSCs were cotransfected with pMIR-R/Tac1/SG and miR-130a, miR-206, and/or miR-302a or pre-miRs using the siPORT NeoFX transfection reagent (Ambion). In parallel studies, cells were transfected with negative control pre-miRs. The negative control molecules were random sequence pre-miRs that had been tested in human cell lines and tissues and validated by the vendor (Ambion) to not produce identifiable effects on known miRNA function. All cells were also transfected with pMIR-REPORT β-gal (pMIR-β-gal) for transfection efficiency. After 48 h, cell-free lysates were prepared as described (36). Lysates were analyzed for luciferase and β-gal levels by using the Luciferase Assay System (Promega, Madison, WI) and β-gal detection kit II (BD Clontech, Palo Alto, CA), respectively. Normalizations were performed with luciferase/β-gal activity in cells transfected with pMIR-R/Tac1/SG alone, arbitrarily assigning a value of 1,500. Transfections were repeated if β-gal activities were <100 relative luciferase units. Total protein was determined with a DC protein assay kit (Bio-Rad, Hercules, CA).
SP ELISA.
D0 and D12 MSCs were transfected with miR-130a, miR-206, and miR-302a miRNA inhibitors (anti-miRs) or pre-miRs using the siPORT NeoFX transfection reagent (Ambion) or left untransfected. In parallel studies, cells were transfected with negative control anti-miRs or pre-miRs. The negative control anti-miRs were random sequence molecules that had been tested in human cell lines and tissues and validated by the vendor (Ambion) to not produce identifiable effects on known miRNA function. Cells were stimulated for 16 h with various concentrations of IL-1α or 10 ng/ml IL-1α. This concentration of IL-1α was found to be optimal for SP quantification. Cell growth media were collected, and competitive ELISA was used to quantify SP production as described (28). SP levels were calculated from a standard curve developed with OD at 405 nm vs. 12 serial dilutions of known SP concentrations.
Data Analysis.
Statistical analyses were performed with ANOVA and Tukey-Kramer multiple comparisons test. P < 0.05 was considered significant.
Acknowledgments
We thank Ambion and Asuragen for providing research service and technical support during this study. This work was supported by a grant from the F. M. Kirby Foundation.
Abbreviations
- miRNA
microRNA
- MSC
mesenchymal stem cell
- BM
bone marrow
- SP
substance P
- pre-miRNA
precursor miRNA
- D0
uninduced MSCs
- D6
day 6 neuronal induction
- D10
day 10 neuronal induction
- D12
day 12 neuronal induction.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Novina C-D, Sharp PA. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 2.Grosshans H, Slack F. J Cell Biol. 2002;156:17–21. doi: 10.1083/jcb.200111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin G-A, Sevignani C, Dumitru C-D, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce C-M. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Jeon K, Lee JT, Kim S, Kim V-N. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim V-N. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Lund E, Guttinger S, Calado A, Dahlberg J-E, Kutay U. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Zamore P-D. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 8.Doench J-G, Petersen C-P, Sharp P-A. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Y, Yi R, Cullen B-R. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai E-C. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 11.Lewis B-P, Burge C-B, Bartel D-P. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutz P-L, Chawla G, Stoilov P, Black D-L. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell D-K, Kaur S, Stanislaw S, Konieczka J-H, Yatskievych T-A, Antin P-B. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 15.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krichevsky A-M, King K-S, Donahue C-P, Khrapko K, Kosik K-S. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin G-A, Croce C-M. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer S, Zavolan M, Grasser F-A, Chien M, Russo J-J, Ju J, John B, Enright A-J, Marks D, Sander C, Tuschl T. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 19.Bianco P, Riminucci M, Gronthos S, Robey P-G. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 20.Deans R-J, Moseley A-B. Exp Hematol. 2000;28:875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 21.Cho K-J, Trzaska K-A, Greco S-J, McArdle J, Wang F-S, Ye J-H, Rameshwar P. Stem Cells. 2005;23:383–391. doi: 10.1634/stemcells.2004-0251. [DOI] [PubMed] [Google Scholar]
- 22.Greco S-J, Zhou C, Ye J-H, Rameshwar P. Stem Cells Dev. 2007 doi: 10.1089/scd.2007.0011. in press. [DOI] [PubMed] [Google Scholar]
- 23.Greco S-J, Rameshwar P. J Immunol. 2007;179:3342–3350. doi: 10.4049/jimmunol.179.5.3342. [DOI] [PubMed] [Google Scholar]
- 24.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, et al. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams B-D, Furneaux Dagger H, White B. Mol Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 26.Anderson C, Catoe H, Werner R. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge X, Wu Q, Wang S-M. BMC Genomics. 2006;7:285. doi: 10.1186/1471-2164-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rameshwar P, Gascon P. Blood. 1995;86:482–490. [PubMed] [Google Scholar]
- 29.Kim J, Krichevsky A, Grad Y, Hayes G-D, Kosik K-S, Church G-M, Ruvkun G. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 31.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flynt A-S, Li N, Thatcher E-J, Solnica-Krezel L, Patton J-G. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esau C, Kang X, Peralta E, Hanson E, Marcusson E-G, Ravichandran L-V, Sun Y, Koo S, Perera R-J, Jain R, et al. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Belasco J-G. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn F-G. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 36.Qian J, Yehia G, Molina C, Fernandes A, Donnelly R-J, Anjaria D-J, Gascón P, Rameshwar P. J Immunol. 2001;166:2553–2561. doi: 10.4049/jimmunol.166.4.2553. [DOI] [PubMed] [Google Scholar]
- 37.Potian J-A, Aviv H, Ponzio N-M, Harrison J-S, Rameshwar P. J Immunol. 2003;171:3426–3434. doi: 10.4049/jimmunol.171.7.3426. [DOI] [PubMed] [Google Scholar]