Abstract
• Scope In the second part of a two-part review, the ubiquity and universality of epigenetic systems is emphasized, and attention is drawn to the key roles they play, ranging from transducing environmental signals to altering gene expression, genomic architecture and defence.
• Key Issues The importance of transience versus heritability in epigenetic marks is examined, as are the potential for stable epigenetic marks to contribute to plant evolution, and the mechanisms generating novel epigenetic variation, such as stress and interspecific hybridization.
• Future Prospects It is suggested that the ramifications of epigenetics in plant biology are immense, yet unappreciated. In contrast to the ease with which the DNA sequence can be studied, studying the complex patterns inherent in epigenetics poses many problems. Greater knowledge of patterns of epigenetic variation may be informative in taxonomy and systematics, as well as population biology and conservation.
Keywords: Epigenetics, ploidy, hybrids, DNA methylation, histones, RNA, chromatin, silencing, transgenes, transposons, genome evolution, plant population biology, plant systematics, variation, heritability, plant development
INTRODUCTION
As will be obvious from the first part of this review (Grant-Downton and Dickinson, 2005), it is now evident that epigenetic systems are essential for genomic function. In terms of genomic structure and its integrity, epigenetic systems appear to be all-controlling. The formation and maintenance of the major structural units of the chromosome defined by their specialized heterochromatic state—the centromeric structures and telomeric structures—appears to be determined by epigenetic systems. Direct evidence for centromeric heterochromatin in plants being determined by transcription of repetitive DNA structures is now accumulating. Transcripts from these repetitive sequences are able to form aberrant RNAs and dsRNA which are then processed to siRNAs and act to direct and maintain sequence-specific heterochromatin formation. The resulting siRNAs appear to be rare and only recently have they been detected directly. It would seem that RNA-based systems are the singular most important mechanism for generating heterochromatin of all types, whether constitutive structural chromatin or heterochromatin directed to other sequences. The RNA-based processing of aberrant transcripts also acts as a surveillance system for unsilenced transposon sequences in the genome, with inactivation of any active elements through DNA methylation and heterochromatin formation. These elements can be deleterious to the genome through their mutagenic effects during movement and their unregulated movement would be damaging over a longer period of time. Already, the activation of such elements in backgrounds such as ddm1 has been shown to produce novel transposon-induced mutants. Naturally, the RNA system for detection of dsRNA viral transcripts is essential to the plant for protection against these pathogens, with its direct trigger of invading RNA virus destruction. Therefore the dual roles of structuring the genome and its defence are united in epigenetic pathways (Waterhouse et al., 2001).
The flip side of control over silencing, i.e. control over gene expression in euchromatic regions, is naturally the domain of epigenetic systems. Structural features of chromatin, from higher-order architecture to positioning of the nucleosome on a specific sequence of a promoter, dictate whether transcriptional machinery such as transcription factors can bind and effect transcription from a sequence. Chromatin states act in conjunction with transcription factors to regulate which genes are transcribed, and to what extent they are transcribed. Even if a sequence is transcribed and transcripts are abundant, post-transcriptional regulation can occur via the miRNA-induced degradation system. What seems the most important factor about these epigenetic systems of control is that the changes are dynamic and typically can be rapidly induced in response to stimuli. Epigenetic systems therefore act as the conduit for information, both developmental and environmental in origin, to initiate short- and long-term gene expression changes. In plants, tracing the pathway from initiating signal to gene expression change via epigenetic systems is rather lagging behind the same kind of work in eukaryotic model systems such as yeast. In terms of developmentally regulated gene expression changes, the most detailed example is the expression of the phaseolin gene in French bean. This gene encodes a major seed storage protein and its expression is under very tight developmental regulation, restricted to the later phase of embryogenesis. A full review of this model system for developmental regulation of a gene via chromatin changes can be found in Li et al. (2001).
EPIGENETICS AS THE CONDUIT FOR ENVIRONMENTAL SIGNALS
Sensing environmental changes and initiating a gene expression response is most important for plants as sessile autotrophs. Responses can be both short term (e.g. synthesis of protectant molecules against stress) and long term (e.g. phenotypic plasticity, with alteration of developmental programmes) because plant tissues typically have the benefit of no rigid or set developmental plan. Epigenetic systems must be part of the relay from sensing a change in the environment to a change in gene expression. Their ability to alter rapidly and reversibly, yet with the potential to keep a stable ‘memory’ through many cell divisions, must be a key to the flexibility of plant responses to the environment. Perhaps this is why plant epigenetic systems are complex and contain some unique constituents not seen in other eukaryotes. Surprisingly, not much is known about epigenetics and plant responses to environmental change, with one major exception. This exception is the regulation of flowering time (i.e. initiating inflorescence development) in Arabidopsis thaliana in response to long periods of cold temperatures, known as vernalization. Some genotypes of this annual species require an extended period of cold temperatures before commitment to flowering, behaving as ‘winter annuals’, with this vernalization requirement preventing flowering until favourable spring weather arrives. Other genotypes do not require cold treatment to induce flowering. The major genetic differences between these genotypes have been identified to two loci, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). Molecular studies have shown that FLC acts as a repressor of flowering, whilst FRI acts to boost the levels of FLC, so these two proteins act synergistically to block flowering. How does exposure to cold block this repression of flowering? Exposure to cold must be ‘recorded and stored’ by the plant, as when temperatures increase and mitotic cell divisions continue, the memory of exposure to cold is still retained as is the competence to flower. Storing this as epigenetic information has now been shown to be the key to the vernalization response. With exposure to cold over a significant length of time, the chromatin at the FLC locus changes, with reduction of acetylation at H3 K9 and K14 sites, and increase in methylation of the H3 K9 and K27 sites, corresponding to the formation of heterochromatin (Bastow et al., 2004; Sung and Amasino, 2004). Furthermore, it has been reported that this locus shows reduced H3 K4 trimethylation, a mark that has been associated with transcriptional activation, after vernalization (Amasino, 2004). It appears that the ultimate outcome of lengthy cold exposure is formation of heterochromatin and silencing of FLC, repressing the main repressor of flowering. Interestingly, FLC alleles with transposable elements inserted in an intron are down-regulated and flower rapidly without vernalization as its presence induces stable heterochromatin formation at the locus, mimicking vernalization-induced heterochromatin (Amasino, 2004). Mutants that block responsiveness to cold have also been identified. Two of the mutants correspond to chromatin-remodelling proteins, with VERNALIZATION2 (VRN2) a Pc-G protein (Bastow et al., 2004) and VERNALIZATION INDEPENDENCE3 (VIP3), forming a repeat-rich protein which is likely to be involved in forming the scaffold for assembling chromatin-remodelling protein complexes (Zhang et al., 2003), fitting with epigenetic pathways regulating vernalization responses. The most significant discovery is that mutations in three Arabidopsis genes homologous to components of the yeast PAF1 complex, a complex which recruits a histone methyltransferase (SET1) catalysing H3K4 trimethylation, all promote vernalization-independent early flowering, block FLC expression and reduce FLC H3K4 trimethylation (Zhang et al., 2002; He et al., 2004; He and Amasino, 2005). Moreover, altered flowering time has been regularly seen as a pleiotropic effect of other mutants in the epigenetic machinery. What is not known is any of the molecular detail of how the cold is sensed over a continuous lengthy period and the signal relayed to effect heterochromatization of FLC. By some mechanism, a long duration of cold is measured and committed to ‘epigenetic memory’, as short durations of cold are insufficient to acquire competence to flower; Amasino (2004) has speculated that this cold sensing may be via titration effects of antagonistic enzymes in a signalling pathway where one enzyme has different temperature-response kinetics. Clearly there is still much work to be done in revealing the full working detail of the pathway from environmental stimulus to gene expression change even in this model system.
EPIGENETIC HERITABILITY IN PLANTS: LAMARCK'S LAST LAUGH?
Epigenetic states and their heritability in plants
How the plasticity of plant growth and development may be a consequence of sophisticated epigenetic systems that allow sensitive and rapid gene expression changes to be effected has been discussed above. Another interesting consequence of plant developmental programmes is that there is no dedicated germline set up, segregated and maintained from early development onwards. Instead, cells giving rise to the germline develop de novo from the somatic tissues. This allows the opportunity for any stable epigenetic information acquired by the chromatin–DNA structures of the somatic tissues to be transmitted to the next generation, provided no epigenetic resetting system that deletes such acquired information is active. Clearly, the structurally important epigenetic information of the centromeres and telomeres is not deleted in this way during reproduction but this is just the start. How in paramutation epigenetic states at some loci can be heritably transmitted and influence susceptible (paramutable) alleles in the next generation, has been discussed in previous paragraphs. Spontaneous epialleles, such as those identified at SUPERMAN and Lcycloidea loci, and epialleles at many other loci induced by events such as interspecific hybridization and a background with compromised epigenetic machinery, have been shown to be heritable with considerable fidelity. Silent (heterochromatic) transposable elements and other repetitive sequences (such as transgene structures) located in euchromatic regions also retain their state with great stability from generation to generation. This could be explained by either stable transmission of epigenetic marks from generation to generation, or a short phase in the meiotic-gametophytic development in which there is an altered epigenetic state (e.g. release of silencing by loss of heterochromatization) followed by rapid and consistent re-establishment. Evidence for both scenarios exists. Perhaps the most interesting evidence comes from analysis of Arabidopsis met1 mutants in the male gametophyte. Heterozygotes for the mutation have been shown to accrue changes to DNA methylation in first-generation offspring, even though the mutation behaves recessively (Saze et al., 2003). This is because after meiosis the male germ line undergoes further mitotic divisions; those meiocytes lacking functional MET1 undergo cell division without the maintenance methyltransferase to preserve DNA methylation with absolute fidelity, leading to sperm cells with aberrant methylation patterns. By uncovering evidence for an essential role for MET1 in keeping epigenetic order through gametophytic development, this work implies that DNA methylation could be the major repository of epigenetic information in plants from generation to generation, as at this phase of the life cycle no ‘back-up’ of chromatin information (such as histone marks) exists. This discovery implies at least some chromatin therefore has to be set up and reprogrammed de novo from the DNA sequence and methylation marks with the next generation. This is fitting with observations of gross, global changes in chromatin during gametophytic development in plants, though detail of this gametophytic reprogramming is scarcely understood and appears to vary considerably from species to species. Interestingly, work in transgene silencing systems has revealed that the methylation in the coding region in PTGS silenced lines is not persistent from generation to generation in the absence of the trigger, whereas in TGS silenced lines there is evidence for some persistence of silencing and retention of symmetrical DNA methylation even after the inducing construct has been segregated away (Aufsatz et al., 2002). It is important to point out that the naturally occurring hypermethylated Lcyc allele in Linaria vulgaris shows quite stable inheritance of DNA methylation in both the promoter and coding regions (Cubas et al., 1999) and the same is true of SUPERMAN epialleles (Kishimoto et al., 2001). However, the exact mechanism of their spontaneous genesis is not known, though conventional transposon insertion events have been ruled out, making these ‘exceptions’ awkward. Further consideration of heritable phenomena will be found in later discussion of epigenetic phenomena in hybridization. This evidence does suggest that some acquired information sealed in the epigenetic state can be passed unperturbed from parent to offspring.
Epigenetic rewriting and the gametophyte
Equally, though, there is evidence in other plant taxa for global changes in DNA methylation in development of the gametophyte. Oakeley et al. (1997) report the global reduction in DNA methylation of the generative nucleus compared with the vegetative nucleus in Nicotiana pollen development after using immunocytological detection. An updated study using antibodies raised to the different histone marks would be hugely informative. Could these changes allow a slight release of the silenced epigenetic state at many loci, with the ‘sensing’ mechanisms that detect ‘aberrant’ transcripts resetting constitutively heterochromatic regions anew for the next generation? Wholesale release of silencing of transposons and repetitive sequences across the genome certainly seems unlikely to take place for two reasons. First, it seems a remote prospect that the epigenetic pathways could fully cope with a deluge of previously suppressed transcripts and, secondly, the mutagenic nature of activated transposable elements would be deleterious, particularly in haploid gametophytes. Indeed, it is tempting to speculate that the presence of specialized gametophytic chromatin states may act to allow partial release of transcription, with a limited increase in transcription from otherwise heterochromatic regions but without an overwhelming derepression of transcription. Using sequences such as transposable elements, centromeric repeats and silenced transgenes to determine their level of transcription and the levels of derived siRNAs in the stages and different cells of gametophytic development would be required to ascertain whether subtle changes in transcriptional potential across the genome are initiated. It is fitting to note that transcriptome analysis of maize sperm cells has revealed that a substantial proportion of sequences identified are transposons, with a published estimate of 8 % being retrotransposons (Engel et al., 2003) and an updated estimate of 19 % (McCormick laboratory website, http://www.pgec.usda.gov/McCormick/McCormick/mclab.html).
Epigenetic memory and environmental responses
The subtle nuances of the heritability of epigenetic states have yet to receive enough attention, although it certainly is not an outright Lamarckian mechanism, even in plants. The work on vernalization is a clear example of plants acquiring and utilizing information from the environment without the epigenetic information becoming heritably stable. Vernalized plants show propagation of the epigenetic memory of cold through mitosis but not meiosis—offspring from cold-exposed plants do not show altered flowering times. A system must exist in reproductive development that re-sets this specific heterochromatic state after the signal to initiate flowering has been conveyed. Indeed, there appears to be no known stable epimutations/epialleles for FLC that affect flowering time, only sequence variants such as transposon insertions. Why is this particular heterochromatic state re-set so efficiently? One reason may be that the FLC locus is part of a domain dedicated to vernalization and it may be distinguished by its own unique chromatin markings. Recent work has shown that FLC and a neighbouring locus (of unknown function) are co-ordinately affected by vernalization treatments (Finnegan et al., 2004). Moreover, transgenes inserted into this region acquire vernalization responsiveness and, when the sequence of this region itself is moved to other parts of the genome, it retains its vernalization responsiveness and can affect the neighbouring sequence. Collectively, this points to the sequence of the FLC region as the key to acquiring a particular heterochromatin information state, which can then spread outwards to the neighbouring sequence. Further dissection of the properties of this domain to explore why and how it subsequently loses this heterochromatic mark would be most valuable.
To what extent is the FLC domain representative of how the plant genome is constructed? It has been argued that other such transcriptional domains are common throughout the genomes of plants (Finnegan, 2001). If each has autonomous mechanisms during the reproductive development for eliminating or resetting epigenetic information acquired during growth, this would limit the amount of epigenetic information passed from generation to generation. However, the apparent rarity of transgene insertions acquiring novel expression profiles from the surrounding sequence (Schubert et al., 2004), and evidence from epigenetics in hybrid systems (discussed below), leaves this subject to debate. The most pressing question is—how many DNA sequences can show metastable epigenetic behaviour, with the potential to occupy more than one epigenetic state (i.e. form epialleles or epimutations) that are heritable and affecting gene expression? As yet, it is only possible to infer answers, but with developments in analysis of transcriptional profiling and epigenetics (such as bisulfite sequencing and CHiP), more information is likely to accrue. Even if a fraction of the genes in plants have the potential to form sets of epialleles, it would be a tremendously important, yet hitherto undetected, source of variation. Even in mammals there are examples of epialleles, such as at the axin and agouti loci, where the normal resetting of the epigenetic state during reproduction is escaped which allows trans-generational inheritance of epigenetic states with its consequences for expression patterns of the gene and subsequent phenotype (Rakyan et al., 2003; Chong and Whitelaw, 2004). If, as seems logical, the capacity to form epialleles is determined to some extent by DNA sequence, this raises new questions about the interplay between sequence evolution and epigenetic evolution.
EVOLUTION AND EPIGENETICS IN PLANTS
Epigenetics of plant genome evolution
In hindsight, it now seems strange that plant biologists have, for many years, discussed evolution of plant genomes and plant populations whilst only considering the DNA and protein sequence as the determinants of phenotype. Prior to the extensive molecular exploration of epigenetics, early ideas on epigenetics and evolution were reviewed and discussed by Jablonka and Lamb (1989). It is now an exciting time for plant biologists interested in evolution as the new data on epigenetic systems in plants can be assimilated into a more complete understanding of the field. In this section it is considered how essential epigenetics have become in understanding various evolutionary problems and where following this avenue to its logical conclusion may eventually lead.
What drives the formation of diversity in the genome? At the level of the DNA sequence, alterations such as deletions, point mutations and chromosomal rearrangements have been the main focus for decades and the prevailing consideration in how genomes and populations evolve over time. There is no dispute that such alterations are essential to generating variation, both ‘neutral’ and ‘non-neutral’ upon which selection can act directly. Interestingly, there is evidence that the mutational frequency of methyl-cytosine is substantially greater than unmodified cytosines (e.g. Rideout et al., 1990). The mutations are formed from the deamination of methyl-cytosine to thymine. This implies that there is a mutational penalty for marking sequences by methylation, which would be more severe for densely methylated sequences such as repetitive DNA. It is tempting to speculate that patterns of DNA methylation can significantly bias the mutational frequency of the marked sequence in plants. Transposons and repetitive DNA targeted for RdDM and heterochromatization may have higher rates of mutation than euchromatic regions, perhaps aiding in defence. This mechanism has been proposed to operate in elimination of ectopic promoters in the coding regions of genes in plants (Tran et al., 2005).
Transposons and epigenetics in genome evolution
This brings us to the next major source of variation in the plant genome—transposon-induced variation. Since the discovery of these selfish mobile elements, their modifying effects on the host genome have become well documented and will be but briefly mentioned here (see Kidwell and Lisch, 1997, 2001; Dimitri and Junakovic, 1999). The first notions of their importance in shaping genomes came from McClintock (see McClintock, 1984). Their effects range from the outright mutagenic, e.g. disruption of coding genes by insertion of a mobile element, the formation of ‘footprints’ in insertion and excision cycles, to more subtle. For instance, ectopic recombination can occur between homologous elements in meiosis, leading to chromosomal rearrangements, and it is possible that ‘macrotransposons’ can form, capable of shifting even relatively large sequences of trapped host DNA to new sites in the genome (Gray, 2000). Clearly, most outcomes of transpositional activity in an evolved, structured genome will be neutral to deleterious, making the defence system described above essential to prevent dangerously high levels of transposition. Of course, the defence system that induces heterochromatin formation of detected transposon sequences may actually cause some of the deleterious effects itself, by silencing neighbouring host sequences. In recent years, there has been a considerable shift in the appreciation of these mobile elements and the view of them as selfish parasites has become mollified to treating them as genomic symbionts. Many plant genomes are enormously rich in these kinds of sequences yet efficient mechanisms to rid genomes of such sequences have evolved in other eukaryotes, suggesting that their co-habitation has some benefits. Nowhere is this more evident than in the structural centromeric and telomeric regions of genomes that are essential for genomic integrity and rich in such sequences. Integration of transposable elements within or close to coding genes has been demonstrated to generate novel phenotypic variation in plants (Kumar and Bennetzen, 1999) with examples from cultivated plants such as maize (Wessler, 1988) and Antirrhinum (Coen et al., 1986) known for many years. However, there is a considerable gulf between populations of cultivated plants and plants in populations where natural selection hones the genome. A boost to the view that transposable elements can shape the genome and gene expression patterns in nature has been the finding that ‘fossil remnants’ of transposable element sequences are ubiquitous in euchromatic gene sequences. Statistical surveys (e.g. Bureau et al., 1996) have found that many plant genes have close associations with transposable elements. White et al. (1994) have identified plant genes that probably have coding regions of genes adopted from copia retrotransposon gene sequences. The same study revealed that many genes have degenerate transposable element sequences in their upstream and downstream flanking sequences; in some cases, this sequence is known to have a direct regulatory effect on the expression of the gene. Perhaps the most interesting and complete example is the imprinted FWA gene, shown to be epigenetically sensitive, which has been shown to be intimately associated with SINE retrotransposon sequences (Lippman et al., 2004). siRNAs corresponding to this sequence were also found in this study and the data points to FWA imprinting and transcription being directly controlled by this retrotransposon sequence. Coding genes may ‘adopt’ integrated or nearby transposable element sequences on a frequent basis, making such elements a major force for generating variation and novelty in the genome; indeed, Kumar and Bennetzen (1999) speculate that perhaps all plant genes will be shown to contain a transposable element legacy. As epigenetic systems are the major regulators of these elements, it is a case for ‘guilt by association’ in their contribution to the molecular and phenotypic evolution of plants.
Another connection between repetitive DNA and the evolution of epigenetic control of gene expression has recently been revealed. By analysing the sequences of microRNA genes in Arabidopsis, it has been shown that these genes are likely to arise from inverted repeats of coding genes. Allen et al. (2004) have identified miRNA genes from Arabidopsis that appear to be intermediate between inverted repeats and typical miRNA genes, as their sequence shows considerable homology to the target gene outside the region of binding of the processed miRNA. Indeed, one of these ‘intermediate’ genes, ASRP1729, did not seem to have behave as an miRNA, as it did not have a targeting function and was insensitive to DCL-1 mutations for its processing, possibly representing an intermediate stage before miRNA function has evolved. It seems plausible that these ‘intermediates’ may become trans-acting siRNAs (Peragine et al., 2004; Vazquez et al., 2004). In this manner, new miRNA genes controlling novel expression patterns of the target genes could arise without much difficulty. Voinnet (2004) commenting on this work suggests that, ‘given their highly evolving nature, young small RNAs are ideally suited to convey rapid adaptation and will probably be more abundant in plants subjected to stress’. In support of this idea, it is interesting to note that a strong candidate for a non-coding RNA involved in transcriptional stress responses was identified by T-DNA tagging in the resurrection plant Craterostigma plantagineum (Furini et al., 1997; Bartels and Salamini, 2001). This sequence is involved in negatively regulating desiccation-tolerance genes and its sequence shows closest similarity to SINE-like retrotransposons. No homologues have been detected in Arabidopsis; it would be interesting to determine how quickly this regulatory RNA evolved by examining whether similar sequences exist in closer relatives in the Scrophulariaceae and Gesneriaceae. At the other extreme, there is evidence for astonishing conservation of miRNAs and their target sequences, indicating that once this regulatory system is in place and controlling transcript abundance it can become highly canalized. The mRNAs of class-III HD-Zip transcription factors from all lineages of land plants have retained an miR165/166 binding site, and evidence for cleavage at this site from several species was also forthcoming (Floyd and Bowman, 2004). The retention of miRNA regulation of these genes may span more than 400 million years. However, there is no evidence for overlap between miRNAs and genes targeted by miRNAs in plants and animals.
Evolution through mutations in genes governing epigenetic systems
Another means by which epigenetic systems could generate novel variation in plant populations is through mutations in genes of major regulatory effect, e.g. genes like MET1 and DDM1. As discussed above, loss of function does not necessarily prove lethal to the plant and their absence can lead to the generation of stable epialleles of genes of developmental and morphological importance, as well as activation of transposable elements. Importantly, there is evidence that some epialleles can be maintained independent of the loss-of-function mutation that provided their genesis; restoration of epigenetic function by crossing back to wild type may rescue epigenetic regulation systems but with the retention of the epiallele. The loss of methylation of the FWA locus in Arabidopsis (Soppe et al., 2000) is heritable and induces ectopic expression of the FWA protein which results in a late-flowering phenotype. In fact, FWA is not expressed at all in adult tissues of the wild type, but instead is restricted to the central cell of the megagametophyte and the resulting endosperm tissue after fertilization (Kinoshita et al., 2004), where expression is correlated with reduced methylation derived from the DNA glycosylase activity of DEMETER in the central cell. The default state of FWA is methylated and silenced, except in terminally differentiated reproductive tissues, but its reactivation and ectopic expression by an indiscriminate demethylation event results in an unrelated and rather unexpected late-flowering phenotype. The floral homeotic genes SUPERMAN and AGAMOUS both show heritable hypermethylation and gene silencing, paradoxically in the hypomethylated backgrounds of met1 and ddm1 mutants. The dramatic phenotypes of their silencing are an excellent illustration of epialleles.
A more enigmatic example is the generation of over-expressing bal epialleles of the CPR1 (constitutive expressor of pathogenesis related proteins 1) locus (located in the chromosome 4 Resistance gene cluster) in ddm1 mutant backgrounds (Stokes and Richards, 2002; Stokes et al., 2002). bal epimutants show phenotypes such as twisted leaves, dwarfing and reduced fertility, similar to cpr1-1 from ethyl methyl sulphonate exposure. Surprisingly, these appear to be two different epialleles and neither associated with nucleotide changes. Intriguingly, selfed offspring from bal/cpr1-1 heterozygotes produce approx. 20 % phenotypically wild-type plants, perhaps derived from epigenetic destabilization and reversion mediated by pairing interactions (Stokes and Richards, 2002). There is also direct evidence that reactivation of transposable elements in the ddm1 background can produce new insertions with phenotypic consequences (Miura et al., 2001). Indeed, once activated by ddm1, transposons can retain their activity even when functional DDM1 is restored (Lippman et al., 2004). It does not seem too dramatic to suggest that saltational changes in the heritable epigenetic marks and phenotype could arise from even short-term exposure to defective activity of one or a few genes in the epigenetic system.
Stress-induced epigenetic change
Environmental and biotic stress may also induce the formation of novel epialleles. Mild inhibition of chaperone activity of the Hsp90 protein in plants by specific pharmacological agents has been shown to produce a diversity of phenotypic changes in Arabidopsis seedlings, such as alterations to plant size, leaf shape and pigmentation (Quietsch et al., 2002). This may be due to the fact that the chaperone folding activity of Hsp90 can partially counteract the effect of mutations in the DNA sequence that generate amino acid changes in proteins, rendering them ‘neutral’ in effect on the protein function. When Hsp90 efficiency is reduced, accumulated variation of this kind is rendered non-neutral and alterations to protein function ensue, exposing them to selection (Sangster and Queitsch, 2005). Definitive genetic proof that the phenotypic effects seen in Arabidopsis are derived in this way has not been presented. Another plausible scenario is that inhibition of full Hsp90 activity can also decrease the functional efficiency of the epigenetic machinery, e.g. decreasing the fidelity of epigenetic marks such as cytosine methylation, that results in a heritable change to epigenetic and gene expression states. Confirmation that this is the case from molecular epigenetic studies is still lacking. It follows that any environmental insult, such as extreme heat stress, could perturb the activity of epigenetic regulation and heritable effects on epigenetic marks and gene expression changes could arise.
Biotic stress in the form of pathogen attacks may also generate epigenetic aberrations. There is evidence that the Bs1 transposable element of maize has been reactivated by viral infection (Johns et al., 1985). Viral infections may bring about epigenetic changes of this ilk by the virtue of proteins they encode to counteract the defensive RNA pathway machinery of the host plant. The role of these proteins is to inhibit the activity of the defence pathway that destroys viral transcripts, but it also appears that the other RNA pathways are affected. Some of these viral suppressors of silencing bind small RNAs and affect their processing, whilst others such as P1/Hc-Pro may operate by inhibiting the protein components of the pathway. For example, there is evidence that the P1/HC-Pro suppressor of viral silencing interferes with miRNA pathways in Arabidopsis where it causes aberrant processing of some miRNAs and developmental aberrations (Kasschau et al., 2003); in tobacco accumulation of siRNAs from transgenes and endogenous miRNAs are differentially affected by P1/Hc-Pro (Mallory et al., 2002). Dunoyer et al. (2004) expressed five different viral silencing suppressors in Arabidopsis, all of which blocked transgene PTGS, but only three (P1/Hc-Pro, P19 and P15) affected plant morphology. Of these three, P1/Hc-Pro and P19 altered miRNA accumulation, whilst all three prevented degradation of miRNA targets. It seems probable that many of the gross abnormalities of plants infected with viruses forming dsRNA intermediates may be due directly to the upset of miRNA processing that regulates normal development. It is just conceivable that, in a manner akin to the formation of pseudogenes, viral transcripts may be incorporated into the host genome and ‘adoption’ of a viral gene encoding a silencing suppressor could bring about dramatic, heritable phenotypic effects by affecting the regulation of miRNAs.
Is epigenetic change important in adaptation?
Evidence for the influence of single epimutations or epialleles on the stable evolution of plant characteristics remains absent, perhaps because changes in DNA sequence during any length of evolutionary time will obfuscate the epigenetic contribution. Speculation is irresistible though. Chandler and Stam (2004) have speculated that paramutation may be a mechanism for transmitting environmentally adapted expression patterns. On a more specific level, it is fascinating that the differences in floral symmetry and floral organ number between Antirrhinum and the related Mohavea is derived from altered CYCLOIDEA expression (Hileman et al., 2003). The adaptive basis of this morphological change is likely to be pollination-based, with Mohavea flowers mimicking the symmetrical flowers of the unrelated Mentzelia in the same desert habitat. Could epigenetic phenomena have initiated this dramatic shift between two adaptive peaks? Epigenetic explanations may be irresistible for other morphological changes that have repeatedly occurred in plant evolution. The timing of transition from the juvenile vegetative to adult vegetative to reproductive development can differ considerably even between closely related taxa. Screens for mutations in Arabidopsis genes that alter the timing of these phase changes have revealed several key genes confirmed to be involved in epigenetic processing such as HASTY (Bollman et al., 2003) and SDE1/RDR6 (Peragine et al., 2004). Not only does this implicate a number of endogenous small RNA species as essential for controlling the timing of key developmental phase transitions in the life of the plant (Peragine et al., 2004), it also hints that mutations in these loci have the potential to heritably alter how a plant proceeds through its life cycle. For instance it may take just a few key mutations to generate plants that proceed very rapidly through their life cycle (as seen in many desert ephemerals) or retain persistently the features of juvenile vegetative growth.
Perhaps the most likely situation where epigenetic changes assist in the generation of an advantageous phenotype is the evolution of apomixis. The evolution of apomixis, in particular autonomous apomixis where an endosperm develops with no paternal genomic contribution, has been seen as problematic to the parent–offspring conflict model (Haig and Westoby, 1991). As apomixis has evolved repeatedly in the angiosperms, there must be pathways that allow its genesis. Experimental evidence suggests that alterations in the epigenetic machinery that control gametophyte development, imprinting and endosperm development are the basis of endosperm development without fertilization. Mutations in several Arabidopsis genes [two of the Polycomb group, MEDEA (MEA) and FERTILIZATION-INDEPENDENT SEED1 (FIE1), as well as FERTILIZATION-INDEPENDENT ENDOSPERM1 (FIS2)] promote proliferation of the central cell prior to fertilization (reviewed in Spielman et al., 2003). Yet none of these mutants promote any substantial development or differentiation of endosperm-like structures. However, loss of function of MET1, leading to hypomethylation of the maternal gametophyte in fie1 mutants, leads to the production of endosperm more closely resembling sexually derived endosperm (Vinkenoog et al., 2000). In the absence of fertilization, the spontaneous formation of endosperm-like syncytial structures has also been reported from mutants in the Arabidopsis MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) gene (Köhler et al., 2003). The MSI1 protein is a WD40 repeat protein that forms part of Polycomb group complexes (discussed in Part 1). Recent work by Guitton and Berger (2005) has confirmed that these endosperm-like structures are derived from central cells, and report that msi1 mutants also initiate the development of non-viable but polarized, parthenogenetic embryos from the egg cell. It seems conceivable that full-blown apomixis could evolve from just a few mutations in genes regulating key epigenetic processes.
EPIGENETICS OF PLANT HYBRIDIZATION
Perhaps by virtue of the highly flexible development allowed by their responsive epigenetic systems, plants are particularly adept at hybridization. Intriguingly, the ability to hybridize does seem to have a taxonomic bias with some families showing far greater rates of hybrid formation than others (Ellstrand et al., 1996). What does seem common in hybridization events in plants is that it can unlock variation not seen in either parental species. This has been exploited in cultivated plants in particular, but there is also evidence that hybridization could be of major importance in the evolution and diversification of plants in nature (for general reviews of plant hybridization see, Stebbins, 1950; Grant, 1971). Rather than being ‘dead-ends’, fertile plant hybrids may make an important contribution to the evolution of plant populations in many taxa, though the extent of their importance remains subject to debate.
Where does all the novel variation come from in hybrids? This has been attributed in the past to the effect of combining the genomes and proteomes of two previously separate, distinct entities; divergence in DNA and protein sequences would allow novel combinations in hybrids with the outcome being altered phenotypes with characteristics different to either parent. In fertile hybrid populations, ‘transgressive segregation’ has become a popular hypothesis to explain the generation of characteristics that are extreme, in both positive and negative directions, compared with the parents (Rieseberg et al., 1999, 2003). The genetic model that explains the generation of ‘extreme’ phenotypes is the segregation of distinct alleles, derived from the genetically divergent parents, of different quantitative effects on a characteristic, in hybrid populations. The combinations of these quantitative trait loci (QTL) alleles from both parents that have additive effects in the same direction generate characteristics in excess of either parent. These combinations, of course, are not possible in the isolated parental populations and only become possible in the hybrids.
The Arabidopsis hybrid model system in epigenetic studies
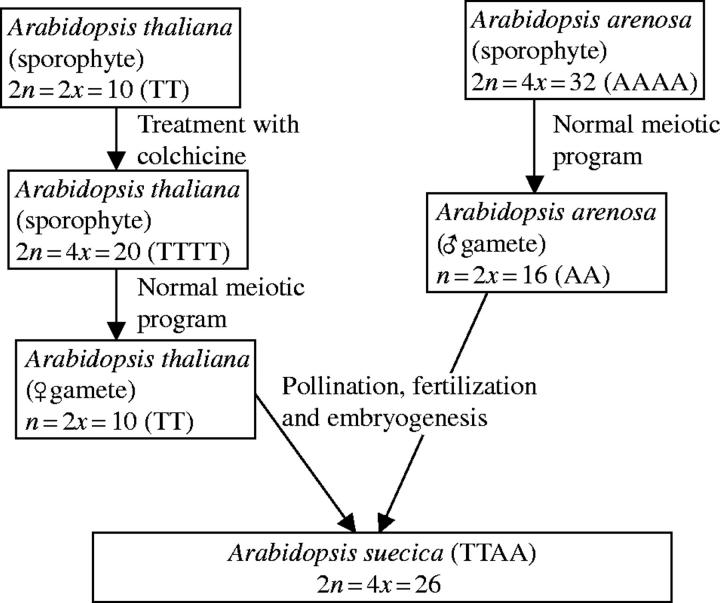
However, investigation of the epigenetics of hybrids, in particular the use of allopolyploid hybrids between Arabidopsis thaliana and A. arenosa as a convenient model system, has been a revelation. This model system is useful as A. thaliana is already the major model plant and the hybrid formed is known to occur in nature to form a stable, fertile hybrid allotetraploid, A. suecica. This facilitates comparison of established wild allotetraploids to synthetic laboratory populations. A diagram showing the chromosomal genetics of this model system is shown in Fig. 1.
Fig. 1.
Formation of synthetic Arabidopsis suecica allotetraploids (T = A. thaliana genome, A = A. arenosa genome).
Early studies revealed that formation of hybrids between tetraploid A. thaliana and A. arenosa in the laboratory yielded F1 hybrids with a range of phenotypes, not necessarily intermediate between the parents. A striking observation was that some of the altered phenotypes—such as meristematic fasciation and pigmentation—were unstable in hybrid individuals, indicating that dynamic epigenetic changes affecting gene expression had taken place. Gene expression analysis using the AFLP-cDNA method to sample expression profiles from both parental genomes has revealed that approx. 11 % of a 2430 cDNA fragment sample showed changes in expression in hybrid populations relative to the parents (Wang et al., 2004). This confirms earlier studies where both gene silencing and gene activation were observed, from both parental genomes (Comai et al., 2000). To confirm that the gene expression changes are due to epigenetic differences from the parents, and could not be attributed to the DNA sequence, the cDNAs displaying these changes have been further analysed.
The first and most compelling question answered by these studies was: what do these arbitrarily identified cDNAs encode? As expected, a range of genes were found even in the first small samples analysed by Comai et al. (2000), from transposon-related sequences probably derived from a heterochromatic region to a coding gene associated with repetitive DNA, to coding genes with no apparent connection to such sequences. Determination of DNA methylation of these sequences in the hybrids showed that they exhibited major differences to their parental counterparts. Later work with a broader sampling has shown that many coding genes with different functions, and many with unknown functions, show differential expression in newly synthesized hybrids (Wang et al., 2004). Interestingly, some of the cDNAs isolated have not shown simple, stable deactivation or reactivation patterns after hybrid formation; some have shown patterns of reactivation and deactivation across selfed generations of the newly synthesized hybrid (Comai et al., 2000; Wang et al., 2004). This is corroborated by work by Madlung et al. (2002) where methylation-sensitive amplified polymorphism (MSAP), assessing cytosine methylation at specific restriction sites throughout the genome, showed variation in methylation patterns in selfed hybrid populations. Are the same patterns of gene expression changes, including this instability, also found in established natural populations of A. suecica? This has been addressed by Lee and Chen (2001). By using the same AFLP-cDNA technique, it was shown that natural hybrids show differential patterns of gene expression compared with their parental species. Later data from Wang et al. (2004) has shown, as expected, that there is an overlap in the gene sets differentially expressed in lines of newly synthesized A. suecica and natural A. suecica. However, these patterns of gene expression are probably much more stable in natural A. suecica than in the synthetic laboratory populations, as the wild plants do not exhibit phenotypic instabilities; however, these instabilities return when the plants are treated with the cytosine demethylating agent azacytidine (Madlung et al., 2002) or when levels of DDM1 and MET1 are reduced by transgenic RNAi techniques (Wang et al., 2004). Both these treatments cause reactivation of gene expression at some of the silent loci examined.
Models and mechanisms for epigenetic change in hybrids
There is no doubt that this molecular work is hugely impressive but many questions still remain. The most disappointing shortcoming has been the inability to connect an epigenetically derived gene expression change with the generation of a phenotypic alteration in the hybrid. This will certainly be challenging in hybrid systems which are not so easy to work with, but essential to confirm the importance of epigenetics. Such work would also give an indication of whether epigenetic changes induced by hybridization could be of immediate survival or adaptive value. Our opinion is that both questions will be answered in the affirmative; the current estimates of the proportion of genes in parental genomes affected by hybridization, even if too high, and the fact that coding genes are highly represented supports our view.
Two further fundamental questions also spring to mind. First, is this epigenetic instability in hybrids taxonomically widespread in plants? The answer is almost certainly ‘yes’. Complex gene expression changes (even more subtle than those found in the Arabidopsis hybrids) have been found in the homeologous gene pairs of 40 protein-coding genes in Gossypium allopolyploid hybrids (Adams et al., 2003), although confirmation that these are epigenetically derived is still lacking. However, in a large sample of allopolyploid hybrids involving Triticum and Aegilops species, Shaked et al. (2001) have shown that methylation changes were widespread using AFLP, MSAP and Southern blotting techniques. In one newly synthesized wheat allotetraploid, gene silencing and methylation changes at multiple loci were observed (Kashkush et al., 2002). A rough estimate of 1–5 % of genes silenced by allopolyploid hybrid formation has been put forward following this work. Evidently monocotyledons also exhibit epigenetic instability in hybrid formation. The other essential question is: what mechanism generates epigenetic instability in hybrids? There is no evidence that the affected genes in A. thaliana are clustered as they map to different locations on the five chromosomes (Wang et al., 2004). Clearly, some kind of genome-wide effect is taking place that affects the epigenetic stability of various loci, in some cases over multiple generations. Comai et al. (2003) have put forward a model to explain this phenomenon. Their hypothesis is that during evolutionary time, as two populations diverge and develop into different species, there is a slight divergence in the protein components of chromatin regulatory complexes. The sub-units of multi-protein complexes that interact with each other would evolve co-ordinately. When the two species then form a hybrid, the alteration in the structure of these complexes through the combination of protein products encoded by different parental genomes may impair their function to an extent. This impairment of function in a hybrid could generate genome-wide alterations in chromatin structure, with changes in DNA methylation as a secondary effect. A second component of this hypothesis is that the changes in chromatin regulation allow derepression of silencing of transposable elements, with the hybridization event inducing a burst of transposition. Re-silencing of these sequences via RNA-based RdDM systems will take place; however, if a homologous sequence is found ‘naturalized’ in a functionally important region such as in a promoter or enhancer, this could also lead to coding gene expression changes. The finding that genes similar to transposable elements and coding genes associated with repetitive DNA show altered expression in hybrids is fitting with this explanation. However, Wang et al. (2004) found that repetitive/transposable element sequences were not abundant in the samples of differentially expressed genes. Perhaps this reflects the efficiency of silencing systems, though if the epigenetic systems of hybrids are to an extent deregulated this is rather surprising. In the future, it may be desirable to clone siRNA populations from hybrid and parental genomes to provide more supportive evidence. In our view, this model may not be a comprehensive explanation of gene expression changes for several reasons. For example, it seems rather incongruous that genes such as FWA, SUPERMAN and BAL/CPR1 that show great sensitivity to perturbation of components of the epigenetic regulatory system, e.g. ddm1 mutants, have yet to appear amongst the lists of genes with altered expression in hybrids. The model of Comai et al. (2003) also does not sit comfortably with data from studies of differences between species in components of the epigenetic system. CenH3, the centromere-specific H3 variant, has been cloned from both A. arenosa and A. thaliana and, although the sequences show divergence in the amino terminal region, whether the amino acid changes have functional relevance is unknown. The CenH3 of the A. thaliana genome, which was immunologically distinguishable from the A. arenosa CenH3, was capable of binding to centromeres of both genomes. As mitotic and meiotic divisions are not greatly compromised in A. suecica, this suggests that these two homeologues are interchangeable in function. More remarkably, in oat–maize addition lines with stable maize chromosomes, only the oat CenH3 is incorporated at maize centromeres (Jin et al., 2004).
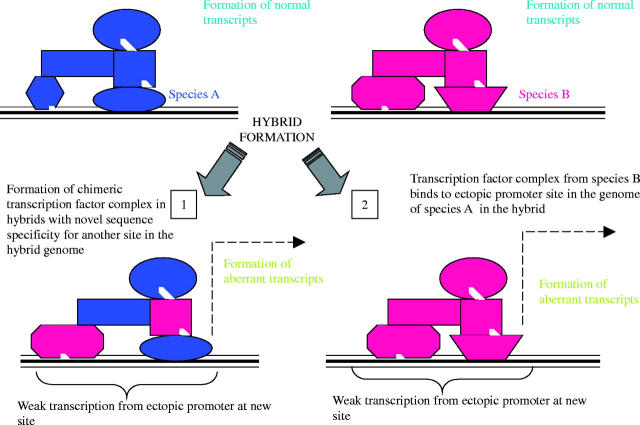
Other explanations for reactivation of previously silenced transposable elements seem possible. It has been shown in Nicotiana that some transposable elements are exquisitely sensitive to the physiology and biochemistry of the plant (such as levels of phytohormones); in a hybrid environment, such elements from the parental genomes may be exposed to new conditions that elicit their re-activation (reviewed in Grant-Downton, 2003). Equally, it seems plausible that coding genes may have altered expression induced through generation of ectopic promoters in hybrids (Grant-Downton, 2003). Ectopic promoters could be generated through divergence between the different species in genomic DNA sequences, their epigenetic modifications and the transcription factors that recognize them (see Fig. 2). When combined in a single genomic context, initiation of transcripts from ectopic sites may result and these would form aberrant RNAs recognized by the RNA surveillance system. The downstream result will be DNA methylation and chromatin modifications at these ectopic promoter sites and any regions of homology in the hybrid genome. Empirical support for this theory comes from work by Tran et al. (2005), where analysis of DNA methylation patterns in the Arabidopsis thaliana genome has revealed evidence for ectopic promoters. Unexpectedly, Tran et al. (2005) found clusters of dense CpG methylation within genes that may have been stably maintained for generations. The patterns of these CpG clusters within genes suggests they may be formed after rare ectopic transcription events, resulting in formation of aberrant RNAs that generate RdDM. Maintenance of CpG sites by MET1 ensures that the methylation pattern is preserved even in the absence of inducing transcripts.
Fig. 2.
How hybrid formation may lead to the generation of ectopic promoters and aberrant transcripts.
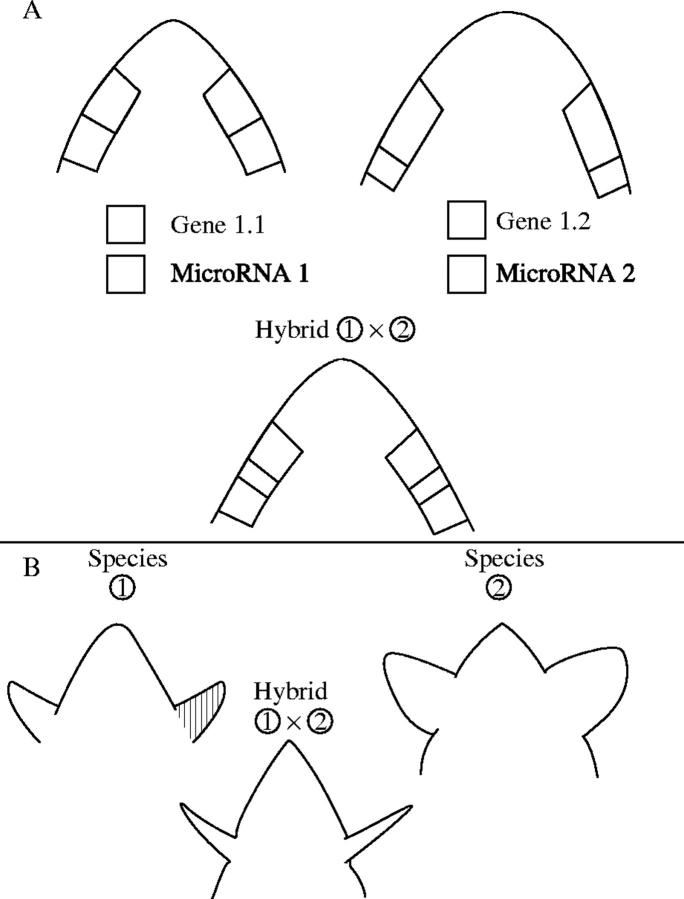
Another conceivable source of gene expression changes in hybrids may come from divergence in miRNAs and their targets, and divergence in patterns of miRNA expression and miRNA target gene expression, between different species. A hypothetical outcome is shown in Fig. 3, where the developmental outcome on the growth of a tissue is shown in parental species, and in their hybrid.
Fig. 3.
Hypothetical model showing outcome of divergence in miRNA expression patterns in two plant species and their hybrid. (A) Two species, 1 and 2, share the same target gene (gene 1.1 and 1.2) that promotes cell divisions and outgrowth of a tissue (expression domain indicated by lines). However, the two species differ in the expression pattern of microRNA genes (microRNA1 and microRNA2, respectively), that both target this gene (expression domain marked by lines). (B) After miRNA-induced degradation, the phenotypic outcome on the tissue is seen. The hybrid differs markedly from both parents as the actions of both miRNA1 and miRNA2 have limited expression to only a small group of cells in the tissue.
As discussed above, miRNAs can evolve with some rapidity from inverted repeats. Of course, miRNAs down-regulating different genes from their original targets would not necessarily produce a heritable epigenetic mark. Both Li and Chen (2001) and Wang et al. (2004) provide interesting evidence that certain sequences are inherently susceptible to expression changes in allopolyploids, indicating that genes are not stochastically selected but have some inherent feature predisposing them to the effects. However, some sequences show an immediate, stable expression change in the first generation hybrid that depends on their species of origin, whereas other sequences establish their silencing over several generations, and which parental sequence of the two is silenced is determined stochastically (Wang et al., 2004).
Epigenetics and other hybrid phenomena
Can epigenetics be invoked to explain other phenomena seen in hybrid plants? Explaining the molecular basis of cases of ‘hybrid vigour’ or ‘heterosis’ has been taxing and an epigenetic perspective may prove to be useful and complementary to conventional genetic explanations (see Grant-Downton, 2003; Chandler and Stam, 2004; Grant-Downton and Dickinson, 2004). Even more strange is the evidence for rapid genomic changes in plants after polyploidization and hybridization events. Ozkan et al. (2001) and Shaked et al. (2001) have presented extensive evidence for elimination of DNA sequences from a large number of hybrids involving Triticum, Aegilops and Secale, both diploid and allopolyploid. Elimination of DNA sequences was permanent and no chimeric tissues were detected, but more strange still was the discovery that the sequence elimination was consistent, with the same elimination pattern for a sequence seen in all the individuals in the same generation of the same hybrid combinations. These patterns were unchanged even when the crosses were duplicated with parents of different accession. Eliminated sequences represented not only high-copy sequences but also low-copy sequences. Another bizarre twist from the work by Shaked et al. (2001) was the demonstration that a bias in the amount of sequence elimination from the different parental genomes can occur in some crosses. Perhaps most significant of all, this work did show that a substantial number of DNA sequences from both parental genomes could be eliminated in just a few generations after a hybridization event. Kaskush et al. (2002) analysed transcriptome changes in hybrids and, although it was not possible to provide an accurate quantitative estimate attributable to elimination, these authors assert that this elimination process could be responsible for a significant amount of transcript loss. However, this work did show the identity of one of the eliminated sequences as a coding gene, Acc-2 (acetyl-coenzyme A carboxylase). The exact mechanism that induces these elimination patterns remains unknown. It seems possible that these genomic changes could be the result of transposon activation in hybrids, resulting in translocations and rearrangements; less dramatic genomic changes of a similar kind occur in Arabidopsis suecica by this mechanism (Madlung et al., 2005). The consistency of sequence elimination in these grasses still remains difficult to explain by this mechanism. Other explanations such as more co-ordinated programmed genomic rearrangements as seen in the programmed DNA excision in ciliates, a mechanism that depends on the RNA-based pathways (reviewed in Matzke and Birchler, 2005) should also be considered.
More dramatic elimination processes are also seen in some hybrid plants. Uniparental loss of chromosomes is frequently seen in wide hybrids of grasses (reviewed in Bennett, 1995), and has also been recorded from other plants (e.g. Solanum; Clulow et al., 1991). The elimination processes typically eliminates all the chromosomes of one parent during mitotic divisions early in somatic development of the hybrid, though examples where elimination is incomplete (such as oat–maize hybrids; Riera-Lizarazu et al., 1996) or biparental are also known. Cytological studies have revealed that the position of chromosomes within the nucleus is strongly correlated with their retention or elimination. However, nothing is known about the molecular basis of the elimination process. In some remarkable cases it has been associated with somatic recombination between eliminated and retained chromosomes (Wilkinson et al., 1995; Pasakinskine et al., 1997). Epigenetic dysfunction at centromeres of eliminated chromosomes in hybrids, affecting the stability of chromosomes on the mitotic spindle, appears to be the most plausible explanation (Grant-Downton, 2003). Position effects of chromosomes in the nucleus may also be responsible for ‘uniparental dominance’ effects in hybrids (Bennett, 1988; Heslop-Harrison, 1990). Fl hybrids between Hordeum vulgare and Secale africanum show this phenomenon, as most morphological and physiological characteristics resemble the Secale parent. It has been suggested that extreme distortion in the Hordeum × Secale hybrid arises from the positional effects of the two different genomes in the nucleus, with cytological studies showing that the Secale chromosomes are always displaced peripherally relative to the Hordeum chromosomes (Heslop-Harrison, 1990). Again, the molecular basis is unknown and even a transcriptomic study of these hybrids is needed. It seems possible that plants also have distinct nuclear territories associated with transcriptional activity or inactivity, akin to those in mammalian nuclei (reviewed in Zink and Cremer, 1998; William, 2003).
EPIGENETICS OF PLOIDY CHANGES
The observant reader may notice that the hybrid examples discussed above, where epigenetic changes and gene expression changes have been confirmed, are all allopolyploids. Whether this reflects a genuine difference between diploid and allopolyploid hybrids, or whether it represents an experimental bias in current research trends and model organisms, cannot be determined from the paucity of published papers. There is no doubt that some useful diploid hybrid systems exist that could be readily examined using the same kinds of techniques. For example, the hybrids between Arabidopsis thaliana and A. lyrata would make a fascinating subject for comparative studies. Equally, the diploid hybrids in the genus Helianthus, where hybrid formation has been shown to have the potential to lead to very rapid speciation by formation of ecophysiologically and morphologically distinct populations, have been extensively examined at the molecular level by Rieseberg and co-workers (e.g. Rieseberg et al., 1995, 1996, 2003; Rieseberg, 2000; Lexer et al., 2003). Here, the architecture of hybrid traits was explored by employing DNA sequence markers and QTL methods, but as these do not have the capacity to resolve gene expression changes and epigenetic differences in the same way as the techniques employed in the studies above, the relative contribution of epigenetic changes to diploid hybrid speciation is unknown. It would be exciting to integrate data on epigenetics with the fine molecular mapping and QTL studies employed before, as the relative contribution of epigenetic variation and sequence variation to the ‘transgressive segregation’ phenomena of hybridization in Helianthus could be dissected. Indeed, our awareness of work in epigenetics begs the question whether some QTL are in fact epialleles, and also highlights the importance of always distinguishing between enzymes that are methylation-sensitive and methylation-insensitive in molecular marker studies.
In the work on allopolyploid hybrids, controls were employed to ensure that gene expression changes were not derived from the change in ploidy alone, in the case of Arabidopsis suecica the autotetraploid A. thaliana parent. Ploidy changes eliciting stable epigenetic changes and gene expression alterations from transgene loci have been recorded on a couple of occasions in A. thaliana (Mittelsten Scheid et al., 1996, 2002). However, Wang et al. (2004) have shown that even endogenous genes can be dramatically affected by ploidy change. Genes encoding a DNA binding protein (DBP) and a putative protein (PP1) were silenced in newly synthesized autotetraploids, but when allopolyploid hybrids were formed from them these genes were reactivated. This suggests that at an epigenetic level some genes are differentially sensitive to a simple ploidy change and an allopolyploidy event. This begs the question of what is the trigger for silencing in autotetraploids, and what allows their subsequent reactivation in an allopolyploid background. There is also evidence of sensitivity of maize gene expression to changes in chromosome number giving rise to aneuploidy. As changes in ploidy in flowering plants appears to be quite a frequent and easily achieved event, such gene expression changes may have some evolutionary importance. However, in Arabidopsis the total number of genes affected in a measurable way appears to be rather small.
Changes in ploidy also affect the ability of many plant species to form viable hybrids as the endosperm appears to be particularly sensitive to ploidy balance. For example, maize hybrids between diploid and autotetraploid maize result in degenerate endosperm, and seed development is usually blocked at an early stage. Indeed, even in formation of allopolyploids where there is greater ploidy balance in the endosperm, there can be failure in early development of this nature. For example, in the formation of A. suecica a large number of F1 embryos do not develop or the seeds do not show viability, probably in part due to defective endosperm development (Comai et al., 2000; Bushell et al., 2003). In many cases of failure of interploidy or interspecific crosses, the embryo can be rescued and developed in vitro. In both situations, the cause of defective development is most likely rooted in aberrant expression of imprinted genes necessary for endosperm development. In the case of interploidy crosses, the effect of imprinted genes is simply quantitative, the outcome of endosperm development in the cross dependent on copy number of the genes (with ploidy level directly determining copy number) and their parent-of-origin. In interspecific crosses, the effects are much more complex; differences between species (even of the same chromosome number) may exist in copy number and sequence/function of imprinted loci, and even presence or absence of some imprinted genes, will affect endosperm development, in addition to parent-of-origin effects. Overlaid on this may be further complexities in gene expression patterns as discovered in the 2n sporophytic tissues. Epigenetic misregulation in endosperm at imprinted loci, leading to abortion of the developing seed, must be a powerful isolating mechanism in plant speciation events. As it can be set up by a simple ploidy change, it could be a hugely effective mechanism to achieve reproductive isolation and initiate speciation events. Even changes in copy number or expression levels of one or two imprinted loci have the potential to set up strong reproductive isolation barriers between different populations. Equally, as allopolyploidy events show, a ploidy change can bring down barriers between species. It is important to note that some plants do not have this reproductive barrier, e.g. hybrids between Papaver somniferum and different species of Papaver in Section Oxytona which show different ploidies, as well as hybrids within section Oxytona, form normal, viable seeds (Ojala and Rousi, 1986; Ojala et al., 1990). Furthermore, it seems more than a coincidence that some plant families which readily form ‘wide’ hybrids (such as intergeneric hybrids) also show a highly pared down endosperm—the Orchidaceae and Gesneriaceae, for example.
THE INTERFACE BETWEEN EPIGENETICS AND OTHER FIELDS
As can be seen from the previous section, epigenetics and the evolution of plants is inextricable. Yet despite this now obvious statement, epigenetics is seldom discussed in a context of taxonomy and systematics, or population biology and conservation. Only one recent review, by Kalisz and Purugganan (2004), has considered the importance of epialleles in population genetics and evolution. Here, an attempt is made to redress the balance.
Taxonomy and systematics
The first encounter between epigenetics and systematics occurred at the birth of modern systematics when Linnaeus became aware of the ‘monstrous’ peloric variant of Linaria vulgaris, now known to be caused by hypermethylation of Lcyc. So different in basic floral morphology was this spontaneous variant, illustrated in Fig. 4, it caused Linnaeus considerable concern (for historical review, see Gustaffson, 1979). The fact that epigenetic variants can generate stable morphological changes in plants, to the point of homeotic transformations, cannot be ignored any longer by those studying plant systematics. As much taxonomy is still based on analysis of morphological features, it would seem that an obvious deduction is that some plant species may have been misclassified, and merely represent epigenetic variants of one species. This problem may be particularly acute in taxa where hybridization is frequent and from locations where sampling is patchy. Conversely, by virtue of the ease of reproductive isolation through ploidy change and, perhaps, through alterations in imprinted genes without associated ploidy changes, there may be more ‘cryptic’ species in plants, difficult or impossible to distinguish by morphology, than previously assumed.
Fig. 4.
Epigenetic control of floral symmetry in Linaria vulgaris (common toadflax). Right: wild-type L. vulgaris where the Lcycloidea locus controlling dorso-ventral patterning is expressed normally. Note the zygomorphic flower with the ventral petal lobe displaying a spur. Left: the peloric epimutant shows hypermethylation of the Lcyc sequence and loss of expression of Lcyc. The flowers show greater radial symmetry as the ventral petal lobe is repeated five times. The plant shown here is homozygous for the Lcyc epiallele; heterozygotes show normal floral development.
Another consideration dictated by the epigenetic revolution relates to the evolution of plant characteristics. If, in a given taxonomic group, some genes controlling certain characteristics (e.g. stamen number, floral symmetry) are initially (or become) unusually labile at the epigenetic level, it would be expected that these characteristics would be more subject to repeated modification, loss or gain in the clade. In phylogenies of certain taxa, the frequent change or reversibility of some characteristics could be rooted in epigenetics. This idea will be illustrated by two different examples. A gene susceptible to stable silencing by hypermethylation of the promoter may become furtherentrenched in an inactive state by virtue of increased rate of mutation of the methyl-cytosine leading to functional change. Alternatively, when a gene is regulated by a miRNA, just a few nucleotide changes in the site of complementary binding to the miRNA could render the mRNA less susceptible or completely immune to regulation. Ectopic or over-expression by this mechanism could result in modification, loss or gain of a characteristic.
We have already hypothesized that rare saltatory changes in multiple characteristics could come about by virtue of aberrations in the epigenetic machinery. To reiterate, there is also supportive evidence that allopolyploid hybridization can lead to widespread epigenetic changes, altered gene expression and phenotypic changes, so allopolyploid speciation can be rapid and involve changes in multiple characteristics. More fascinating still is the (frustratingly incomplete) evidence for substantial differences in epigenetic systems in the gametophyte between different angiosperm taxa. Even if such a saltatory event occurs very infrequently in a phylogeny, its manifestation may create an interesting situation. Traditional morphology-based analysis may face considerable problems in resolving the taxonomy and phylogeny of a clade where an event of this kind has happened. The occurrence of such events may assist in the explanation of why phylogenies based on the DNA sequence sometimes show that morphological change is out of phase in relation to change to the DNA sequence. Epigenetics may illuminate and increase the complexity of the debate on molecules versus morphology in plant systematics.
Population genetics and conservation
Implicit in the discussion above is that epigenetic regulation and epigenetic variation has the capacity to generate phenotypic variation that may (or may not) have adaptive value. Unfortunately, measuring epigenetic variation in plant genomes is much more complex than determining variation in DNA sequences. For example, digestion of genomic DNA with isoschizomers can reveal DNA methylation patterns, whilst bisulfite sequencing can reveal in detail the methylation patterns of individual sequences. However, unlike the relatively static DNA sequence, methylation patterns can vary dramatically upon even the same DNA sequence, depending on factors such as environment and developmental stage, making sampling more complex. For instance, genomic DNA methylation levels have been shown to increase with developmental age, from seedling to mature plant, even in ephemeral species such as Arabidopsis thaliana (Ruiz-García et al., 2005). This adds another new set of problems to generating data on DNA methylation patterns. At least there are some data for variation in populations of A. thaliana, with considerable variation between ecotypes in methylation detected at rRNA gene repeats (Riddle and Richards, 2002). Detectable differences between ecotypes at the sequence level of the rRNA genes were negligible, though substantial differences in gene copy number were evident. QTL analysis of the control of this trait was performed by Riddle and Richards; unsurprisingly, the major determinant of methylation mapped to the rRNA repeats themselves is probably due to the strong inheritance of parental epigenetic states at these loci. However, this could not account for all of the variation between ecotypes and trans-acting modifier loci affecting methylation whichwere shown to exist. These QTL loci contained strong candidates for genes controlling methylation such as KRYPTONITE and DNA methyltransferases. Of course, DNA methylation patterns are but one facet of epigenetic variation. The technology to assess variation in histones on a sequence-by-sequence basis and siRNA populations is in its infancy.
Measuring sequence variation by methods such as RAPD, RFLP, AFLP and VNTR amplification (for reviews, see Karp et al., 1996; Mueller and Wolfenbarger, 1999) will only give an estimate of variation in the DNA sequence. How much epigenetic variation is stored by the genomes of a population cannot be adequately described by present techniques, and a more detailed picture of the epigenetic variation in a model species, say Arabidopsis, would be a major step forward. How important is this otherwise ‘cryptic’ variation? Epigenetic data superimposed upon sequence data is likely to greatly improve the association of markers and traits, showing both continuous and discontinuous variation, in studies of populations. Another impact is that many repetitive sequences in genomes, previously considered neutral or nearly neutral, may emerge to show effects on traits and fitness. By affecting transcription of key genes, repetitive sequences and the chromatin they form may impact on quantitative variation and even effects on fitness. For example, some diseases in humans are now known to map to an increase in size of a repetitive DNA (reviewed in Sinden et al., 2002) and the structural effects generated by repeat expansion may block transcription of the gene (Sakamoto et al., 1999, 2001). These effects may be local to exceptionally non-local. A superb example of the latter is paramutation at the B locus in maize, where the repetitive region, a tandem array, exhibiting the epigenetic changes that are governing paramutation behaviour is 100 kb upstream of the b1 coding region (Stam et al., 2002). Even by smaller influences on factors such as chromatin structure that affect chromosomal architecture and recombination events, and the distance between enhancers and coding sequence, repetitive DNA can have a non-neutral and non-local effect on other sequences. With repetitive sequences emerging time and time again as a key target of epigenetic modification, selection must always be acting on repetitive sequences and it is quite wrong to dismiss these sequences as ‘junk’ evolving out of tempo with coding sequences.
What happens to epigenetic variation in populations affected by artificial pressures exerted by humans, more precisely in wild populations subjected to disruption by man and those in cultivation exposed to artificial selection (plant domestication and plant breeding)? Conservation biologists interested in conserving variation in plant populations should now heed the fact that epigenetic variation may be significant but cannot be readily measured. For example, in plant populations, epigenetics has been shown again and again to have a tremendous influence on plant fertility. In disrupted environments or through exposure to over-exploitation, small or fragmented wild populations may be subjected to in-breeding; fixation of mutations in the epigenetic machinery that reduce fertility (amongst other effects) may have an impact on the long-term population genetics and even survival of the sub-populations. On the other hand, epigenetic variation induced in such backgrounds may have the opposite effect, allowing survival of the population in the face of abiotic and biotic adversity such as higher grazing pressure and climate change, by altering traits of selective value such as plant stature and flowering time. Metastable alleles at some loci may allow temporary escape of unfavourable conditions with considerable rapidity and without the permanence of nucleotide changes. Consequently, carrying or generating an ‘epimutational load’ may have advantages and disadvantages. On the other hand, epigenetic changes may assist in explaining why hybrids between native and non-native species may be unusually adaptable and become highly invasive. The invasive allopolyploid Spartina anglica, formed less than 150 years ago between a native and an introduced species, would be an interesting candidate for investigation. Surprisingly, genetic analysis of wild S. anglica clones showed no significant genetic changes compared with the parental species, even showing relative quiescence of transposable elements (Baumel et al., 2001, 2002). Recently, significant epigenetic changes have been revealed using MSAP analysis (Ainouche et al., 2003). In the invasive Senecio hybrids formed between S. squalidus (2n) and S. vulgaris (4n), the infertile S. baxteri (3n) and the derived fertile S. cambrensis (6n), there are significant changes to the transcriptome in the hybrids compared with the parental species (Hegarty et al., 2005). Unfortunately, these have yet to be associated with epigenetic changes.
The pool of epigenetic variation, and the comparative ease with which it can be modified, may help to explain why some naturalized species can establish and become invasive in a short time, even though the naturalization process is accompanied by a massive bottleneck in genetic variation. An epigenetic perspective may also be useful in understanding how natural population bottlenecks in founder events can lead to formation of viable colonies and (eventually) speciation in isolated environments such as islands. In conservation biology, epigenetics may have much use, and questions such as whether epigenetic diversity is eroded in a similar way to genetic diversity in plant populations under pressure should be addressed in the future.
In plant domestication and plant breeding, population bottlenecks are also a feature, often together with hybridization and ploidy changes. A notable feature of artificial selection is that novel or extreme traits are frequently the object of selection, whether the effort is deliberate or unconscious. Are epigenetic variants selected for under these conditions? In clonally propagated plants, selection may particularly favour epigenetic variants as even non-heritable or metastable epigenetic states could be propagated indefinitely. Efforts are definitely needed to address this question, as it may help to explain why genetic diversity can drop dramatically yet phenotypic variation and phenotypic plasticity remains high. An outstanding example is analysis of AFLP variation in cultivated hybrids in Hemerocallis (Tompkins et al., 2001). This genus is particularly useful as hybridization and selection from the handful of progenitor species, such as H. citrina and H. altissima, is only just over a century old but the original clones of the species remain in cultivation. As an intense level of selection has proceeded, producing cultivars increasingly novel and phenotypically divergent from the progenitor species, genetic diversity has decreased dramatically. This drop in genetic diversity is particularly sharp in tetraploids, a ploidy state derived artificially by plant breeders using colchicine and similar compounds. It would be interesting to determine whether epigenetic diversity in hybrid cultivars compared with progenitor species has decreased, remained stable or increased during this selection.
CONCLUSION
In this two-part review, a basic introduction to epigenetics in plants has been given. It has highlighted that beyond the intrinsic interest in molecular biology, an epigenetic perspective on plant evolution, systematics, population biology and conservation is now needed. There is hope that the ‘epigenetic revolution’ will initiate and influence thinking and experiments in these fields to a much greater extent in the future.
FOOTNOTE ADDED IN PROOF
During the production of this review, an outstanding and important paper has emerged which highlights how little is still known about the interactions between epigenetic and genetic systems in plants. Homozygous loss-of-function mutations in the HOTHEAD locus of Arabidopsis were shown by Lolle et al. (2005) to spontaneously correct the nucleotide change generating the mutant hothead allele in question, and consequently revert to wild type. This correction was specific and well beyond the frequency expected from background mutations. Furthermore, HOTHEAD mutants showed the same enhanced correction of other mutations in disparate parts of the genome, both coding and non-coding regions. Although the mechanism remains obscure, it is proposed that plants must carry a cache of replica genomic information, possibly encoded by RNA species that is passed across generations and subsequently used as a template. Correction of mutations by these ‘ghosts of genomes passed’ is held to be effected by some part of the repair system, and may be useful in adapting to various stresses such as abiotic stress and in-breeding depression. How this tantalizing discovery relates to the epigenetic systems described above, and how it relates to more complex genomic situations in ploidy change and hybridization, remains a fascinating prospect for future research.
LITERATURE CITED
- Adams KL, Cronn R, Percifield R, Wendel JF. 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proceedings of the National Academy of Sciences of the USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainouche M, Baumel A, Salmon A, Yannic G. 2003. Hybridization, polyploidy and speciation in Spartina (Poaceae). New Phytologist 161: 165–172. [Google Scholar]
- Allen E, Xie Z, Gustafson A, Sung G-H, Spatafora J, Carrington J. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genetics 36: 1282–1290. [DOI] [PubMed] [Google Scholar]
- Amasino R. 2004. Vernalization, competence, and the epigenetic memory of winter. The Plant Cell 16: 2553–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette M, van der Winden J, Matzke, M, Matzke A. 2002. RNA-directed DNA methylation in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 99 (Suppl. 4): 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Salamini F. 2001. Desiccation tolerance in the resurrection plant Craterostigma plantagineum. A contribution to the study of drought tolerance at the molecular level. Plant Physiology 127: 1346–1353. [PMC free article] [PubMed] [Google Scholar]
- Bastow R, Mylne J, Lister C, Lippman Z, Martiennsen R, Dean C. 2004. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche ML, Levasseur JE. 2001. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France). Molecular Ecology 10: 1689–1701. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche M, Kalendar R, Schulaman AH. 2002. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Molecular Biology and Evolution 19: 1218–1227. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1988. Parental genome separation in F1 hybrids between grass species. In: Brandham PE, ed. Proceedings of Kew Chromosome Conference III. London: HMSO, 195–208.
- Bennett MD. 1995. Losing genomes in hybrid plants In: Oono K and Takaiwa F, eds. Modification of gene expression and non-Mendelian inheritance. Proceedings of the US–Japanese joint meeting. Tsukuba: Japan, NIAR.
- Bollman K, Aukerman M, Park M, Hunter C, Berardini T, Poethig R. 2003. HASTY, the Arabidopsis ortholog of exportin-5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504. [DOI] [PubMed] [Google Scholar]
- Bureau TE, Ronald TC, Wessler SR. 1996. A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proceedings of the National Academy of Sciences of the USA 93: 8524–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell C, Spielman M, Scott R. 2003. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. The Plant Cell 15: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V, Stam M. 2004. Chromatin conversations: mechanisms and implications of paramutation. Nature Reviews Genetics 5: 532–544. [DOI] [PubMed] [Google Scholar]
- Chong S, Whitelaw E. 2004. Murine metastable epialleles and transgenerational epigenetic inheritance. Cytogenetics and Genome Research 105: 311–315. [Google Scholar]
- Clulow SA, Wilkinson MJ, Waugh R, Baird E, Demaine MJ, Powell W. 1991. Cytological and molecular observations on Solanum phureja induced dihaploid potatoes. Theoretical and Applied Genetics 82: 545–551. [DOI] [PubMed] [Google Scholar]
- Coen ES, Carpenter R, Martin C. 1986. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell 47: 285–296. [DOI] [PubMed] [Google Scholar]
- Comai L, Tygai AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens, Y, et al. 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. The Plant Cell 12: 1551–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Madlung A, Joseffson C, Tygai A. 2003. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philosophical Transactions of the Royal Society of London, Series B 358: 1149–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Vincent, C, Coen E. 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161. [DOI] [PubMed] [Google Scholar]
- Dimitri P, Junakovic N. 1999. Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends in Genetics 15: 123–124. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier C-H, Parizotto E-A, Himber C, Voinnet O. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. The Plant Cell 16: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ellestrand NC, Whitkus R, Rieseberg LH. 1996. Distribution of spontaneous plant hybrids. Proceedings of the National Academy of Sciences of the USA 93: 5090–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Chaboud A, Dumas C, McCormick S. 2003. Sperm cells of Zea mays have a complex complement of mRNAs. The Plant Cell 34: 697–707. [PubMed] [Google Scholar]
- Finnegan EJ. 2001. Is plant gene expression regulated globally? Trends in Genetics 17: 361–365. [DOI] [PubMed] [Google Scholar]
- Finnegan E, Sheldon C, Jardinaud F, Peacock W, Dennis E. 2004. A cluster of Arabidopsis genes with a coordinate response to environmental stimulus. Current Biology 14: 911–916. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. 2004. Gene regulation: ancient microRNA target sequences in plants. Nature 428: 485–486. [DOI] [PubMed] [Google Scholar]
- Furini A, Koncz C, Salamini F, Bartels D. 1997. High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. EMBO Journal 16: 3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R. 2003. Epigenetics and the understanding of the generation of plant hybrid phenotypes. DPhil Thesis, University of Oxford.
- Grant-Downton R, Dickinson H. 2004. Plants, pairing and phenotypes: two's company? Trends in Genetics 20: 188–195. [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Dickinson H. 2005. Epigenetics and its implications for plant biology. 1. The epigenetic network in plants. Annals of Botany 96: 1143–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YHM. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends in Genetics 16: 461–468. [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M. 1991. Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philosophical Transactions of the Royal Society of London: Biological Sciences 333: 1–13. [Google Scholar]
- Grant V. 1971. Plant speciation. New York: Columbia University Press.
- Guitton A-E, Berger F. 2005. Loss of function of MULTICOPY SUPPRESSOR OF IRA1 produces nonviable parthenogenetic embryos in Arabidopsis. Current Biology 15: 750–754. [DOI] [PubMed] [Google Scholar]
- Gustaffson Å. 1979. Linnaeus' Peloria: the history of a monster. Theoretical and Applied Genetics 54: 240–248. [DOI] [PubMed] [Google Scholar]
- He Y, Amasino R. 2005. Role of chromatin modification in flowering-time control. Trends in Plant Science 10: 30–35. [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. 2004. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes and Development 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, Jones JM, Wilson ID, Coghill JA, Sanchez-Baracaldo P, Liu G, et al. 2005. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Molecular Ecology 14: 2493–2510. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS. 1990. Gene expression and parental dominance in hybrid plants. Development Suppl.: Genomic Imprinting, 21–28. [PubMed]
- Hileman L, Kramer E, Baum D. 2003. Differential regulation of floral symmetry genes and the evolution of floral morphologies. Proceedings of the National Academy of Sciences of the USA 100: 12814–12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Lamb M. 1989. The inheritance of acquired epigenetic variations. Journal of Theoretical Biology 139: 69–83. [DOI] [PubMed] [Google Scholar]
- Jin W, Melo J, Nagaki K, Talbert P, Henikoff S, Dawe R, et al. 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. The Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns MA, Mottinger J, Freeling M 1985. A low copy number copia-like transposon in maize. EMBO Journal 4: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisz S, Purugganan M. 2004. Epialleles via DNA methylation: consequences for plant evolution. Trends in Ecology and Evolution 19: 309–314. [DOI] [PubMed] [Google Scholar]
- Karp A, Seberg O, Buiatti M. 1996. Molecular techniques in the assessment of biological diversity. Annals of Botany 78: 143–149. [Google Scholar]
- Kashkush K, Feldman M, Levy AA. 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K, Xie Z, Allen E, Llave C, Chapman E, Krizan K, et al. 2003. P1/Hc-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 1997. Transposable elements as sources of variation in plants and animals. Proceedings of the National Academy of Sciences of the USA 94: 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Transposable elements, parasitic DNA and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen S, et al. 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Kishimoto N, Sakai H, Jackson J, Jacobsen SE, Meyerowitz EM, Dennis ES, et al. 2001. Site specificity of the Arabidopsis MET1 DNA methyltransferase demonstrated through hypermethylation of the superman locus. Plant Molecular Biology 46: 171–183. [DOI] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus, U, Gruissem W. 2003. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO Journal 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. 1999. Plant retrotransposons. Annual Review of Genetics 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Lee HS, Chen ZJ. 2001. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proceedings of the National Academy of Sciences of the USA 98: 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Welch ME, Durphy JL, Rieseberg LH. 2003. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Molecular Ecology 12: 1225–1235. [DOI] [PubMed] [Google Scholar]
- Li G, Chandrasekharan MB, Wolffe AP, Hall TC. 2001. Chromatin structure and phaseolin gene regulation. Plant Molecular Biology 46: 121–129. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn M, Dedhla N, McComble R, et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476. [DOI] [PubMed] [Google Scholar]
- Lolle S, Victor J, Young J, Pruitt R. 2005. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434: 505–509. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 266: 792–801. [DOI] [PubMed] [Google Scholar]
- Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L. 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiology 129: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Tygai A, Watson B, Jiang H, Kagochi T, Doerge RL. 2005. Genomic changes in synthetic Arabidopsis polyploids. The Plant Journal 41: 221–230. [DOI] [PubMed] [Google Scholar]
- Mallory A, Reinhart B, Bartel D, Vance V, Bowman L. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and microRNAs in tobacco. Proceedings of the National Academy of Sciences of the USA 99: 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Birchler J. 2005. RNAi -mediated pathways in the nucleus. Nature Genetics 6: 24–35. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Jakovleva L, Asfar K, Maluszynska J, Paskowski J. 1996. A change in ploidy can modify epigenetic silencing. Proceedings of the National Academy of Sciences of the USA 93: 7114–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Probst AV, Afsar K, Paszkowski J. 2002. Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proceedings of the National Academy of Sciences of the USA 99: 13659–13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Mueller U, Wolfenbarger L. 1999. AFLP genotyping and fingerprinting. Trends in Ecology and Evolution 14: 389–394. [DOI] [PubMed] [Google Scholar]
- Oakeley EJ, Podestà A, Jost J-P. 1997. Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proceedings of the National Academy of Sciences of the USA 94: 11721–11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala A, Rousi A. 1986. Interspecific hybridisation in Papaver. 1. F1 hybrids of P. somniferum with perennial species of sect. Oxytona. Annales Botanici Fennici 23: 289–303. [Google Scholar]
- Ojala A, Rousi A, Lewing E, Pyysalo H, Widén C-J. 1990. Interspecific hybridisation in Papaver. III. F1 hybrids between species of sect. Oxytona. Hereditas 112: 221–230. [Google Scholar]
- Ozkan H, Levy AA, Feldman M. 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. The Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasakinskine I, Anamthawat-Jónsson K, Humphreys MW, Jones RN. 1997. Novel diploids following chromosome elimination and somatic recombination in Lolium multiflorum×Festuca arundiancea hybrids. Heredity 78: 464–469. [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht H, Poethig R. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes and Development 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Rakyan V, Chong S, Champ M, Cuthbert P, Morgan H, Luu K, et al. 2003. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proceedings of the National Academy of Sciences of the USA 100: 2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Richards EJ. 2002. The control of natural variation in cytosine methylation in Arabidopsis. Genetics 162: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout III W, Coetzee G, Olumi A, Jones P. 1990. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science 249: 1288–1290. [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Rines HW, Phillips RL. 1996. Cytological and molecular characterisation of oat×maize partial hybrids. Theoretical and Applied Genetics 93: 123–135. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2000. Crossing relationships among ancient and experimental sunflower hybrid lineages. Evolution 54: 859–865. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Van Fossen C, Desrochers AM. 1995. Hybrid speciation accompanied by genomic reorganisation in wild sunflowers. Nature 375: 313–316. [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer M, Arias DM. 1996. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372. [DOI] [PubMed] [Google Scholar]
- Rieseberg L, Widmer A, Arntz A, Burke J. 2003. The genetic architecture necessary for transgressive segregation is common in natural and domesticated populations. Philosophical Transactions of the Royal Society of London, Series B 358: 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-García L, Cervera M, Martínez-Zapater J. 2005. DNA methylation increases throughout Arabidopsis development. Planta 222: 301–306. [DOI] [PubMed] [Google Scholar]
- Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, et al. 1999. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedrich's ataxia. Molecular Cell 3: 465–475. [DOI] [PubMed] [Google Scholar]
- Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. 2001. Sticky DNA, a self-associated complex formed at long GAA·TTC Repeats in intron 1 of the Frataxin gene, inhibits transcription. Journal of Biological Chemistry 276: 27171–27177. [DOI] [PubMed] [Google Scholar]
- Sangster T, Queitsch C. 2005. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Current Opinion in Plant Biology 8: 86–92. [DOI] [PubMed] [Google Scholar]
- Saze H, Mittlesten Scheid O, Paszkowski J. 2003. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nature Genetics 34: 65–69. [DOI] [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R. 2004. Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. The Plant Cell 16: 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridisation and allopolyploidy in wheat. The Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R, Potaman V, Oussatcheva E, Pearson C, Lyubchenko Y, Shlyakhtenko L. 2002. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. Journal of Bioscience 27 (Suppl. 1): 53–65. [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SJ, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, et al. 2000. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Molecular Cell 6: 791–802. [DOI] [PubMed] [Google Scholar]
- Spielman M, Vinkenoog R, Scott R. 2003. Genetic mechanisms of apomixis. Philosophical Transactions of the Royal Society of London, Series B 358: 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL. 2002. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes and Development 16: 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press.
- Stokes TL, Richards EJ. 2002. Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proceedings of the National Academy of Sciences of the USA 99: 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes T, Kunkel B, Richards E. 2002. Epigenetic variation in Arabidopsis disease resistance. Genes and Development 16: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. [DOI] [PubMed] [Google Scholar]
- Tompkins J, Wood T, Barnes L, Westman A. 2001. Evaluation of genetic variation in the daylily (Hemerocallis spp.) using AFLP markers. Theoretical and Applied Genetics 102: 489–496. [Google Scholar]
- Tran R, Henikoff J, Zilberman D, Ditt R, Jacobsen S, Henikoff S. 2005. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Current Biology 15: 154–159. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory A, et al. 2004. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Vinkenoog R, Spielman M, Adams S, Dickinson HG, Scott RJ. 2000. Demethylation promotes autonomous endosperm development and rescues post-fertilization lethality in fie mutants. The Plant Cell 12: 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2004. Shaping small RNAs in plants by gene duplication. Nature Genetics 36: 1245–1246. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee, H-S, Chen M, Lee J, et al. 2004. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang M-B, Lough T. 2001. Gene silencing as an adaptive defence against viruses. Nature 411: 834–841. [DOI] [PubMed] [Google Scholar]
- Wessler SR. 1988. Phenotypic diversity mediated by the maize transposable elements Ac and Spm. Science 242: 399–405. [DOI] [PubMed] [Google Scholar]
- White SE, Habera LF, Wessler SR. 1994. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proceedings of the National Academy of Sciences of the USA 91: 11792–11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MJ, Bennett ST, Clulow SA, Allainguillaume J, Harding K, Bennett MD. 1995. Evidence for somatic translocation during potato diahploid induction. Heredity 74: 146–151. [DOI] [PubMed] [Google Scholar]
- William RRE. 2003. Transcription and the territory: the ins and outs of gene positioning. Trends in Genetics 19: 298–302. [DOI] [PubMed] [Google Scholar]
- Zhang H, van Nocker S. 2002. The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant Journal 31: 663–673. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ransom C, Ludwig P, van Nocker S. 2003. Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of flowering-time switch FLOWERING LOCUS C. Genetics 164: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Cremer T. 1998. Cell nucleus: chromosome dynamics in nuclei of living cells. Current Biology 8: R321–R324. [DOI] [PubMed] [Google Scholar]