Abstract
Functional telomeres are required for the replicability of cancer cells. The G-rich strand of telomeric DNA can fold into a 4-stranded structure known as the G-quadruplex (G4), whose stabilization alters telomere function limiting cancer cell growth. Therefore, the G4 ligand RHPS4 may possess antitumor activity. Here, we show that RHPS4 triggers a rapid and potent DNA damage response at telomeres in human transformed fibroblasts and melanoma cells, characterized by the formation of several telomeric foci containing phosphorylated DNA damage response factors γ-H2AX, RAD17, and 53BP1. This was dependent on DNA repair enzyme ATR, correlated with delocalization of the protective telomeric DNA–binding protein POT1, and was antagonized by overexpression of POT1 or TRF2. In mice, RHPS4 exerted its antitumor effect on xenografts of human tumor cells of different histotype by telomere injury and tumor cell apoptosis. Tumor inhibition was accompanied by a strong DNA damage response, and tumors overexpressing POT1 or TRF2 were resistant to RHPS4 treatment. These data provide evidence that RHPS4 is a telomere damage inducer and that telomere disruption selectively triggered in malignant cells results in a high therapeutic index in mice. They also define a functional link between telomere damage and antitumor activity and reveal the key role of telomere-protective factors TRF2 and POT1 in response to this anti-telomere strategy.
Introduction
Telomere maintenance is important for all dividing cells, including cancer cells. Functional telomeres are essential for genomic stability, and without mechanisms maintaining telomeres, cells activate pathways leading to cell-cycle arrest or apoptosis (1). Indeed, when telomeres become deprotected, an ataxia telangiectasia mutated–dependent (ATM-dependent) response pathway leads to phosphorylation of damage factors such as γ-H2AX, 53BP1, and the Mre11 complex and to activation of downstream kinases, resulting in a rapid growth arrest or apoptosis (2–5).
Activation of telomerase is crucial in telomere maintenance for most cancer cells (6). In preclinical studies, some telomerase inhibitors have shown promise as effective agents for a wide variety of malignancies. However, with many but not all telomerase therapeutic approaches, senescence or apoptosis has been observed only when telomeres reach a critically short length (7, 8). Conversely, directly targeting telomeric chromatin might have immediate and profound effects on cell proliferation. This might be achieved by antagonizing the protective mechanisms acting at telomeres.
Human telomeres consist of tandem repeats of the hexanucleotide sequence TTAGGG in double-stranded DNA, except for a terminal 3′ G-rich overhang (9, 10). Telomeres can form a loop structure (t-loop), with the 3′ G-rich strand invading the duplex telomeric repeats (11, 12), involved in telomere protection. The 3′ G-rich overhang can also fold into a 4-stranded DNA structure, termed G-quadruplex (G4). An unfolded 3′ overhang is required for an optimal telomerase reaction, and G4 formation has been shown to inhibit telomere elongation in vitro (13). Several classes of G4 ligands have been designed to counteract telomerase, making these compounds attractive anticancer agents (14, 15).
The protection of human telomeres involves telomeric repeat binding factor 2 (TRF2), a protein binding the duplex part of telomeric DNA and participating in t-loop formation. TRF2 is essential for protecting telomeres from being repaired by nonhomologous end-joining (16) and recognized as DNA damage by ATM (17). Accordingly, TRF2-depleted telomeres appear deprotected or uncapped and can undergo end-to-end fusions and subsequent chromosome abnormalities (4, 18, 19). Along the same line, we recently showed that, in checkpoint-compromised telomerase-positive human fibroblasts, TRF2 inhibition promotes heritable changes, associated with a burst of telomere instability, that increase the ability to grow in soft agar (20). Strikingly, we also reported that the expression of a dominant-negative form of TRF2 in a melanoma cell line markedly inhibits its tumorigenic ability (21), suggesting that TRF2 inhibition can also counteract some of the steps involved in tumorigenesis. Protection of telomeres 1 (POT1) is another telomere protein that binds the single-stranded telomeric DNA and other telomeric proteins (22). In POT1-compromised cells, the 3′ overhang of telomeric DNA is reduced, and the telomeres elicited a transient DNA damage response (23).
RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is a pentacyclic acridine that binds G4 DNA and inhibits telomerase activity at submicromolar levels (24, 25). Long-term exposure of cells to low RHPS4 concentration causes an irreversible cessation of growth and telomere erosion, demonstrating that this compound possesses classical telomerase-inhibitory properties (24, 26). However, when used at higher doses, RHPS4 triggers short-term apoptosis/senescence in human melanoma lines (27). This effect does not result from telomere shortening and correlates with an increased incidence of telomere fusions (27), suggesting that this compound is more than a simple telomerase inhibitor. In this context, our study aims at understanding the mechanism(s) by which the G4 ligand RHPS4 rapidly limits cell growth and at determining tumor sensitivity to G4 pharmacological stabilization. We found that RHPS4 rapidly disrupts telomere architecture without affecting the length of telomeric DNA, resulting in a potent DNA damage response. We further show that telomere injury plays a critical role in the antitumor properties of this compound.
Results
RHPS4 induces damage foci at telomeres.
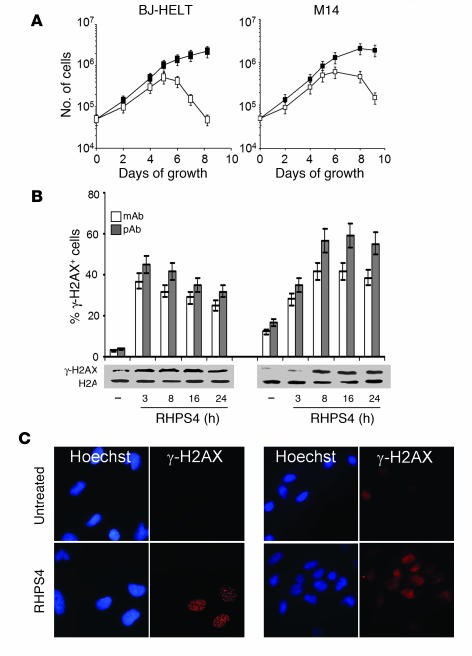
In order to investigate whether RHPS4 uncaps telomeres, human transformed BJ-EHLT fibroblasts and M14 melanoma cells were exposed to the drug at a concentration of 1 μM, a dose causing apoptosis when cells are chronically exposed to the drug for 4 days (Figure 1A and ref. 27). Strikingly, 3–8 hours of treatment was enough to induce a strong phosphorylation of H2AX, a hallmark of DNA double-strand break response in both transformed and tumor cells (Figures 1, B and C), indicating that RHPS4 is a DNA damage inducer. Notably, H2AX was never phosphorylated in primary culture of normal and telomerized fibroblasts (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32461DS1), even at long exposure time or when exposed up to a 5-fold higher RHPS4 dose (data not shown). Consistent with these results, the growth of these cells was not affected by the drug (Supplemental Figure 1).
Figure 1. RHPS4 induces phosphorylation of H2AX in transformed and tumor cells.
(A) Human transformed BJ-EHLT and M14 melanoma cells were treated with RHPS4 for 4 days. At the indicated times, cells were counted and the viability determined. The data represent the number of untreated (filled squares) and RHPS4-treated cells (open squares) during the growth in culture. The mean of 3 independent experiments with comparable results is shown. Error bars indicate SD. (B) Percentage of cells containing γ-H2AX foci (upper panel) and Western blot analysis (lower panel) of γ-H2AX in BJ-EHLT and M14 cells treated with RHPS4 for the indicated times. γ-H2AX foci were quantified using mouse mAbs and rabbit pAbs. On average, more than 200 cells were screened per time point in 3 independent experiments. Error bars indicate SD. The levels of H2A were used as loading control. (C) Representative immunofluorescence images of BJ-EHLT and M14 cells treated with RHPS4 for 8 hours. Original magnification, ×40.
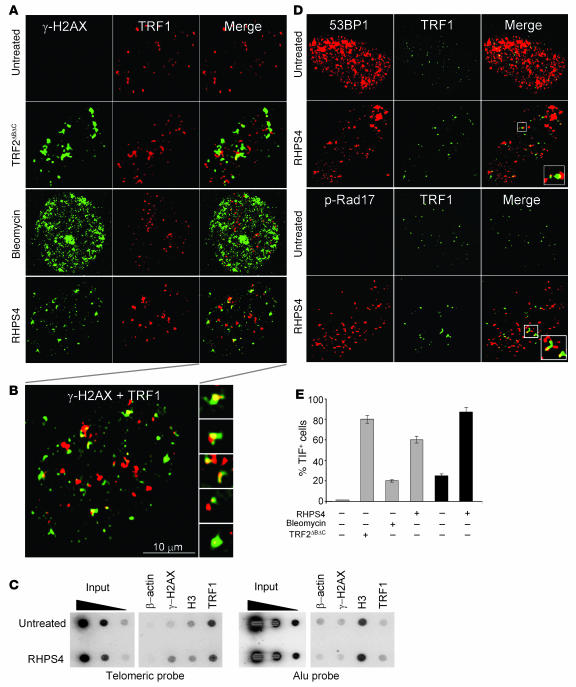
To ascertain whether γ-H2AX was phosphorylated in response to dysfunctional telomeres, double immunofluorescence experiments were performed in BJ-EHLT cells. Confocal microscopy revealed that most of the γ-H2AX foci induced by RHPS4 colocalized with TRF1, a good marker for interphase telomeres (28), forming the so-called telomere dysfunction–induced foci (TIFs; ref. 3) (Figure 2, A and B). Scoring of 80 RHPS4-treated cells, and examining 1 focal plane, revealed a mean ± SD of 31 ± 8.5 γ-H2AX foci per nucleus, and of these, more than 70% were largely coincident with TRF1 (Figure 2B). These results were confirmed by chromatin immunoprecipitation (ChIP), which showed that anti–γ-H2AX associated to telomeres only in treated cells, demonstrating that RHPS4 induced the phosphorylation of H2AX specifically at telomeric regions (Figure 2C). As control, anti-TRF1 antibody precipitated telomeric DNA in both untreated and treated cells, and this enrichment was not observed in the presence of β-actin control antibody. Other DNA damage response factors, such as phosphorylated Rad17 and 53PB1, were also recruited to the telomeres upon RHPS4 exposure (Figure 2D), confirming that RHPS4 triggers a genuine DNA damage response at telomeres. Telomere damage caused by RHPS4 appeared to be very potent, since the percentage of cells with more than 4 TIFs approached that measured in cells expressing the dominant-negative TRF2ΔBΔC allele (Figure 2E). Bleomycin, a genotoxic drug that induces double-strand breaks, did not induce TIFs (Figures 2, A and E), implying that telomeres are specific targets of RHPS4.
Figure 2. RHPS4 triggers DNA damage response at telomeres.
(A) BJ-EHLT fibroblasts expressing TRF2ΔBΔC or treated with either bleomycin or RHPS4 for 8 hours were fixed and processed for immunofluorescence using antibodies against γ-H2AX (green) and TRF1 (red). Representative confocal images are shown. (B) Higher-magnification view of A, bottom right. Shown are the nucleus and γ-H2AX foci, magnified ×2 and ×4, respectively, relative to the panel in A. (C) Formaldehyde-cross-linked chromatin fragments were immunoprecipitated with antibodies against TRF1, γ-H2AX, H3 (positive control of ChIP assay), or β-actin (negative control). To verify that an equivalent amount of chromatin was used in the immunoprecipitates, serial dilutions of sample the total chromatin (Input) were included in the blot. Specific (telomeric) and nonspecific (Alu) probes were used. (D) Untreated and RHPS4-treated BJ-EHLT cells stained with TRF1 (green) and 53BP1 or p-Rad17 (red). Boxes in the merged images of the RHPS4-treated samples indicate the location of the enlarged views. Original magnification, ×63; ×126 (enlarged panels). (E) TIF index, defined as foci of DNA damage response factors that coincide with TRF1, was calculated as the percentage of TIF-positive cells in the BJ-EHLT fibroblasts expressing TRF2ΔBΔC or treated with either bleomycin or RHPS4. Cells with 4 or more γ-H2AX/TRF1 (gray bars) or 53BP1/TRF1 (black bars) foci were scored as TIF positive. The mean of 3 independent experiments is reported. Error bars indicate SD.
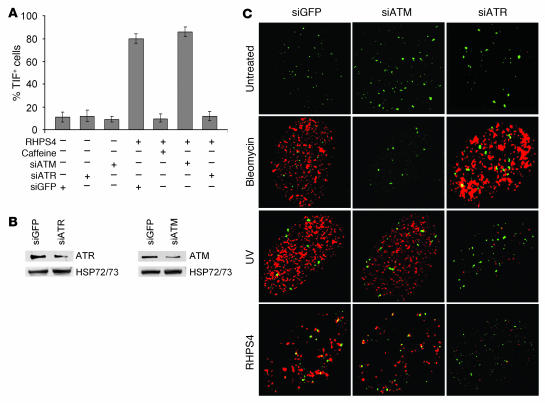
The PI3K inhibitor caffeine completely prevented the induction of TIFs by RHPS4, indicating that telomere response closely depends on ATM and/or ataxia telangiectasia and Rad3–related (ATR) kinases (Figure 3A). Moreover, RNA interference experiments (Figure 3B) revealed that ATR is the kinase involved in the phosphorylation of H2AX (Figure 3, A and C).
Figure 3. RHPS4-induced DNA damage at the telomeres depends on ATR kinase.
Human transformed BJ-EHLT cells exposed to the PI3K inhibitor caffeine or transfected with siATM or siATR SMARTpool or siGFP (as control) and treated with RHPS4 were fixed and processed for immunofluorescence using antibodies against γ-H2AX and TRF1. (A) TIF index was calculated as the percentage of TIF-positive cells under the indicated conditions. Cells with 4 or more γ-H2AX foci were scored as TIF positive (n = 72). The mean of 3 independent experiments is reported. Error bars indicate SD. (B) Western blot analysis of ATR and ATM protein expression in siGFP and siATR- or siATM-transfected cells. (C) Representative confocal images of merged TRF1 (green) and γ-H2AX (red) staining of the indicated samples. Original magnification, ×63.
POT1 is rapidly removed from telomeres upon RHPS4 treatment.
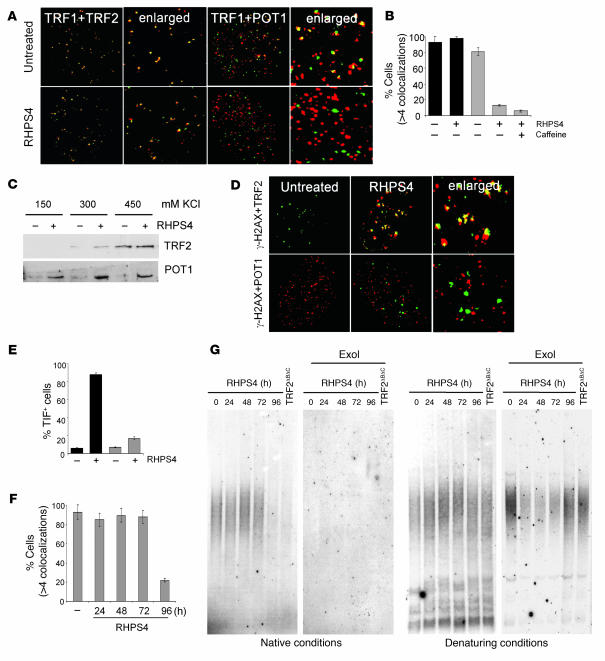
To determine the cause of telomere uncapping, we investigated the effect of RHPS4 on the localization of TRF2 and POT1, 2 telomeric proteins inducing telomere dysfunction and evoking DNA damage signaling when their levels are reduced at telomeres (18, 19, 23). Confocal microscopy showed that RHPS4 rapidly delocalized POT1 from TRF1 foci, while TRF2 remained associated to the telomeres early on in RHPS4 treatment (Figure 4A). Quantitative analysis revealed that nearly all untreated and treated cells had clearly detectable TRF1/TRF2 foci, while the percentage of nuclei with more than 4 POT1/TRF1 colocalizations was markedly reduced in cells exposed to RHPS4 (Figure 4B). Caffeine did not antagonize this effect, indicating that loss of POT1 from telomeres is not the consequence of the damage response but is likely to be a very early event caused by RHPS4 (Figure 4B). These findings were confirmed by Western blot analysis using differential salt concentrations upon RHPS4 treatment (Figure 4C). Consistent with a specific removal of POT1 from telomeres, TRF2, but not POT1, colocalized with the γ-H2AX foci induced by RHPS4 (Figure 4, D and E). Of note, a longer treatment (96 hours) triggered the removal of TRF2 from telomeres and the loss of 3′ overhang (Figure 4, F and G).
Figure 4. RHPS4 specifically and rapidly delocalizes POT1 from telomeres.
Transformed BJ-EHLT fibroblasts were treated with RHPS4 and double stained with the indicated antibodies. (A) Representative confocal images showing merged TRF1 (green) and TRF2 or POT1 (red) staining in untreated and treated cells. (B) Percentages of cells with more than 4 colocalizations per nucleus of TRF1 and TRF2 (black bars) and of TRF1 and POT1 (gray bars). Error bars indicate SD. (C) Western blot analysis of TRF2 and POT1 in untreated (–) and RHPS4-treated cells (+) lysed with various concentrations of KCl (150, 300, and 450 mM). (D) Representative confocal images showing merged γ-H2AX (red) and TRF2 (green) or γ-H2AX (green) and POT1 (red) staining in untreated and RHPS4-treated cells. Original magnification, ×63; ×126 (enlarged panels). (E) Percentage of TIF-positive cells using TRF2 (black bars) and POT1 (gray bars) as telomeric proteins. On average, more than 80 cells were screened per point in 4 independent experiments. Error bars indicate SD. (F) Percentages of cells with more than 4 TRF1/TRF2 colocalizations. Error bars indicate SD. (G) BJ-EHLT fibroblasts were treated with RHPS4 for the indicated times or transfected with an expression vector carrying TRF2ΔBΔC cDNA and processed for telomeric overhang assay under native conditions (left). Subsequently, the DNA was denatured in the gel and rehybridized with the same probe (right). Where indicated, the DNA was incubated with Exonuclease I (ExoI). Signals were measured using ImageQuant software by integration of the entire lane.
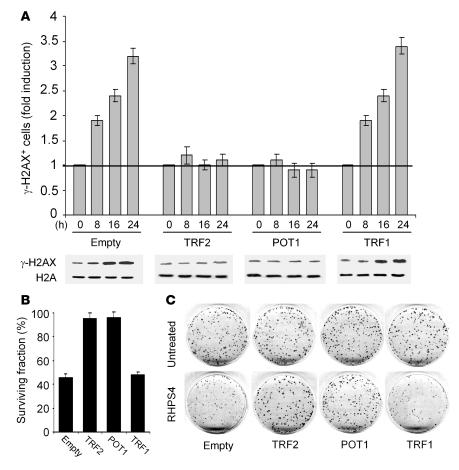
The functional relevance of POT1 and TRF2 dissociation upon RHPS4 treatment was directly revealed by the finding that either POT1 or TRF2 overexpression led to the absence of damage foci (Figure 5A and Supplemental Figure 2). By contrast, overexpression of TRF1 did not interfere with RHPS4-induced damage, the behavior of phosphorylated H2AX being similar in control and TRF1-overexpressing cells (Figure 5A and Supplemental Figure 2). Analysis of cell viability revealed that overexpression of either TRF2 or POT1, in contrast to TRF1, made cells resistant to RHPS4 (Figure 5, B and C). Overall, these results show that reinforcement of telomere capping functions by either TRF2 or POT1 counteracts the effects of RHPS4 on telomeres and tumor cells.
Figure 5. Overexpression of TRF2 or POT1 antagonizes RHPS4-induced damage response.
(A) M14 human melanoma cells overexpressing TRF1, TRF2, POT1 or puromycin-resistant gene only (empty) were treated with RHPS4 for the indicated times and processed for immunofluorescence and Western blotting. Histogram showing the increase in γ-H2AX–positive cells compared with the untreated group (upper panel) and a representative Western blot (lower panel) are shown. On average, more than 200 cells were screened per point in independent experiments. Error bars indicate SD. The levels of H2A were used as loading control. (B) M14 cells overexpressing TRF1, TRF2, POT1, or drug resistance (empty) were treated with RHPS4 for 4 days. The surviving fraction was calculated as the ratio of absolute survival of the treated sample to absolute survival of the control sample. (C) Representative images of colony assay.
RHPS4 induces telomere injuries and apoptosis in xenografted tumors.
The above results raise the interesting possibility that telomere damage induced by RHPS4 in malignant cells may rapidly promote growth inhibition and cell death.
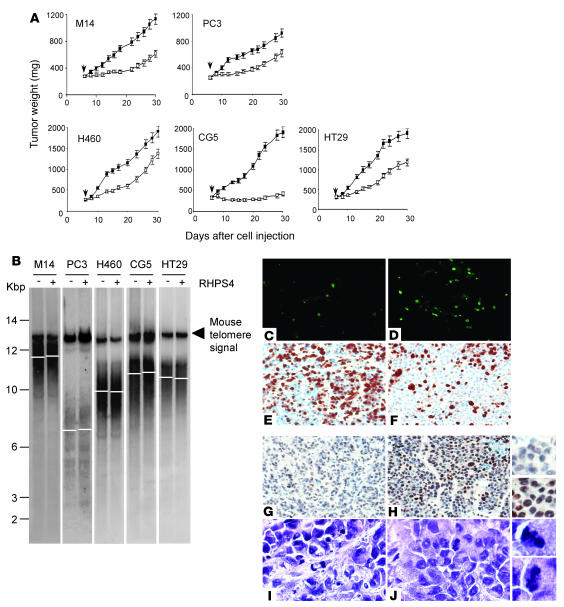
We therefore evaluated the antitumor activity of RHPS4 on tumors of different histotypes by injecting the maximum tolerated dose (15 mg/kg), chosen on the basis of an acute toxicological study on healthy mice (Supplemental Figure 3). Moreover, based on plasma pharmacokinetic analysis, we decided to administer the drug daily (Supplemental Figure 3), and the treatment, given for 15 consecutive days, was not toxic, as revealed by evaluation of mouse fitness, blood counts (Supplemental Figure 4), and histological analysis of bone marrow (Supplemental Figure 4) and of major organs (data not shown). By treating the animals with 15 mg/kg/d for 15 consecutive days, the most effective administration schedule (Supplemental Figure 5), we found that RHPS4 is active as a single agent on all the tumors analyzed. Specifically, CG5 breast xenografts resulted in the most sensitive tumor, as the treatment produced a marked inhibition of tumor weight (tumor weight inhibition [TWI] about 80%; P < 0.001) in a very short time, and this effect persisted for at least 30 days (Figure 6A). A complete tumor response was observed in 80% of mice, and 40% were cured. In all the other tumor xenografts, RHPS4 treatment produced about 50% (P < 0.001) TWI at the nadir of the effect and, more important, resulted in a delay of tumor growth of about 15 (M14 and PC3) and 10 (HT29 and H460) days.
Figure 6. RHPS4 is active as single agent in xenograft tumors by inducing telomere injury and apoptosis.
(A) Mice were injected i.m with M14 melanoma, PC3 prostate, H460 non–small cell lung, CG5 breast, and HT29 colon carcinoma cells and, starting from day 6 after cell injection, when a tumor mass of about 300 mg was observed, treated i.v. with RHPS4 at 15 mg/kg for fifteen consecutive days (days 6–21). Mean tumor weights in untreated (filled squares) and RHPS4-treated (open squares) mice. Points represent mean (bars, SD). Arrow indicates the start of treatment. (B) Terminal restriction fragment measured by Southern blotting in the indicated tumors untreated (–) and treated with RHPS4 (+). (C–F) In situ apoptosis and proliferation measured by TUNEL (C and D) and Ki-67 (E and F) staining, respectively, in untreated (C and E) and RHPS4-treated M14 tumors (D and F). Original magnification, ×40. (G–J) Immunohistochemical analysis of γ-H2AX (G and H) and H&E (I and J) staining in untreated (G and I) and RHPS4-treated M14 tumors (H and J). Original magnification, ×40. Enlarged views (original magnification, ×80) showing γ-H2AX reactivity and atypical mitoses are reported on the right. Analyses performed on tumor tissues were carried out at the end of treatment (day 22 after tumor cell injection) and were repeated 3 times using 3 different tumors for each point. Representative images of 3 independent experiments with comparable results are shown.
Consistent with the short-term antitumor effect and in agreement with our previously reported data (27), no telomere erosion was found in RHPS4-treated compared with untreated tumors at the end of drug administration, when the antitumor effect was already evident (Figure 6B). Pharmacodynamic monitoring of effects in M14 melanoma xenograft tissues showed that the antitumor activity of RHPS4 resulted from the activation of apoptosis (apoptotic index [AI]: 9.2% ± 2.2% versus 2.6% ± 1.6%; P = 0.01) and decrease in proliferation (proliferation index [PI]: 82% ± 7.2% versus 37% ± 6.9%; P < 0.0001) as revealed by TUNEL and Ki-67 staining (Figure 6, C and D). Untreated tumors showed a high level of γ-H2AX regardless of treatment (Figure 6G), a phenomenon already described for other tumors (29, 30). Importantly, RHPS4 markedly increased the percentage of γ-H2AX–positive cells (30% ± 6.5% versus 80% ± 10.3%; P < 0.01), as well as the immunostaining intensity of the positive cells (Figure 6H). Consistent with these results, RHPS4 significantly induced the presence of atypical mitotic figures, which are indicative of telomere dysfunction (atypical mitotic index [AMI]: 0.60 ± 0.08 versus 0.25 ± 0.11; P < 0.0001). Moreover, anaphase/telophase bridges were also seen in tumors after treatment with RHPS4 (data not shown).
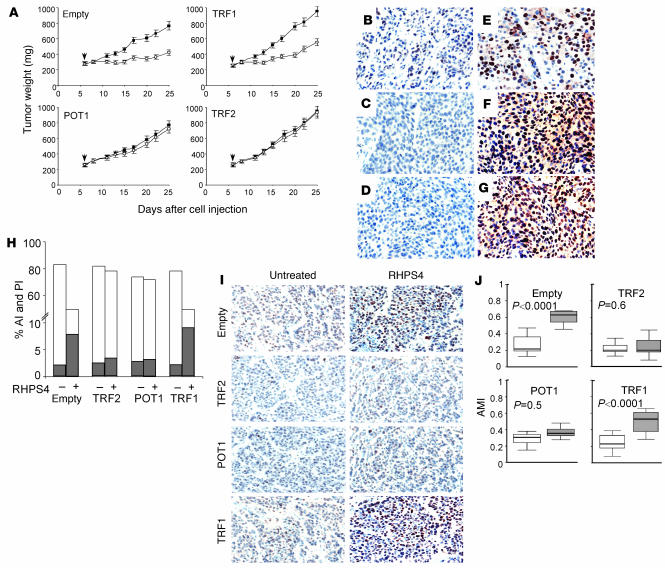
Next, we investigated whether telomere damage is responsible for the antitumor activity of the drug by testing the effect of telomeric proteins. Overexpression of TRF2, POT1, and TRF1 was maintained during the in vivo tumor growth (Figure 7, B–G), and the analysis of antitumor activity demonstrated that RHPS4 was effective to the same extent on control and TRF1-overexpressing xenografts (TWI about 50%), while both TRF2- and POT1-overexpressing tumors were completely resistant to the treatment (Figure 7A). Moreover, no significant increase in the percentage of apoptosis or reduction of proliferation was found in the TRF2- or POT1-overexpressing tumors (Figure 7H). More interestingly, RHPS4 induced H2AX phosphorylation in TRF1-overexpressing tumors to the same extent as in controls, while a low reactivity of γ-H2AX antibody was found in TRF2 or POT1 xenografts, regardless of treatment (Figure 7I). These data indicate that RHPS4 does not induce damage in vivo when POT1 and TRF2 are overexpressed. Moreover, compared with untreated specimens, TRF2- and POT1-overexpressing tumors did not shown an increase in atypical mitotic figures (Figure 7J), demonstrating a functional link between damage and antitumor effect of RHPS4.
Figure 7. TRF2- or POT1-overexpressing tumors were resistant to RHPS4, as TRF2 and POT1 antagonized telomere dysfunction–mediated effects of this ligand.
Mice were implanted intramuscularly with M14 melanoma cells overexpressing TRF1, TRF2, POT1, or puromycin-resistant gene only (Empty). (A) Antitumor activity of RHPS4 in tumors overexpressing TRF1, TRF2, POT1, or the empty vector. Mean tumor weights in untreated (filled squares) and RHPS4-treated (open squares) mice. Points represent mean (bars, SD). Arrow indicates the start of treatment. (B–G) Immunohistochemical analysis of TRF2, POT1, and TRF1 in sections of empty vector– (B, C, and D), TRF2- (E), POT1- (F), and TRF1-transfected (G) tumors excised on day 25 after tumor implantation. TRF2 expression was revealed using anti-TRF2 mAb (B and E), while POT1 (C and F) and TRF1 (D and G) were detected by anti-Flag mAb. Original magnification, ×40. (H) Apoptotic index (AI; gray bars) and PI (white bars; mean percentage) in the indicated groups untreated (–) and treated with RHPS4 (+) as indicated by TUNEL and Ki-67 staining. The analysis was performed at the end of treatment and was repeated 3 times using 3 different tumors for each point. The mean of 3 independent experiments is reported (SD was less than 10%). (I) Immunohistochemical analysis of γ-H2AX in tumor sections from the indicated groups. Original magnification, ×40. (J) Atypical mitotic index (AMI) in tumor sections from the indicated groups untreated (white bars) and treated with RHPS4 (gray bars).
Discussion
G4 ligands were initially designed to counteract telomerase action at telomeres. Surprisingly, their antiproliferative effects can occur in telomerase-negative cells, and their kinetics cannot be explained merely by telomere shortening, suggesting that these compounds affect other pathways not necessarily related to telomere biology (31). The main conclusion of this work is that cancer cells treated with the G4 ligand RHPS4 undergo a unique type of telomere damage that is responsible for the antitumor effects of the drug. These findings reveal the telomere-specific effects of RHPS4 and validate telomeres as promising targets for future anticancer therapies.
The immediate response to RHPS4 combines a potent DNA damage response at telomeres and POT1 removal. These results are in agreement with a recent study showing that an ectopically expressed POT1 protein is delocalized from telomere upon treatment with telomestatin, another G4 ligand (32). Importantly, the RHPS4 effects do not correlate with any obvious change in the length of telomeric DNA and of 3′ overhang. The RHPS4-induced H2AX phosphorylation is ATM independent and ATR dependent. This points to a telomere damage response distinct from the one elicited by TRF2 inhibition, which is mainly ATM dependent (3). Indeed, we were unable to detect any sign of TRF2 removal at the time of appearance of this damage response, and TRF2 still colocalized with the telomeres. By contrast, POT1 did not colocalize with RHPS4-induced damaged telomeres, suggesting a causal relationship between the loss of POT1 and the RHPS4-induced telomere injury. The rapid removal of POT1 induced by RHPS4 is likely to result at least in part from the inability of this protein to bind G4 structures (33), which, indeed, are stabilized by the drug. One might also speculate that RHPS4 alters other telomeric components involved in the recruitment of POT1 at telomeres (22). In agreement with the latter possibility, the high level of the telomere damage induced by RHPS4, which is similar to that triggered by the expression of a dominant-negative allele of TRF2, does not recapitulate the reported phenotype of POT1 depletion (23, 34). In any case, POT1 removal and/or the telomere dysfunctions leading to POT1 removal are likely to be involved in the telomere damage provoked by the drug.
During prolonged treatment of the cells, when they became committed to cell death, TRF2 disappeared from telomeres, and the length of the 3′ overhang was reduced. These results are consistent with the hypothesis that TRF2 removal is only a secondary consequence of RHPS4 treatment, perhaps related to apoptosis. They are in agreement with previous findings showing that telomestatin, another G4 ligand, delocalizes TRF2 (35).
Molecules binding to the telomeric G4 structures can also display potent activity against other G4s, such as c-myc promoter (36). A broad view of the gene expression profile in untreated and treated cells demonstrated that RHPS4 does not modify the transcriptional signature of cells (Supplemental Figure 6), including the expression of c-myc (27). These results together with the demonstration that cells overexpressing either POT1 or TRF2 become resistant to RHPS4 suggest that telomere is the main target of RHPS4, even if we cannot exclude that RHPS4-induced damage could extend beyond the telomeric regions. In particular, it is tempting to speculate that G4 DNA is perceived as damage because it prevents t-loop formation and/or blocks the progression of the replication fork through telomeric DNA. The ATR dependency of the RHPS4 telomere response is in agreement with the latter possibility, since this kinase is the major transducer of replication stress (37). TRF2 overexpression might counteract the deleterious effects of the stabilization of G4 DNA by RHPS4 through several means, including the formation of t-loop that would mask the 3′ overhang against RHPS4 binding (11); the recruitment and stimulation of RecQ helicases, which are expected to resolve G4 DNA or other DNA structures induced by the stabilization of G4 during telomere replication or recombination (38–42); and favoring binding of POT1 at telomeres (43). Similarly, an excess of POT1 might trap the G-tail in an unfolded form (33) and/or stabilize the t-loop–associated D-loop (44).
Interestingly in view of the clinical application, RHPS4 does not induce telomere damage in normal and telomerized human fibroblasts, which are unaffected by the treatment, suggesting that this agent would preferentially kill cancer cells. Moreover, consistent with these results, RHPS4 is effective on tumor cells of different histotype, but not in nonmalignant cells of the same tissue type (data not shown). The lack of telomere damage in normal cells correlates with the inability of RHPS4 to induce POT1 and TRF2 delocalization (Supplemental Figure 7). The RHPS4 resistance of normal and telomerized cells does not seem to be caused by a general inability to respond to DNA damage, since activation of H2AX is triggered by the DNA-damaging agent bleomycin and upon expression of TRF2ΔBΔC (Supplemental Figure 1). The selective effect of RHPS4 on transformed and cancer cells could result from a checkpoint failure in the transformed cells that would allow the accumulation of deleterious damage. In fact, the very early appearance of telomere damage and the subsequent detrimental effects on cell viability suggest that cells have to be chronically exposed to the drug to accumulate enough lethal damages. Moreover, while it is difficult to imagine how telomeres differ between normal and tumor cells, the slow growth of normal versus the rapid proliferation of tumor cells may be involved in the RHPS4 selectivity. However, since highly proliferating human peripheral blood lymphocytes are resistant to the drug (data not shown) at the dose that is toxic to transformed cells, we can conclude that the growth rate is not the main cause of RHPS4 tumor selectivity. Therefore, we cannot exclude the possibility that the protein composition at the telomeres differs, thus providing normal cells with a higher degree of telomere stability. This opens the door to an interesting situation in light of future clinical development of this new class of antitumor agents.
Our work goes one important step further, by demonstrating in xenograft models that RHPS4 is active on tumors of different histotype, without any sign of toxicity. The observation that RHPS4 has a high therapeutic index in xenografts has to be taken with caution considering the different length and structure of telomeres in mice and humans. However, the lack of toxicity in mice should not depend on differences in telomere structure, since RHPS4 does not affect the normal haematological compartment of mice (see Supplemental Figure 4). We also found that the anticancer effects of RHPS4 are associated with telomere dysfunction and are antagonized in tumors overexpressing either TRF2 or POT1. These results clearly demonstrate that telomere damage is the cause of antitumor effect and define a strong correlation between the occurrence of telomeric DNA damage foci and the antitumor effect. Overall, these data are consistent with a model whereby RHPS4 exerts specific antitumor effects by disrupting telomere chromatin rather than by provoking telomeric DNA erosion. Our results seem to be in contradiction to previously published data demonstrating that telomere length of tumor xenografts influences the response to growth inhibition by RHPS4 (26). This discrepancy might depend on differences in the drug administration.
A number of G4-interactive compounds have been previously employed in xenografted tumors, but their antitumor activity is revealed generally only after a chronic administration for several weeks. By contrast, the in vivo activity of RHPS4, reported here, appears very rapid in all the tumors employed. Although the antitumor effect of this compound does not suggest a clinical application in monotherapy, the short-term reduction in tumor growth observed after only few days of treatment indicates that further studies aimed at evaluating RHPS4 in combination therapy are warranted.
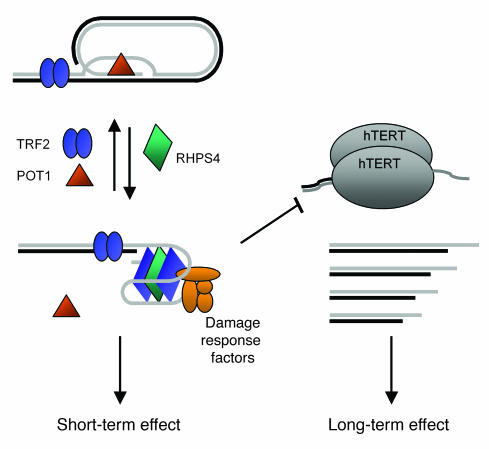
In conclusion, the data reported in this article provide mechanistic insights into the short-term effect of RHPS4 and evidence that, in addition to its telomerase-inhibitory properties, this drug exerts an anticancer effect by altering telomeric chromatin (Figure 8). They also validate telomeres as potential therapeutic targets and identify TRF2 and POT1 as compelling targets for anticancer drugs, suggesting that the combined use of G4 ligands, such as RHPS4, and TRF2- or POT1-inhibitory molecules may have a synergistic effect on tumor response, offering new opportunities for cancer therapy.
Figure 8. Model for the short- and long-term effects of RHPS4.
G4 stabilization by RHPS4 can induce short-term telomere damage and release of POT1. Overexpression of either TRF2 or POT1 can prevent G4 formation, for example, by favoring t-loop formation or G4 unwinding, and render cells resistant to RHPS4-induced damage. RHPS4 can also induce long-term effects through telomerase inhibition and consequent telomere shortening.
Methods
Cells and culture conditions.
Transformed BJ fibroblasts expressing hTERT and SV40 early region (BJ-EHLT) were maintained as previously described (20). Human M14 melanoma, CG5 breast carcinoma, and HT29 colon carcinoma lines were previously described (45–47). Human non–small cell lung carcinoma and PC3 prostate cancer cells were obtained from ATCC and maintained in RPMI-1640 supplemented with 10% FCS, 2 mM l-glutamine and antibiotics (Invitrogen).
Cells were transfected with epitope-tagged Flag-hPOT1 (48), Flag-hTRF1, and Myc-hTRF2 (4) in a pWPIR-GFP lentiviral vector backbone (obtained from Didier Trono, Swiss Institute of Technology, Lausanne, Switzerland) or vector alone using Lipofectamine 2000 reagent (Invitrogen). Analysis of cell viability demonstrated that overexpression of TRF1, TRF2, or POT1 does not modify the growth rate of BJ-EHLT and M14 cells (data not shown). For RNA interference experiments, cells were transfected with 200 pmol of siATM and siATR SMARTpool (Dharmacon) or with siGFP as negative control (Cell Signaling Technology) in a 35-mm Petri dish using Lipofectamine 2000 reagent.
Antibodies.
The following antibodies were used: mAb and polyclonal antibody (pAb) anti–γ-H2AX (Upstate), pAb anti-pRad17 (Cell Signaling Technology), mAb anti-TRF2 (Imgenex), pAb anti-53BP1 (Novus Biologicals), mAb anti-TRF1 (Abcam), pAb anti-mPOT1 (the antibody, generated by us cross-reacts with human POT1); pAb anti-hPOT1 (generated and validated for its specificity by Western blotting and immunofluorescence; generated by us); pAb anti-H2A (Upstate); mAb anti–Ki-67 (clone MIB-1; Dako); mAb anti-Flag M2 (Sigma-Aldrich); mAb anti-ATM (a generous gift from D. Delia, Istituto Nazionale Tumori, Milan, Italy); mAb anti-ATR (Abcam) and anti–HSP72/73 (Calbiochem).
In vitro treatments.
RHPS4 was synthesized as described previously (49). The drug, used at a concentration of 1 μM for various times, was added to the cells 24 hours after plating. Cell counts (Coulter Counter; Kontron Instruments) and viability (trypan blue dye exclusion) were determined in each experiment. Caffeine (Sigma-Aldrich) at 10 mM, a dose with no toxic effect on cell viability, was left in the medium for 24 hours. Bleomycin (Aventis Pharma S.p.A) was used at a concentration of 30 mM for 30 minutes.
Immunofluorescence.
Cells were fixed in 2% formaldehyde and permeabilized in 0.25% Triton X-100 in PBS for 5 minutes at room temperature. For immunolabeling experiments, cells were incubated with the primary antibody, then washed and incubated with the secondary antibody. Nuclei were visualized using DAPI or Hoechst (Sigma-Aldrich). Fluorescence signals were analyzed either in stained samples recorded using a CCD camera (Zeiss) and IAS2000/H1 software (Delta Sistemi) or in confocal vertical (x-z) sections (interval: 0.5 mm) captured with a Zeiss Confocal Laser Scanning Microscope.
ChIP.
Formaldehyde-cross-linked chromatin fragments were immunoprecipitated with mAb anti–γ-H2AX (Upstate). mAb anti–β-actin and mAb anti-H3 were used as negative and positive controls, respectively, for the ChIP assay. After precipitation with each antibody, the precipitants were blotted onto Hybond-N membrane (Amersham), and telomeric repeat sequences were detected with a TeloTAGGG probe. A nonspecific probe (Alu) was also used. To verify that an equivalent amount of chromatin was used in the immunoprecipitates, samples representing the 0.1%, 0.01%, and 0.001% of the total chromatin (input) were included in the teloblot. The filter was exposed to a PhosphorImager screen (Bio-Rad), and the signals were measured using ImageQuant software (Quality One; Bio-Rad).
Clonogenic assay.
Cells were seeded in 60-mm Petri dishes (Nunc) at a density of 2 × 105 cells per dish and exposed to RHPS4 for 4 days. Cell colony–forming ability was determined as previously described (50). All the experiments were repeated 4 times in triplicate.
Western blotting.
Western blot on total extracts was performed as previously reported (50). To obtain the nucleoplasmic fractions, cells were resuspended in cell lysis buffer (5 mM PIPES pH 8, 85 mM KCl, 0.5% NP40, 1 mM DTT, and protease inhibitors). After 10 minutes of incubation on ice, cells were homogenized with a glass Dounce, and cell homogenates were centrifuged at 5,000 g for 5 minutes at 4°C. Nuclei were resuspended in buffer prepared with various concentrations of KCl (150, 300, and 450 mM) in lysis buffer (20 mM HEPES-KOH pH 7.5, 25% glycerol, 0.1 mM EDTA, 5 mM MgClB2B, 0.25% NP40, 1 mM DTT, and protease inhibitors). After 10 minutes of incubation on ice, the nuclear suspension was centrifuged at 14,000 g for 10 minutes at 4°C. Thirty micrograms of the resulting supernatant, containing the nucleoplasmic fraction, was separated by SDS-PAGE and used for immunoblot analysis.
In vivo treatment.
CD-1 male nude (nu/nu) mice, 6–8 weeks old and weighing 22–24 g, were purchased from Charles River Laboratories. All procedures involving animals and their care were approved by the ethical committee of the Regina Elena Cancer Institute. Each experimental group included 10 mice.
To evaluate the antitumor efficacy of RHPS4, mice were implanted intramuscularly with the tumor cells at 2 × 106 (H460), 3 × 106 (CG5 and HT29), and 5 × 106 (PC3 and M14) cells/mouse. In these experimental conditions, a tumor mass of about 300 mg was evident in all the animals at day 6 after cell injection. Mice received the maximum tolerated dose (15 mg/kg) i.v. for 15 consecutive days. As control, tumor-bearing mice were treated with saline solution (vehicle). The tumor weights were measured by caliper. Percent TWI was calculated as [1 – (mean tumor weight of treated mice / mean tumor weight of untreated mice)] × 100. Tumor growth delay was evaluated by comparing the median times for treated and untreated tumors, respectively, to achieve equivalent size.
Telomere assays.
Terminal restriction fragment determination was performed as previously reported (50). To measure telomeric overhang, 5 μg of DNA were digested with 20 U of Hinf1 and resuspended in hybridization buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA) and mixed with [CCCTAA]B4 probe end-labeled with [γ-32P]ATP (6,000 Ci/mmol; Amersham) and T4 polynucleotide kinase. After incubation for 16 hours at 50°C in a PerkinElmer thermocycler, samples were size fractionated on 0.8% agarose gel. As control for the 3′ overhang assay, DNA was also incubated overnight at 37°C with 50 U of Exonuclease I (New England BioLabs). The gel was dried on Whatman DE-81 paper at 50°C and exposed to a PhosphorImager screen. Signals were measured using ImageQuant software by integration of the entire lane. Subsequently, the paper was removed and the gel was denatured in 0.5 M NaOH, 1.5 M NaCl for 30 minutes; neutralized in 0.5 M Tris-HCl pH 7.5, 3 M NaCl for 20 minutes; and hybridized with the same [CCCTAA]B4 probe in Church mix (0.5 M Na2HPO4 pH 7.2, 1 mM EDTA, 7% SDS, 0.1% BSA) overnight at 55°C. The gel was washed 3 times in 4× SSC for 30 minutes at room temperature and once with 4× SSC, 0.1% SDS at 50°C and exposed to a PhosphorImager screen.
Immunohistochemical analyses.
In situ detection of apoptosis in formalin-fixed, paraffin-embedded tumor tissues was performed by TUNEL assay (Enzo, Dako), as previous reported (51). PI, activation of damage response, and expression of telomeric proteins were determined immunohistochemically on formalin-fixed, paraffin-embedded tumor sections (4 different experiments). Antigen retrieval was achieved by pretreating dewaxed sections in a microwave oven at 750 W for 5 minutes in citrate buffer (pH 6) and then processing them with a Super Sensitive Link-Label Detection System (BioGenex). The enzymatic activity was developed using 3-amino-9-ethylcarbazole (Dako) as a chromogenic substrate. Following counterstaining with Mayer hematoxylin, slides were mounted in aqueous mounting medium (Glycergel; Dako). PI was determined by counting positive nuclei in 8 high-power fields (magnification, ×400) per section. Negative controls consisted of parallel sections in which the primary antibody was omitted. In addition, H&E-stained sections were prepared. Slides were observed by 2 independent observers with no knowledge of the experimental protocol.
Statistics.
The experiments were repeated 3–5 times, and the results are presented as mean ± SD. Significant changes were assessed using 2-tailed Student’s t test for unpaired data, and P values of less than 0.05 were considered significant. To determine the significant differences between group means in an ANOVA setting (PI, AI, AMI), the Bonferroni test was used. Differences were considered statistically significant when P was less than 0.05.
Supplementary Material
Acknowledgments
This work was supported by grants from the Italian Association for Cancer Research (AIRC) and Ministero della Salute. A. Biroccio thanks the European Molecular Biology Organization for short-term fellowship. A. Rizzo is the recipient of a fellowship from the Italian Foundation for Cancer Research (FIRC). Work in Eric Gilson’s laboratory was supported by La Ligue Nationale contre le Cancer (“équipe labellisée”) and by the Institut National du Cancer (program EPIPRO). Work in Mario Blasco’s laboratory was supported by the Spanish Ministry of Education and Culture, the Regional Government of Madrid, the European Union, and the 2003 Josef Steiner Cancer Research Award. Work in Maurizio D’Incalci’s laboratory was supported by the European Union “Sixth Framework Programme, Priority 1: Life Sciences, Genomics and Biotechnology for Health” (MOL CANCER MED, project LSHC-CT-2004-502943). We thank Mike Bibby, Amanda Race, and Paul Loadman for pharmacokinetic studies and Giorgia Cardinale for her excellent assistance with confocal microscopy. We are also grateful to Steve Jackson and Silvia Soddu for critical reading of the manuscript. Christoph Geserick in Mario Blasco’s laboratory generated the pAb anti-mPOT1.
Footnotes
Nonstandard abbreviations used: ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3–related; G4, G-quadruplex; pAb, polyclonal antibody; PI, proliferation index; POT1, protection of telomeres 1; RHPS4, 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate; TIF, telomere dysfunction–induced focus; TRF2, telomeric repeat binding factor 2; TWI, tumor weight inhibition.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3236–3247 (2007). doi:10.1172/JCI32461
References
- 1.de Lange T., Jacks T. For better or worse? Telomerase inhibition and cancer. Cell. 1999;98:273–275. doi: 10.1016/s0092-8674(00)81955-8. [DOI] [PubMed] [Google Scholar]
- 2.d’Adda di Fagagna F., et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 3.Takai H., Smogorzewska A., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 4.Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. . 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A., de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 7.Mergny J.L., Riou J.F., Mailliet P., Teulade-Fichou M.P., Gilson E.2002Natural and pharmacological regulation of telomerase . Nucleic Acids Res. 30839–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biroccio A., Leonetti C. Telomerase as a new target for the treatment of hormone-refractory prostate cancer. Endocr. Relat. Cancer. 2004;11:407–421. doi: 10.1677/erc.1.00764. [DOI] [PubMed] [Google Scholar]
- 9.Wright W.E., Tesmer V.M., Huffman K.E., Levene S.D., Shay J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. . 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarov V.L., Hirose Y., Langmore J.P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 11.Griffith J.D., et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 12.Nikitina T., Woodcock C.L. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han H., Hurley L.H. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000;21:136–14. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 14.Mergny J.L., Hélène C. G-quadruplex DNA: a target for drug design. Nat. Med. . 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 15.Cuesta J., Read M.A., Neidle S. The design of G-quadruplex ligands as telomerase inhibitors. Mini Rev. Med. Chem. 2003;3:11–21. doi: 10.2174/1389557033405502. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. . 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 17.Karlseder J., et al. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 19.Ancelin K., Brun C., Gilson E. Role of the telomeric DNA-binding protein TRF2 in the stability of human chromosome ends. Bioessays. . 1998;20:879–883. doi: 10.1002/(SICI)1521-1878(199811)20:11<879::AID-BIES2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 20.Brunori M., et al. TRF2 inhibition promotes anchorage-independent growth of telomerase-positive human fibroblasts. Oncogene. . 2006;25:990–997. doi: 10.1038/sj.onc.1209135. [DOI] [PubMed] [Google Scholar]
- 21.Biroccio A., et al. TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur. J. Cancer. 2006;42:1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Loayza D., De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. . 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 23.Hockemeyer D., Sfeir A.J., Shay J.W., Wright W.E., de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gowan S.M., Heald R., Stevens M.F., Kelland L.R. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 25.Gavathiotis E., Heald R.A., Stevens M.F.G., Searle M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Cookson J.C., et al. Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol. Pharmacol. 2005;68:1551–1558. doi: 10.1124/mol.105.013300. [DOI] [PubMed] [Google Scholar]
- 27.Leonetti C., et al. Biological activity of the G-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol. Pharmacol. . 2004;66:1138–1146. doi: 10.1124/mol.104.001537. [DOI] [PubMed] [Google Scholar]
- 28.van Steensel B., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 29.Gorgoulis V.G., et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 30.Bartkova J., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 31.Riou J.F. G-quadruplex interacting agents targeting the telomeric G-overhang are more than simple telomerase inhibitors. Curr. Med. Chem. Anticancer Agents. 2004;4:439–443. doi: 10.2174/1568011043352740. [DOI] [PubMed] [Google Scholar]
- 32.Gomez D., et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 33.Zaug A.J., Podell E.R., Cech T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veldman T., Etheridge K.T., Counter C.M. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Tahara H., et al. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3ι telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 36.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward I.M., Chen J.2001Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress . J. Biol. Chem. 27647759–47762. [DOI] [PubMed] [Google Scholar]
- 38.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 39.Fry M., Loeb L.A. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 40.Lillard-Wetherell K., et al. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 41.Stavropoulos D.J., et al. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 42.Opresko P.L., et al. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. . 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 43.O’Connor M.S., Safari A., Xin H., Liu D., Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stansel R.M., de Lange T., Griffith J.D.2001. . T-loop assembly in vitro involves binding of TRF2 near the 3ι telomeric overhang. EMBO J. 205532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonetti C., et al. In vivo administration of liposomal vincristine sensitizes drug-resistant human solid tumors. Int. J. Cancer. 2004;110:767–774. doi: 10.1002/ijc.20174. [DOI] [PubMed] [Google Scholar]
- 46.Del Bufalo D., et al. 2004Lonidamine causes inhibition of angiogenesis-related endothelial cell functions . Neoplasia. 6513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laudonio N., Zupi G., Erba E., Leonetti C., D’Incalci M. Synergism between 5-fluorouracil and N-methylformamide in HT29 human colon cancer line. Br. J. Cancer. 1990;61:377–381. doi: 10.1038/bjc.1990.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armbruster B.N., et al. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heald R.A., et al. Antitumor polycyclic acridines. 8. Synthesis and telomerase-inhibitory activity of methylated pentacyclic acridinium salts. . J. Med. Chem. . 2002;45:590–597. doi: 10.1021/jm011015q. [DOI] [PubMed] [Google Scholar]
- 50.Biroccio A., et al. Reconstitution of hTERT restores tumorigenicity in melanoma-derived c-Myc low-expressing clones. Oncogene. 2002;21:3011–3019. doi: 10.1038/sj.onc.1205415. [DOI] [PubMed] [Google Scholar]
- 51.Leonetti C., et al. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin. Cancer Res. 1999;5:2588–2595.. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.