Abstract
Recent work has shown that the nature of hydration of pure hydrophobic surfaces changes with the length scale considered: water hydrogen-bonding networks adapt to small exposed hydrophobic species, hydrating or “wetting” them at relatively high densities, whereas larger hydrophobic areas are “dewetted” [Chandler D (2005), Nature 29:640–647]. Here we determine whether this effect is also present in peptides by examining the folding of a β-hairpin (the 14-residue amyloidogenic prion protein H1 peptide), using microsecond time-scale molecular dynamics simulations. Two simulation models are compared, one explicitly including the water molecules, which may thus adapt locally to peptide configurations, and the other using a popular continuum approximation, the generalized Born/surface area implicit solvent model. The results obtained show that, in explicit solvent, peptide conformers with high solvent-accessible hydrophobic surface area indeed also have low hydration density around hydrophobic residues, whereas a concomitant higher hydration density around hydrophilic residues is observed. This dewetting effect stabilizes the fully folded β-hairpin state found experimentally. In contrast, the implicit solvent model destabilizes the fully folded hairpin, tending to cluster hydrophobic residues regardless of the size of the exposed hydrophobic surface. Furthermore, the rate of the conformational transitions in the implicit solvent simulation is almost doubled with respect to that of the explicit solvent. The results suggest that dehydration-driven solvent exposure of hydrophobic surfaces may be a significant factor determining peptide conformational equilibria.
Keywords: hydrophobicity, hydration density, molecular dynamics simulation, explicit and implicit solvent model, β-hairpin
Hydrophobic interactions are among the main thermodynamic driving forces in self-assembly of soft and biophysical matter (1–3). Recent work indicates that the nature of hydrophobicity differs depending on whether small molecules or large clusters are considered (4–7). A small number of methane molecules in water show little tendency to aggregate (8, 9). On such small length-scales, water can host and “wet” the hydrophobic species without significant modification of its intermolecular organization (7, 10). In contrast, when the nucleus of a hydrophobic species extends beyond a critical length (11), a liquid–vapor-like “dewetted” interface will form at the solute surface (4, 12, 13), involving a higher thermodynamic cost (7, 14) and inducing hydrophobic collapse (12, 15, 16).
Although quantitative theories exist regarding the role played by hydrophobicity in the clustering processes of pure hydrophobic systems in water (14, 17), our understanding of hydrophobicity in the context of protein and peptide folding in which amphiphilic species are concerned is still incomplete. Solution-scattering experiments on aqueous solutions of small amphiphiles, such as methanol (10, 18), or single hydrophobic amino acids, such as N-acetyl-leucine-amide (19, 20), have revealed that, even at high concentrations, the solutes form only small clusters and do not segregate themselves completely from the aqueous solvent to form a large hydrophobic core. However, there is no clear consensus on the nature of hydration of such amphiphilic species. The organization of water molecules might be enhanced around the exposed hydrophobic portions involving, for example, clathrate-like structures (20–22), or alternatively may remain bulk-like (18, 23). For folded proteins, there is evidence that the local hydration density varies with the surface topography and local electrostatic field, rather than directly with the hydrophobicity of the surface residue considered (24).
Here we perform a simulation study of the folding of a β-hairpin peptide, to address the role played by solvent hydration density variations in peptide conformational equilibrium. To do this, two simulation models are compared, one using explicit water molecules and one with implicit solvent. In the explicit model, water molecules may adapt to the local structural properties of the peptide. In contrast, in the implicit model, which is computationally less expensive, the solvent is treated as a homogeneous isotropic dielectric continuum. The implicit solvent model used here is the generalized Born/surface area (GB/SA) model (25), which is among the most widely applied implicit solvent models. In GB/SA, the solvation free energy is decomposed into a nonpolar (cavity formation and dispersion interaction) component, typically approximated by a linear term that is proportional to the solvent-exposed surface area (26, 27), and an electrostatic term corresponding to the free energy of charging the solute (25). By using GB/SA, structures in agreement with experiment have been computed for some proteins (28, 29) and peptides that possess hydrophobic cores (30–35).
The peptide studied here is an amyloidogenic β-hairpin peptide (residues 109–122 of the Syrian hamster prion protein, H1 peptide). This peptide was chosen because it exhibits large solvent exposure of the hydrophobic residues, typical of aggregation-prone molecules, but the size scale of the exposed hydrophobic patches is not large enough to actually induce hydrophobic collapse. Analysis of both the explicit and implicit solvent simulations enables us to examine the role played by hydration density variations in the thermodynamic stability of the peptide and points to dehydration solvent exposure of hydrophobic surfaces as being a driving force in the folding of the peptide.
Results and Discussion
A structural, thermodynamic, and kinetic analysis of the folding/misfolding of the H1 peptide at room temperature, obtained by long time-scale (1-μs) atomistic molecular dynamics (MD) simulation in explicit water has been provided previously (36–38). The peptide, when initially modeled as an α-helix, preferentially adopts a β-hairpin conformation that undergoes several unfolding/refolding events. An example of the observed β-hairpin is given in Fig. 1a. The hairpin is consistent with chemical shifts measured by NMR (39) and with a low-resolution structure derived using x-ray diffraction (40). For the present study, an additional 2-μs simulation of the H1 peptide was performed in equivalent physicochemical conditions but with the implicit solvent model (GB/SA simulation).
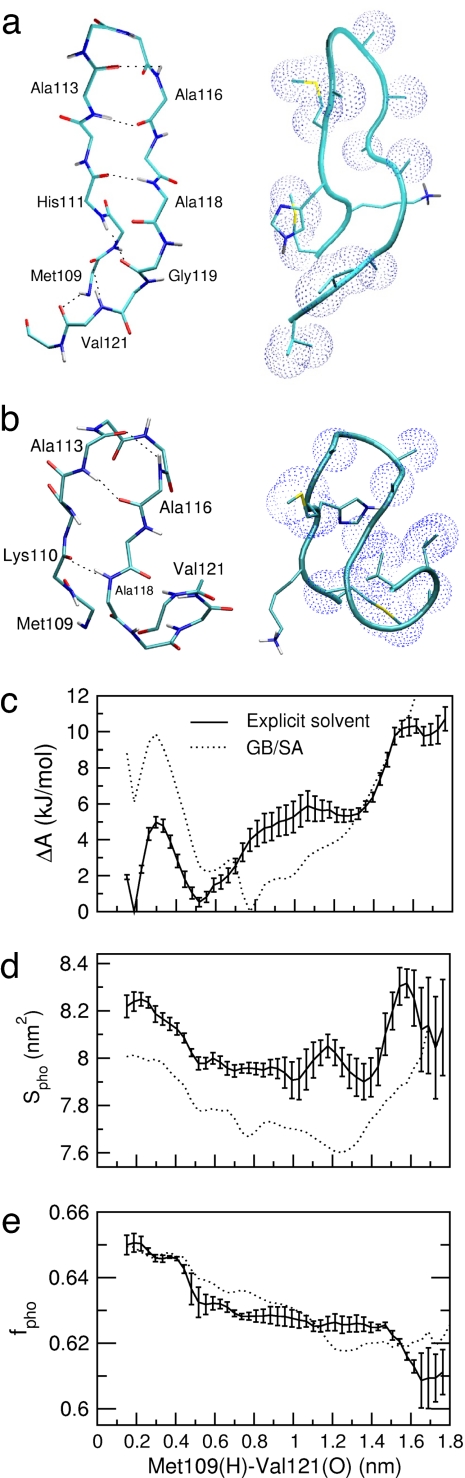
Fig. 1.
Peptide structure and thermodynamics. (a) Representative fully folded β-hairpin structure observed in the reference explicit solvent simulation. (b) Representative structure of the most populated state observed in the implicit solvent simulation, with the β-turn formed (Ala113–Ala116) and with the C terminus bent so as to collapse a nucleus of hydrophobic residues. (c) Free energy profiles along the terminal β-hairpin interstrand hydrogen bond distance, Met109(H)–Val121(O). (d and e) Hydrophobic solvent-accessible surface area, Spho (d), and the fraction of the solvent-accessible surface area that is hydrophobic, fpho (e), as a function of hydrogen bond distance. Error bars shown throughout the article correspond to a standard deviation of the corresponding property as obtained by considering five subsets of the trajectory.
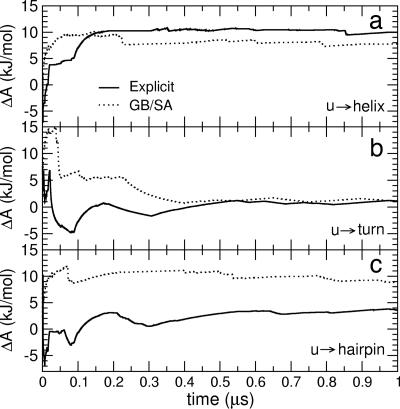
To quantitatively compare the structural thermodynamics in the two simulation models, the stability of the unfolded state relative to that of the helical and β secondary structure motifs was calculated. Given thermodynamic equilibrium, at constant temperature and volume, the stability is given by the free energy difference between the unfolded state, u, and the given folded state, f, ΔAu→f = −RT ln pf/pu, where R is the ideal gas constant, T is the temperature, and pf and pu are the probabilities of being in states f and u, respectively. In Fig. 2 is presented the sampling dependence of the free energy change estimate for the transition from the unfolded state to the following secondary structure states: (i) the “β-turn” state, which comprises all configurations in which the Ala113–Ala116 turn is formed (u → turn); (ii) the “fully folded β-hairpin” state, comprising all configurations in which the turn is formed and the termini are close together in space, i.e., the hydrogen-bonding distance between Met109(H) and Val121(O) is <0.3 nm (u → hairpin); (iii) helical structures (u → helix). The plots show that in both simulations convergence of the free energy values is achieved within ≈1 μs, such that free energy changes associated with the secondary-structure conformational transitions are obtained to within 0.5 kJ/mol statistical accuracy. Extending the GB/SA simulation up to 2 μs confirmed the convergence for this method (data not shown).
Fig. 2.
Convergence with time of the free energy change, ΔA, corresponding to the transition from the unfolded condition to helical (a), β-turn (b), and fully folded β-hairpin (c) structures for the two simulations. The final values (at 1 μs) are the equilibrium free energies and show statistical accuracy within 0.5 kJ/mol.
The free energy changes for the u → turn and u → helix transitions are very similar in the explicit and implicit solvent simulations (ΔAu→turn ≈1 kJ/mol and ΔAu→helix ≈ 8–10 kJ/mol), both exhibiting the higher stability of the β-turn structures relative to helical geometries. However, the fully folded hairpin is destabilized in the implicit solvent relative to the explicit solvent simulation by ≈6.5 kJ/mol (ΔAu→hairpin ≈ 9 kJ/mol and ≈3.5 kJ/mol, respectively). The 3.5 ± 0.5 kJ/mol value of ΔAu→hairpin can be compared with experimental values ranging from ≈ −7 to ≈3 kJ/mol for various β-peptides (41). In the 1-μs explicit solvent simulation, six refolding events were observed (vs. five in the implicit solvent simulation). This yields a mean folding time, as defined by the mean residence time of the unfolded state, of ≈150 ns, which compares with experimental folding times of various β-peptides ranging between 52 μs and 260 ± 20 ns (41–43). Therefore, both the rate of folding/unfolding and the stability are consistent with the experimental data available on fast-folding β-hairpin peptides.
To further explore the difference in stability of the hairpin found in the GB/SA and explicit solvent simulations, the free energy profiles as a function of the distance between the hydrogen and oxygen atoms involved in the β-hairpin interstrand hydrogen bonds were calculated. The profile showing the largest difference is that of the terminal hydrogen bond, between Met109(H) and Val121(O), shown in Fig. 1c. The profiles from both models possess a narrow minimum at ≈0.2 nm (corresponding to fully folded structures, as in Fig. 1a), a significant barrier to separation of the hydrogen bond at ≈0.3 nm, and other minima at ≈0.5–0.8 nm. In both the explicit solvent and GB/SA simulations, the fully extended hairpin state (at ≈0.2 nm) is characterized by high solvent exposure of hydrophobic residues, as shown by the corresponding average hydrophobic solvent-accessible surface area, Spho, plotted in Fig. 1d. Although this state is the global free energy minimum in explicit solvent, it is highly destabilized in the implicit solvent. In contrast, structures populating the minima at 0.5–0.8 nm, i.e., corresponding to the global GB/SA free energy state, mostly have the turn formed, but with disordered termini and a lower Spho, consistent with more collapsed structures. A representative structure of the global GB/SA free energy minimum is illustrated in Fig. 1b.
High Spho, as observed for the fully folded hairpin, is also observed for unfolded conformations [Met109(H)–Val121(O) distance ≥1.4 nm]. However, a more instructive property differentiating the folded and unfolded structures is the fraction of the exposed surface area that is hydrophobic, fpho, i.e., the ratio of the hydrophobic solvent-accessible surface area to the total solvent-accessible surface area. This ratio is lowest in the unfolded state (Fig. 1e).
The question arises as to the role played by the solvent in the different arrangements of hydrophobic residues observed in the two solvent models. To investigate solvent effects in the context of the hydration properties proposed in refs. 7, 11, 14, 15, and 44, we compare the hydration in the explicit-solvent simulation of the H1 peptide with the hydration of a hypothetical pure hydrophobic polymer of the same size as the H1 peptide, modeled by setting all partial charges to zero and simulated for 500 ns under the same conditions as the explicit solvent simulation of the H1 peptide.
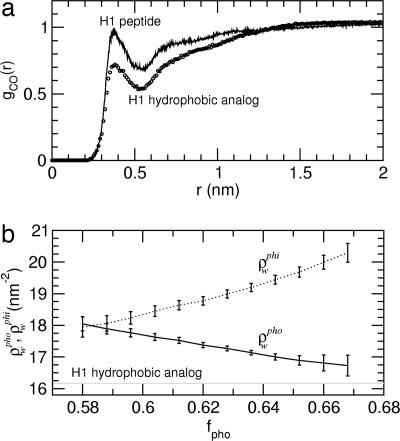
The H1 hydrophobic analog was found to collapse into a globular structure, with a radius of ≈0.9–1.0 nm, that remains stable over the whole simulation time. The corresponding radial distribution of the distance of the water oxygen atoms to the peptide surface hydrophobic-residue carbon atoms exhibits a first hydration shell density ≈25% lower than that of the bulk (Fig. 3a, circles), indicating solvent depletion at the surface, as has also been observed previously for simulations of hard spheres with radii ≥1.0 nm (14). The hydration density around the exposed hydrophobic surface, ρwpho, remains constant over the whole simulation time, with an average value of ≈16.22 nm−2 (shown in Fig. 3b as a gray line). These data show that the behavior of the H1 hydrophobic analog is consistent with the “dry” regime of purely hydrophobic solutes for which drying-induced hydrophobic collapse is observed (15).
Fig. 3.
Hydration properties. (a) Radial distribution, gCO(r), of the distance, r, between the water oxygen atoms and the peptide surface hydrophobic-residue carbon atoms (circles, H1 hydrophobic analog; solid line, H1 peptide calculated for structures with ρwpho ≤ 16.75 nm−2). (b) H1 peptide hydration density around hydrophobic, ρwpho (solid line), and hydrophilic, ρwphi (dotted line), residues as a function of the fraction of the solvent-accessible surface area that is hydrophobic, fpho. Nearly identical results are found when Spho is used instead of fpho. ρwpho (ρwphi) is defined here as the number of water molecules, Nw, within a 0.55-nm (0.45-nm) radius cutoff from every hydrophobic (hydrophilic) atom on the peptide surface per unit of exposed hydrophobic (hydrophilic) surface area (ρwpho = Nw/Spho; ρwphi = Nw/Sphi). ρwpho was also calculated for the H1 hydrophobic analog, and the average value is shown as a gray line.
For the H1 peptide, which contains both hydrophilic and hydrophobic surface sites, the radial distribution corresponds to a first-shell hydration density comparable to the bulk density, suggestive of a wet regime (Fig. 3a). Furthermore, analysis of the local hydration densities around the hydrophobic and hydrophilic residues, ρwpho and ρwphi, as a function of the exposed hydrophobic surface area fraction, fpho, reveals clear trends (Fig. 3b). When fpho is at its minimum (≈0.58), the hydration density is similar around hydrophobic and hydrophilic sites (ρwpho ≈ ρwphi ≈ 18.0 nm−2). With increasing fpho, the local hydration density around hydrophobic residues decreases, while increasing around hydrophilic sites. These trends suggest that the surface polar atoms may aid in depletion of water around neighboring hydrophobic surfaces by taking up hydration water.
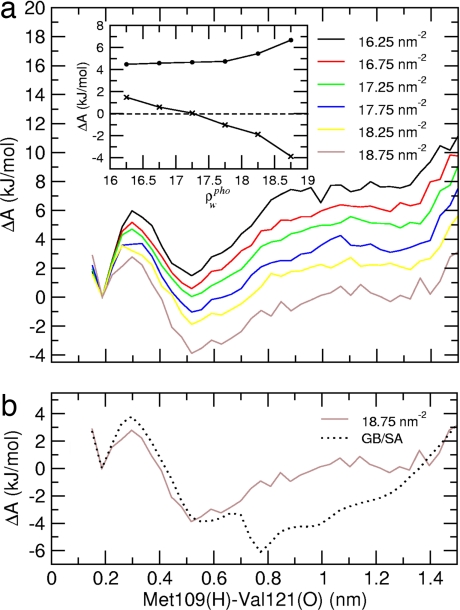
To study the effects of local hydration density variations on folding thermodynamics, the structures generated from the explicit solvent simulation were clustered according to their ρwpho values, and for each cluster the free energy profile along the Met109(H)–Val121(O) distance was calculated. The results are shown in Fig. 4a. In the Inset, the free energy differences between the solvent-separated (at 0.55 nm) and hydrogen-bonded (at 0.2 nm) minima, and between the transition state and solvent-separated minimum, are shown for the different profiles. Although the barrier height shows little dependence on ρwpho, a crossover from positive (+2 kJ/mol for ρwpho = 16.25 ± 0.25 nm−2) to negative (−4 kJ/mol for ρwpho = 18.75 ± 0.25 nm−2) values of the free energy difference between the solvent-separated and global minimum is observed, i.e., the fully folded hairpin is destabilized with respect to the collapsed, partially unfolded state by ≈6 kJ/mol when ρwpho increases by 13%.
Fig. 4.
Free energy profiles. (a) Free energy profiles along the terminal interstrand hydrogen bond distance, Met109(H)–Val121(O), for the explicit solvent simulation. The different profiles are calculated by using subpopulations with different hydrophobic surface hydration density, ρwpho. The values indicated are the central values of each ρwpho range (±0.25 nm−2). (Inset) Free energy difference between the solvent-separated minimum and the hydrogen bond formation minimum (crosses), shown for the different profiles. A crossover from positive to negative values of the free energy difference can be observed at a value ρwpho of ≈17.25 ± 0.25 nm−2. The free energy barrier to forming the hydrogen bond is also shown (filled circles). (b) Free energy profile, as obtained from the GB/SA simulation, compared with the explicit solvent profile calculated on the subpopulation with the highest possible hydration density (ρwpho = 18.75 ± 0.25 nm−2). In both profiles, the free energy difference between solvent-separated and the hydrogen bond formation minimum is the same, namely approximately −4 kJ/mol.
Existing implicit solvent models treat the water as a high-dielectric continuum. This leads to the continuum models being unable to properly describe phenomena such as local water depletion around hydrophobic residues. Indeed, comparison of the free energy profiles obtained from the GB/SA simulation and the explicit solvent simulation calculated on the subpopulation with the highest surface hydration, which is the hydration condition closest to the uniform dielectric implicit solvent model, confirms this hypothesis: the free energy difference between the solvent-separated and hydrogen-bonded minima is the same for both models in this case (Fig. 4b). Therefore, because the implicit solvent model does not represent local dewetting, the collapse of hydrophobic residues is favored, with a consequent strong effect on the equilibrium conformational distribution.
Finally, we examine solvent effects on the peptide dynamics. Following previous work (37, 38), we consider the dynamics along a single backbone collective degree of freedom, the conformational coordinate, q, defined as the slowest relaxing backbone collective coordinate resulting from a principal component analysis of the backbone atomic fluctuations (45–47).
We model the mean square displacement along the coordinate q, 〈Δq2(t)〉, with a multiexponential decay of the corresponding velocity autocorrelation function (37, 38). The diffusion along the coordinate q is described by two processes: (i) short-time diffusion (typically up to a few picoseconds) within a configurational region, which may correspond to a single harmonic well, followed by (ii) long-time diffusion between such regions. The model has the following functional form:
where τi are the relaxation times, Ai are parameters defined by the integrals of the velocity autocorrelation function (see Appendix of ref. 37), D0 is the diffusion constant describing the initial fast diffusion process, and D∞ is the diffusion constant characterizing the long-time diffusion process.
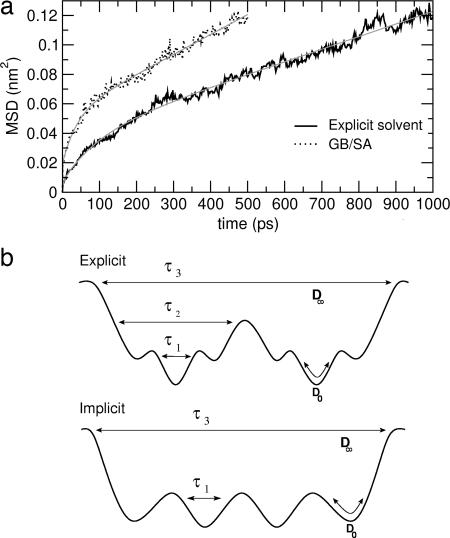
Fig. 5a shows 〈Δq2(t)〉 obtained by the simulations, together with fits of Eq. 1 over time ranges of 1,000 ps and 500 ps for the explicit and implicit solvent simulations, respectively. The different time intervals are chosen such that, for all of the profiles, the same final 〈Δq2(t)〉 value is reached, corresponding to ≈0.12 nm2. Diffusion coefficients and relaxation times obtained from the models are listed in Table 1.
Fig. 5.
Dynamics. (a) Mean square displacement, as a function of time, along the main conformational coordinate, q, which is the eigenvector with the highest eigenvalue obtained by diagonalizing the covariance matrix of positional fluctuations of the 15 Cα atoms of the peptide (black line). The fit of Eq. 1 is also shown (gray curves). The correlation coefficients are >0.992. (b) Schematic drawing of the potential energy surface for the explicit (Upper) and implicit (Lower) solvent systems, with associated model parameters evaluated from Eq. 1.
Table 1.
Conformational diffusion constants and corresponding relaxation times (Eq. 1)
| Solvent simulation | Diffusion constant, nm2·ps−1 |
Relaxation time, ps |
Avg. H-bond lifetime, ps |
||||
|---|---|---|---|---|---|---|---|
| D0 | D∞ | τ1 | τ2 | τ3 | τwp | τpp | |
| Explicit | 0.02 (<0.01) | 2.6 (0.5) 10−5 | <1 | 7 (2) | 102 (5) | 8 (3) | 132 (21) |
| GB/SA | 0.05 (0.01) | 5.5 (0.5) 10−5 | <1 | — | 43 (4) | — | 49 (10) |
D0 is the short-time diffusion constant; D∞ is the long-time diffusion constant; and τ1, τ2, and τ3 are the relaxation times. The data are calculated over a time range of 1,000 ps and 500 ps for the explicit and implicit solvent simulations, respectively. In the last two columns are reported the average hydrogen bond lifetimes for two different kinds of hydrogen bonds: water–peptide, τwp, and intrapeptide, τpp. Noise is indicated in parentheses and corresponds to a standard deviation.
The model functions reproduce 〈Δq2(t)〉 over the whole time range. For the implicit solvent simulation, 〈Δq2(t)〉 is found to be accurately modeled with only two exponentials, whereas three are necessary for the explicit solvent simulations. Furthermore, the implicit solvent simulation undergoes faster conformational diffusion. The reason for these differences is again found in the analysis of the specific peptide–solvent interactions. The average lifetime of the water–peptide hydrogen bonds (Table 1) calculated from the explicit solvent simulations almost coincides with the τ2 relaxation time (≈10 ps). This relaxation is absent in the GB/SA simulation. Another striking result is that in both simulations the average intrapeptide hydrogen bond lifetime (last column of Table 1) and the slowest conformational relaxation time are found to almost coincide, indicating that with both the explicit and implicit solvent models the slowest conformational relaxation is driven by intrapeptide hydrogen bond dynamics.
The above data indicate that the use of the implicit solvent results in significantly faster conformational diffusion because of lower complexity and a less-hierarchical character of the underlying energy surface (as depicted in Fig. 5b), due to the absence of water–protein hydrogen bond breaking and forming.
Conclusions
The present work investigates solvent effects on the folding/misfolding process of a β-hairpin peptide, the 14-residue amyloidogenic prion protein H1 peptide (39, 40). To distinguish between continuum and explicit solvent effects, an implicit solvent simulation based on the generalized Born method (GB/SA simulation) is compared with explicit solvent molecular dynamics simulations. This comparison is made on the microsecond time scale, 2 to 3 orders of magnitude longer than in previous comparative studies (32, 48–50) and within reach of experimental peptide folding times (51).
The implicit solvent model reproduces the explicit solvent simulation conformational thermodynamics (i.e., the free energy differences between specific β-structures and the unfolded state) only up to the formation of the β-turn. In the implicit solvent simulation, low free energy structures are always associated with low exposure of hydrophobic surface area. Therefore, the conformation of lowest free energy in the explicit solvent simulation, the fully folded extended hairpin, is destabilized by the implicit solvent model as it is characterized by high exposure of hydrophobic residues and the absence of a hydrophobic core.
A consistent explanation for the thermodynamic behavior arises from the analysis of how local water hydration density changes accompany the exposure of hydrophobic patches in the explicit solvent simulation. Local variations of the first hydration shell density are found to correspond to partial drying around exposed hydrophobic residues, with a simultaneous increase in hydration density around the hydrophilic residues. If the depletion of water were to be uniform around the peptide, hydrophobic interactions would be expected to dominate (15), with resulting destabilization of the fully folded hairpin. Instead, the local drying allows hydrophobic patches to be hosted by the solvent, thus stabilizing the noncollapsed form. The implicit solvent model is unable to reproduce the local water density variations and therefore does not accommodate small hydrophobic nucleii at the surface, resulting in a tendency to collapse, irrespective of the size scale of the exposed hydrophobic surface area.
The possibility does exist of errors in the computed GB/SA structures and relative thermodynamics arising from inaccurate effective Born radii. However, the parameters used here in the approximate analytical formula for computing the Born radii (25) have been optimized to minimize the differences between the effective Born radii calculated by the finite-difference Poisson–Boltzmann method and by the approximate formula. Moreover, another GB/SA solvent simulation of the H1 peptide (52) performed with a different force field, namely OPLS (53), showed the same structural features of the peptide found in the present GB/SA simulation. Conceivably, however, an implicit solvent model could be devised that would mimic the effect on peptide conformational equilibrium of the hydration shell density variations seen here.
Analysis of the dynamics along the main conformational coordinate indicates that the absence of atomistic details in the implicit solvent simulation results in an enhanced diffusion rate (almost doubled in the present case). The dynamics is found to correlate well with intrapeptide and peptide–water hydrogen bond formation and rupture. In particular, a relaxation process in the peptide, driven by peptide–solvent hydrogen bond forming and breaking, is present in the explicit solvent simulation only.
The present results point to a role of local hydration structure in determining the structural organization of peptides and, therefore, folding/unfolding thermodynamics. Enhanced interactions between water and polar sites at the surface allow modulation of the hydration density of neighboring hydrophobic units and thus, consistent with the principles described in refs. 7, 12, 16, 20, and 54, in the character and strength of the hydrophobic interaction in biomolecular systems.
Methods
The explicit and implicit solvent simulations of the H1 peptide (MKHMAGAAAAGAVV) were performed under the same physicochemical conditions, with the same force field for the peptide (55). Simulations were performed in the NVT ensemble with isokinetic temperature coupling to keep the temperature constant at 300 K (56). The bond lengths were fixed (57), and a time step of 2 fs was used for numerical integration of the equations of motion. Coordinates were stored every 2 ps. The side-chain protonation states reproduce a pH of ≈7. The N and C termini were amidated and acetylated, respectively (39).
For the simulations in explicit solvent, one peptide molecule was placed in a periodic truncated octahedron large enough to contain ≈1.0 nm of water on all sides, modeled with the simple point charge model (58), and one negative counter ion (Cl−) was added to neutralize the system. For the simulations without explicit water molecules, the implicit solvent was modeled by using the generalized Born theory (59), in which the protein is surrounded by a uniform polar solvent with a dielectric constant εw = 80. The Born radii were calculated by using the fast asymptotic pairwise summation of ref. 60. The relevant parameters for the GROMOS96 force field were taken from ref. 61. The nonpolar part of the solvation free energy was modeled by using an effective surface tension associated with the solvent-accessible surface area (60). Instead of a costly calculation of the accurate surface area, a mimic based on the Born radii was used, which has been shown to be very accurate and much faster (30).
The following simulations were performed:
Explicit solvent. MD was performed in explicit water for 1 μs with long-range electrostatic interactions treated with the particle mesh Ewald method (62), with a real-space cutoff of 0.9 nm. The first 240 ns were performed starting from the α-helix conformation obtained in previous work (36) and the following 760 ns starting from the β-hairpin conformation observed during the previous 240-ns simulation, using a new set of initial velocities.
GB/SA. MD was performed with implicit solvent for 500 ns, starting from the α-helix conformation obtained in previous work (36), and for 1.5 μs starting from the β-hairpin conformation observed in the explicit solvent simulation. All pairwise interactions were modeled, i.e., no cutoff was used. Similar results were obtained by an additional Langevin dynamics simulation performed with the same GB/SA implicit solvent model but using a Langevin integrator with a friction coefficient of 90 ps−1.
To obtain the free energy along hydrogen bond distance, d, the MD structures were projected onto 50 grid cells used to divide the overall accessible range. For every cell, the number of points was counted and the relative probability density, ρeq(d), was calculated. The grid cell with the highest probability density, ρeq(dref), i.e., the cell corresponding to the overall free energy minimum, was chosen as the reference state. Finally, the free energy profile was evaluated as ΔA(d) = −RT ln ρeq(d)/ρeq(dref).
To evaluate the average hydrogen bond lifetime of a group of hydrogen bonds, use was made of the bond existence autocorrelation function C(t), which is the probability that a randomly chosen hydrogen bond that was intact at the initial time (t = 0) is also found intact at later time t (63, 64). The average lifetime, τ, is then defined by C(τ) = e−1. Two kinds of average lifetimes are calculated: τwp and τpp for water–peptide and intrapeptide hydrogen bonds, respectively. Intrapeptide hydrogen bonds involved in stable secondary structure elements were not included because they possess significantly longer lifetimes.
Acknowledgments
I.D. was supported by VolkswagenStiftung Project I/80 437; M.B.U. was supported by The Wellcome Trust.
Abbreviations
- GB/SA
generalized Born/surface area model
- MD
molecular dynamics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Kauzmann W. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Tanford C. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 3.Dill K. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 4.Stillinger F. J Solution Chem. 1973;2:141–158. [Google Scholar]
- 5.Lee C, McCammon J, Rossky P. J Chem Phys. 1984;80:4448–4455. [Google Scholar]
- 6.Huang D, Chandler D. Proc Natl Acad Sci USA. 2000;97:8324–8327. doi: 10.1073/pnas.120176397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler D. Nature. 2005;29:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 8.Rapaport DC, Scheraga HA. J Phys Chem. 1982;86:873–880. [Google Scholar]
- 9.Buchanan P, Aldiwan N, Soper A, Creek J, Koh C. Chem Phys Lett. 2005;415:89–93. [Google Scholar]
- 10.Soper A, Finney J. Phys Rev Lett. 1993;71:4346–4349. doi: 10.1103/PhysRevLett.71.4346. [DOI] [PubMed] [Google Scholar]
- 11.Rajamani S, Truskett T, Garde S. Proc Natl Acad Sci USA. 2005;102:9475–9480. doi: 10.1073/pnas.0504089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Margulis C, Berne B. Proc Natl Acad Sci USA. 2003;100:11953–11958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poynor A, Hong L, Robinson I, Granick S, Zhang Z, Fenter P. Phys Rev Lett. 2006;97:266101. doi: 10.1103/PhysRevLett.97.266101. [DOI] [PubMed] [Google Scholar]
- 14.Lum K, Chandler D, Weeks J. J Phys Chem B. 1999;103:4570–4577. [Google Scholar]
- 15.ten Wolde P, Chandler D. Proc Natl Acad Sci USA. 2002;99:6539–6543. doi: 10.1073/pnas.052153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raschke T, Tsai J, Levitt M. Proc Natl Acad Sci USA. 2001;98:5965–5969. doi: 10.1073/pnas.111158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hummer G, Garde S, Garcia AE, Pohorille A, Pratt LR. Proc Natl Acad Sci USA. 1996;93:8951–8955. doi: 10.1073/pnas.93.17.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixit S, Crain J, Poon W, Finney J, Soper AK. Nature. 2002;416:829–832. doi: 10.1038/416829a. [DOI] [PubMed] [Google Scholar]
- 19.Head-Gordon T, Sorenson J, Pertsemlidis A, Glaeser R. Biophys J. 1997;73:2106–2115. doi: 10.1016/S0006-3495(97)78241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorenson J, Hura G, Soper A, Pertsemlidis A, Head-Gordon T. J Phys Chem B. 1999;103:5413–5426. [Google Scholar]
- 21.Cheng Y, Rossky P. Nature. 1998;392:696–699. doi: 10.1038/33653. [DOI] [PubMed] [Google Scholar]
- 22.Despa F, Berry R. Biophys J. 2007;92:373–378. doi: 10.1529/biophysj.106.087023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacs H, Mark A, van Gunsteren W. Proteins Struct Funct Genet. 1997;27:395–404. doi: 10.1002/(sici)1097-0134(199703)27:3<395::aid-prot7>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 24.Merzel F, Smith J. Proc Natl Acad Sci USA. 2002;98:5378–5383. doi: 10.1073/pnas.082335099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 26.Chothia C. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- 27.Sharp K, Nicholls A, Fine R, Honig B. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 28.Pitera J, Swope W. Proc Natl Acad Sci USA. 2003;100:7587–7592. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan H, Mark AE, Zhu J, Honig B. Proc Natl Acad Sci USA. 2005;102:6760–6764. doi: 10.1073/pnas.0408857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer M, Bartels C, Karplus M. J Mol Biol. 1998;284:835–848. doi: 10.1006/jmbi.1998.2172. [DOI] [PubMed] [Google Scholar]
- 31.Ma B, Nussinov R. Proteins Struct Funct Genet. 1999;37:73–87. doi: 10.1002/(sici)1097-0134(19991001)37:1<73::aid-prot8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 32.Bursulaya BD, Brooks CL., III J Phys Chem B. 2000;104:12378–12383. [Google Scholar]
- 33.Felts AK, Harano Y, Gallicchio E, Levy RM. Proteins Struct Funct Genet. 2004;56:310–321. doi: 10.1002/prot.20104. [DOI] [PubMed] [Google Scholar]
- 34.Snow C, Qiu L, Du D, Gai F, Hagen S, Pande V. Proc Natl Acad Sci USA. 2004;101:4077–4082. doi: 10.1073/pnas.0305260101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulmschneider J, Jorgensen W. J Am Chem Soc. 2004;126:1849–1857. doi: 10.1021/ja0378862. [DOI] [PubMed] [Google Scholar]
- 36.Daidone I, Simona F, Roccatano D, Broglia RA, Tiana G, Colombo G, Di Nola A. Proteins Struct Funct Bioinf. 2004;57:198–204. doi: 10.1002/prot.20178. [DOI] [PubMed] [Google Scholar]
- 37.Daidone I, Amadei A, Di Nola A. Proteins Struct Funct Genet. 2005;59:510–518. doi: 10.1002/prot.20427. [DOI] [PubMed] [Google Scholar]
- 38.Daidone I, D'Abramo M, Di Nola A, Amadei A. J Am Chem Soc. 2005;127:14825–14832. doi: 10.1021/ja053383f. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen J, Baldwin MA, Cohen FE, Prusiner SB. Biochemistry. 1995;34:4186–4192. doi: 10.1021/bi00013a006. [DOI] [PubMed] [Google Scholar]
- 40.Inouye H, Kirschner DA. J Struct Biol. 1998;122:247–255. doi: 10.1006/jsbi.1998.3998. [DOI] [PubMed] [Google Scholar]
- 41.Muñoz V, Ghirlando R, Blanco F, Jas G, Hofrichter J, Eaton W. Biochemistry. 2006;45:7023–7035. doi: 10.1021/bi052556a. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Oyola R, Gai F. J Am Chem Soc. 2003;125:15388–15394. doi: 10.1021/ja037053b. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, Purkayastha P, Gai F. J Am Chem Soc. 2006;128:15836–15842. doi: 10.1021/ja064865+. [DOI] [PubMed] [Google Scholar]
- 44.Hummer G, Garde S, Garcia A, Pratt L. Chem Phys. 2000;258:349–370. [Google Scholar]
- 45.Garcia A. Phys Rev Lett. 1992;66:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 46.Amadei A, Linssen ABM, Berendsen HJC. Proteins Struct Funct Genet. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 47.de Groot BL, Amadei A, Scheek RM, van Nuland NA, Berendsen HJC. Proteins Struct Funct Genet. 1996;26:314–322. doi: 10.1002/(SICI)1097-0134(199611)26:3<314::AID-PROT7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Zhou R, Berne B. Proc Natl Acad Sci USA. 2002;99:12777–12782. doi: 10.1073/pnas.142430099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nymeyer H, Garcia A. Proc Natl Acad Sci USA. 2003;100:13934–13939. doi: 10.1073/pnas.2232868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumketner A, Shea JE. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68 doi: 10.1103/PhysRevE.68.051901. 051901. [DOI] [PubMed] [Google Scholar]
- 51.Kubelka J, Hofrichter J, Eaton W. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Ulmschneider J, Ulmschneider MB, Di Nola A. J Phys Chem B. 2006;110:16733–16742. doi: 10.1021/jp061619b. [DOI] [PubMed] [Google Scholar]
- 53.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 54.Hummer G, Garcia A, Garde S. Phys Rev Lett. 2000;85:2637–2640. doi: 10.1103/PhysRevLett.85.2637. [DOI] [PubMed] [Google Scholar]
- 55.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Zürich: Hochschulverlag AG an der ETH Zürich; 1996. [Google Scholar]
- 56.Brown D, Clarke JHR. Mol Phys. 1984;51:1243–1252. [Google Scholar]
- 57.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 58.Berendsen HJC, Grigera JR, Straatsma TP. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 59.Bashford D, Case DA. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 60.Qiu D, Shenkin P, Hollinger F, Still W. J Phys Chem A. 1997;101:3005–3014. [Google Scholar]
- 61.Zhu JY, Shi Y, Liu H. J Phys Chem B. 2002;106:4844–4853. [Google Scholar]
- 62.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 63.Luzar A, Chandler D. Phys Rev Lett. 1996;76:928–931. doi: 10.1103/PhysRevLett.76.928. [DOI] [PubMed] [Google Scholar]
- 64.Luzar A. J Chem Phys. 2000;113:10663–10675. [Google Scholar]