Abstract
Palladin and SPIN90 are widely expressed proteins, which participate in modulation of actin cytoskeleton by binding to a variety of scaffold and signaling molecules. Cytoskeletal reorganization can induced by activation of signaling pathways, including the PDGF receptor and Src tyrosine kinase pathways. In this study we have analyzed the interplay between palladin, SPIN90 and Src, and characterized the role of palladin and SPIN90 in PDGF and Src-induced cytoskeletal remodeling. We show that the SH3 domains of SPIN90 and Src directly bind palladin’s poly-proline sequence and the interaction controls intracellular targeting of SPIN90. In PDGF-treated cells, palladin and SPIN90 co-localize in actin rich membrane ruffles and lamellipodia. The effect of PDGF on the cytoskeleton is at least partly mediated by the Src kinase, since PP2, a selective Src kinase family inhibitor, blocked PDGF-induced changes. Furthermore, expression of active Src kinase resulted in coordinated translocation of both palladin and SPIN90 to membrane protrusions. Knock-down of endogenous SPIN90 did not inhibit Src-induced cytoskeletal rearrangement, whereas knock-down of palladin resulted in cytoskeletal disorganization and inhibition of remodeling. Further studies showed that palladin is tyrosine phosphorylated in cells expressing active Src indicating bidirectional interplay between palladin and Src. These results may have implications in understanding the invasive and metastatic phenotype of neoplastic cells induced by Src.
Keywords: Palladin, SPIN90, Src, cytoskeleton
Introduction
Regulation of the actin microfilament cytoskeleton is important in cellular processes such as adhesion, migration and contraction. For these purposes the cytoskeleton is organized into specialized dynamic structures, such as stress fibers, focal adhesions and lamellipodia. When the cells receive new stimuli from their environment, these structures are disassembled and remodeled to meet the new requirements. The remodeling is tightly controlled by a variety of actin-binding proteins, scaffold proteins and effector proteins. An important effector is Src, a non-receptor tyrosine kinase, which contains SH3, SH2 and kinase domains. Src is activated by several mechanisms including receptor tyrosine kinase pathways[1]. Binding of platelet derived growth factor (PDGF) to its receptor leads to receptor autophosphorylation and association with Src. Activated Src translocates to the plasma membrane where it reorganizes actin cytoskeleton and induces formation of lamellipodia and membrane ruffles. This process requires a direct interaction between Src and actin-associated proteins[1, 2]. Indeed, Src kinase is shown to have several cytoskeletal substrates, including FAK, vinculin, cortactin and ezrin [3, 4]. The molecular and functional connection between Src and the structural and scaffold proteins is, however, incompletely understood. These events are of major biological and medical importance, as activation of Src occurs in several types of malignant tumors and increases tumor cell migration and invasion and hence promotes tumor dissemination[5].
Palladin is a member of the recently characterized palladin-myotilin-myopalladin protein family[6]. All family members associate with actin cytoskeleton and contain characteristic conserved IgI type domains in the C-terminal part of the protein. In striated muscle, the proteins are located in sarcomere Z-discs and control the organization of actin cytoskeleton. For instance, myotilin and myopalladin play an important role in the maintenance of sarcomere integrity and myotilin mutations lead to muscle disorders[7, 8]. The family members have also common binding partners, such as α-actinin[9]. However, the sequence, expression pattern, molecular interactions and functions clearly distinguish palladin from other family members. In contrast to myopalladin and myotilin, palladin is expressed as several alternatively spliced isoforms, termed by the number Ig-domains, in a cell-type specific manner[10–12]. The most widely expressed of the isoforms, the major 3Ig isoform, has two amino-terminal proline-rich sequences, which serve as binding sites for Ena/VASP proteins, SH3 domains and profilin[13–15]. On the other hand, the 4Ig isoform has an additional unique poly-proline sequence, which mediates binding LASP-1, a regulator of cell motility[12]. Palladin also interacts with the ERM-family protein ezrin and with Eps8[11, 16]. Whereas myotilin and myopalladin are mainly expressed in striated muscle, palladin has a wider pattern; it is expressed in for instance neuronal cells, fibroblasts and epithelial cells[10]. In non-muscle cells palladin is typically localized in stress fiber dense bodies, in focal adhesions and in lamellipodial regions. Previous studies have shown that depletion of palladin results in a loss of stress fibers and disruption of cell morphology thus implying that palladin is involved in the maintenance of normal actin cytoskeleton[10, 17]. Palladin is phosphorylated both on serine/threonine and tyrosine residues but the specific kinases have not been identified[10].
In the present study we set out to find new interaction partners for palladin and identified two new associated molecules, SPIN90 and Src. Palladin and SPIN90 are similarly targeted to membrane ruffles after PDGF treatment or Src activation. We show that depletion of SPIN90 does not affect Src induced actin remodeling, whereas knock-down of palladin causes disruption of stress fibers and reduction of membrane ruffle formation and thereby prevents Src-induced cytoskeletal changes. Palladin is physically associated with Src SH3 domain and active Src induces its tyrosine phosphorylation in vivo demonstrating that the interplay between the proteins occurs at several levels.
Materials and Methods
cDNA constructs and antibodies
The palladin constructs have been described before ([14, 22]. The amino acids are numbered based on the 772 residue sequence of palladin 3Ig isoform [11]. For yeast two hybrid (Y2H) experiments palladin fragments were subcloned in pLexA vector, for expression in mammalian cells in pEGFP-C or pAHP vectors and for mitochondrial targeting in pCDNA3-MOM-BiPro vector[23]. The cDNA encoding full-length SPIN90 (amino acids 1–722) and various SPIN90 fragments (ΔSH3 C-term = amino acids 280–722; SH3 = amino acids 1 – 145) were subcloned into pACTII prey vector for years two–hybrid analyses, into pGEX4T-1 (Amersham Pharmacia Biotech, Piscataway, NJ) for expression of recombinant protein in bacteria and into HA pcDNA3 (Invitrogen, Carlsbad, CA) for mammalian expression. The cDNA of the wild-type (WT) Src, constitutively activated Src containing Y529F mutation and the dominant negative kinase-dead Src K296R/Y528F in pUSEamp eukaryotic expression vector were from Upstate Biotechnology.
The SPIN90 and palladin 3Ig antibodies have been described [18,24] .A polyclonal antibody raised against the FL 3Ig mouse isoform was also used. Anti-HA mAb was from Roche and anti-phosphotyrosine antibody clone PT-66 from Sigma. Cortactin was detected with 4F11 monoclonal antibody (Upstate Biotechnology, Lake. Placid, NY).
Yeast two hybrid analysis
The bait and prey vectors (pLexA and pACT2) were cotransformed into the Saccharomyces cerevisae L40 reporter strain using a modified lithium acetate protocol. The transformants were plated onto synthetic medium lacking leucine and tryptophan. Activation of the His 3 reporter gene was assayed by restreaking the cells on plates lacking histidine, leucine, tryptophan, uracil and lysine and incubating them for 3 days at 30°C. His+ colonies were grown on synthetic medium lacking leucine and tryptophan, incubated for 3 days at 30°C and assayed for beta-galactosidase activity by a filter assay.
Cell cultures, transfections and immunofluorescence microscopy
U251mg, COS-7 and HEK293 cells were grown in Modified Eagles Medium (MEM) supplemented with 10% fetal calf serum and gentamycin. Cells were transfected using FuGENE 6 (Roche Diagnostics) transfection reagent. After 24–48 h cells grown on glass coverslips were washed and fixed with 3.5% PFA, permeabilized with 0.1% Triton X-100 and stained. HA-tagged constructs were detected with anti-HA mAb followed by rhodamine-conjugated anti-mouse IgG antibody and specimens were analyzed in Zeiss Axiophot fluorescence microscope equipped with AxioCam cooled CCD-camera.
In vitro translation, affinity/immunoprecipitation and phosphorylation assays
HA-tagged palladin 8–772 (3Ig) was in vitro translated using a rabbit reticulocyte T7 kit according to the manufacturer’s instructions (Promega). GST coupled SPIN90 FL, SH3 and C-term ( SH3) constructs were produced in E. coli strain DH5α and purified using glutathione beads (Pharmacia). In the affinity precipitation assay 5μl of 32S Met-labeled in vitro translated palladin construct was mixed with an equal amount of GST-SPIN90 beads in binding buffer (10mM Tris-HCl, pH7.4, 150mM NaCl, 1mM MgCl2, 0.1% Triton X-100). After 2h incubation at +4°C the beads were washed three times with the binding buffer and harvested in Laemmli buffer. The proteins were resolved in 10% SDS-PAGE and bound proteins were detected by autoradiography.
For co-immunoprecipitations COS-7 cells were co-transfected with EGFP-tagged SPIN90 (FL) and HA-tagged palladin 8–772 (3Ig). 36 h after transfection, the cells were washed in PBS and lysed in ELB buffer (150 mM Nacl, 50 mM HEPES pH 7.4, 5 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol (DTT), 2.5 μg/ml aprotinin, 0.5 mM phenylmethylsulfonyl fluoride, 10 mM ßglycerol-phosphate and 1 μg/ml leupeptin). The lysate was centrifuged at 13 000 g for 10 min at +4°C. The cleared supernatant was incubated with 5 μl of anti-HA mAb for 1 h at +4°C prior to adding protein G-Sepharose beads for 3 h. Subsequently beads were washed four times with ELB and harvested in Laemmli buffer and subjected to Western blot analysis.
For GST-protein binding assays HEK293 cells were transfected with a HA-tagged full length SPIN90 or with a WT Src construct. The cleared lysates were allowed to bind GST or GST-Palladin construct containing glutathione-Sepharose beads. Bound proteins were resolved in a 10% SDS-PAGE gel and detected either with a SPIN90 or a Src antibody.
For phosphorylation assays HEK293 cells were co-transfected either with active Src (CA) or dominant-negative Src (DN) constructs and HA-tagged 8–772 (3Ig) palladin. As a negative control a HA-tagged N-terminal ezrin construct (amino-acids 1–309) with a Y145F mutation was used [4]. 36 h after transfection the cells were extracted in cold RIPA buffer (50 mM Tris-HCl, 1% NP-40, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1 mM orthovanadate supplemented with protease inhibitors, pH 7.4) and centrifuged at 13000 g for 10 min at +4°C. Palladin and Ezrin constructs were immunoprecipitated with anti-HA antibody. Phosphotyrosine was detected with anti-phosphotyrosine mAb PT-66 (Sigma).
siRNA experiments
To knock-down the expression of human palladin by RNA interference, two 19-base 3’-Alexa Fluor 546 labeled oligonucleotides were obtained from Qiagen. RNA sequences were: sense 5’-GCACAAAGGAUGCUGUUAU and antisense 5’-AUAACAGCAUCCUUUGUGC. As a control a non-targeting siRNA from Invitrogen was used. The SPIN90 siRNA oligonucleotides have been described previously[20]. Briefly the SPIN90 targeting si-824 psiRNA-hH1GFPzeo G2 vector was used to knock-down SPIN90 and the empty vector was used as a control.
Results
Palladin interacts with SPIN90 and Src
To identify novel interaction partners for palladin, we focused on actin cytoskeleton associated proteins which contain SH3 domain(s), a possible binding interphase with the N-terminal proline-rich regions of palldin 3Ig isoform. According to this rationale we have previously identified ArgBP2 as a palladin interacting protein [14]. SPIN90, a recently identified actin-associated protein, meets these criteria and in this study we set out to test whether it binds palladin[18]. Since the SH3 domain of SPIN90 is quite homologous (35% identity and 50% similarity) to the Src SH3 domain we also included Src in these analyses.
We first performed yeast two-hybrid analyses with palladin, SPIN90 and Src constructs. In these analyses the SH3 domain containing sequences of both SPIN90 and Src interacted with constructs containing the N-terminal part of palladin 3Ig isoform (for sequence information see Supplementary figure 1). This palladin sequence contains two proline rich stretches (residues 65–85 and 185–220) located between amino acids 8–228 (Fig. 1A). Constructs, which contained these sequences demonstrated a strong reactivity, as judged by β-galactosidase reporter color intensity, whereas constructs that lacked both poly-proline sequences did not interact with SPIN90 or Src. The second sequence was sufficient for interaction with Src but not with SPIN90.
Figure 1. Palladin interacts with SPIN90 and Src.
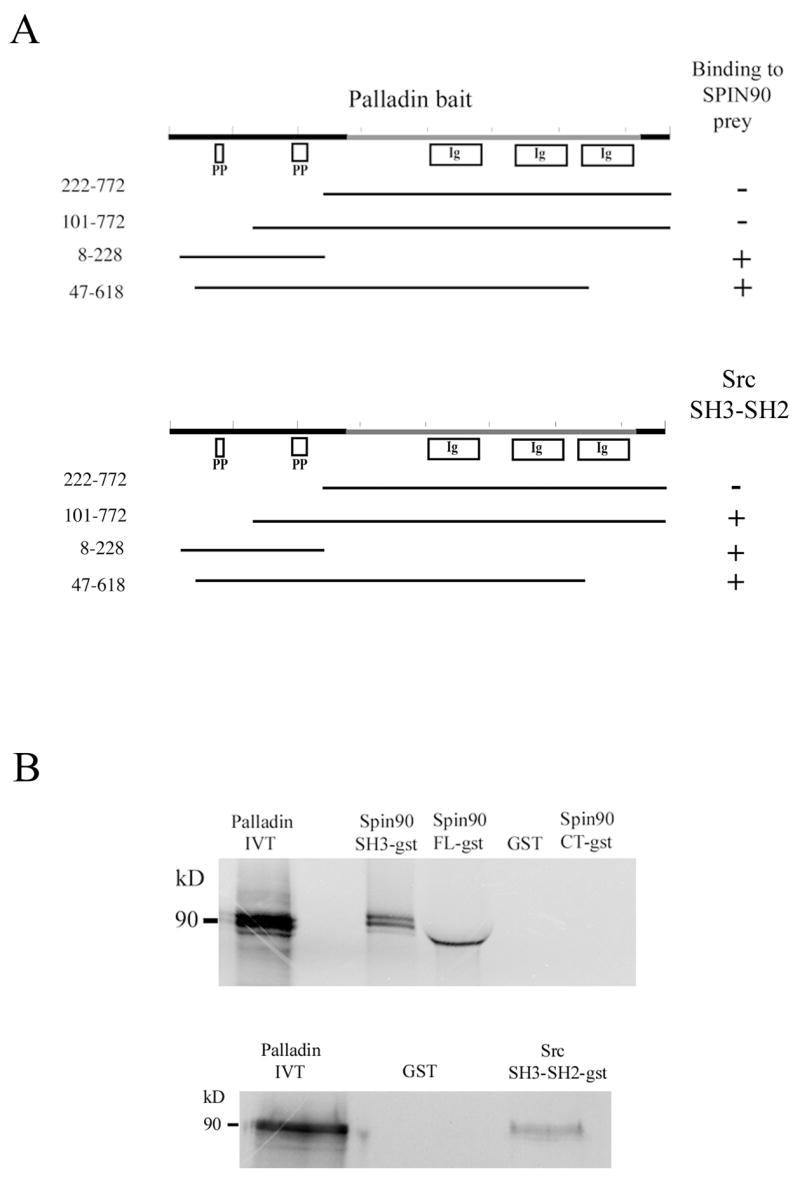
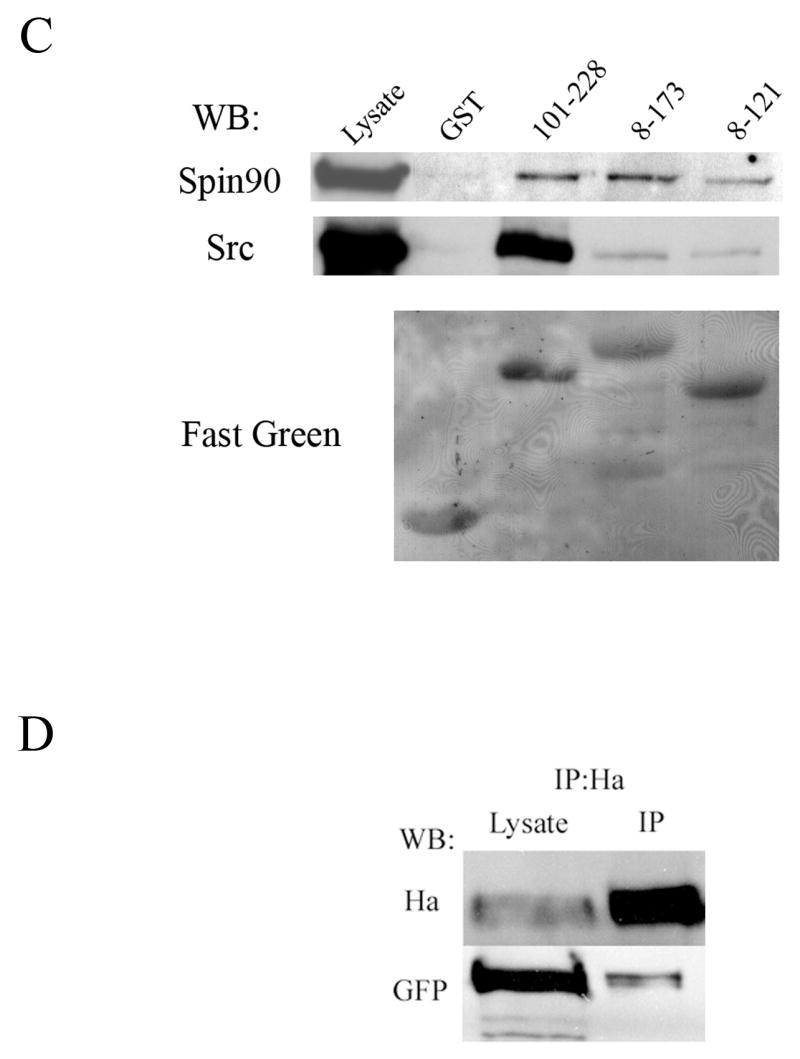
(A) Yeast-two hybrid analysis. Yeast cells were cotransformed with various palladin fragments in bait vector and either full length SPIN90 or Src SH3-SH2 in prey vector. Interaction was monitored by scoring growth on plates lacking histidine and by a filter based β-galactosidase assay. + = positive reaction, − = negative reaction. A scheme of the domains palladin 3Ig isoform is shown on top. The poly-proline (PP) regions are necessary for the interaction with SPIN90 and Src. Ig = immunoglobulin domain. (B) Affinity precipitation assay. In vitro translated (IVT) 35S-labeled palladin 8–772 was allowed to bind GST-SPIN90 and GST-Src constructs coupled to glutathione-Sepharose beads. The panel is an autoradiograph showing the material eluted from beads. The SH3-domain of SPIN90 and SH3-SH2 domain of Src is sufficient in mediating the interaction. GST-FL-SPIN90 and IVT palladin have a similar molecular weight, which explains the slightly differential mobility of IVT protein in the gel. (C). Interaction between palladin poly-proline sequences, SPIN90 and Src. HEK293 cells were transfected with a HA-tagged full length SPIN90 or with wild type Src construct. The cleared lysates were allowed to bind GST (lane 2) or GST-palladin constructs containing the indicated residues on glutathione-Sepharose beads. Bound proteins were resolved in a 10% SDS-PAGE gel and detected either with a SPIN90 or a Src antibody. SPIN90 binding does not show marked preference between the poly-proline sequences, whereas Src binds preferably to the sequence contained within the 101–228 construct. Lane 1 shows immunoblotting of the cell lysates and bottom panel shows Fast Green staining of the membrane to demonstrate the equal loading of GST constructs. (D) Coimmunoprecipitation. COS-7 cells were co-transfected with HA-tagged palladin and GFP-tagged SPIN90. The cleared lysate was immunoprecipitated with an anti-HA antibody and the precipitate was blotted with anti-HA and anti-GFP antibody. The GFP-tagged protein is detected in the precipitate thus verifying the interaction.
In order to verify the interaction with an independent in vitro method, we used an affinity precipitation assay. Palladin was in vitro translated using reticulocyte lysates and allowed to bind to SPIN90 and Src sequences fused to GST and coupled on glutathione-Sepharose beads. Bound proteins were eluted, run in SDS-PAGE and detected by autoradiography. The results show that the SH3 domains of SPIN90 and Src are required and sufficient to mediate the interaction (Fig. 1B). The role of palladin’s proline-rich sequences in the interactions was studied in more detail with an affinity precipitation assay (Fig. 1C). In this assay both halves of the palladin’s N-terminus, containing either the first or the second poly-proline stretch, could mediate binding to SPIN90 and Src. While SPIN90 bound roughly equally to either half, Src seemed to have a preference to palladin’s second proline-rich stretch but the binding constants were not experimentally quantitated.
To study whether palladin binds to SPIN90 also in vivo, an immunoprecipitation assay was used. COS-7 cells were transiently co-transfected with HA-tagged palladin and GFP-tagged SPIN90. After 24 h the cells were lysed and HA-palladin was precipitated with an anti-HA antibody and protein G-Sepharose beads and the precipitates were immunoblotted for HA and GFP. Both HA-palladin and GFP-SPIN90 could be detected in the precipitate indicating that a complex could be formed in vivo (Fig. 1D).
Phosphorylation of palladin in Src CA transfected cells
To further investigate the interplay between palladin, SPIN90 and Src, we tested, whether palladin can serve as a substrate for Src, as previously shown for SPIN90 (also known as DIP) [32]. An HA-tagged construct of palladin (3Ig) was co-expressed with constitutively active (CA) Src in HEK293 cells and the transfected proteins were subsequently immunoprecipitated with an anti-HA antibody. Blotting of the precipitates with a phospho-tyrosine antibody demonstrated a clear reactivity in palladin precipitate (Fig. 2). When palladin was co-expressed with a dominant negative (DN) Src construct, tyrosine phosphorylation of palladin was not detected (Fig. 2).
Figure 2. Palladin is tyrosine phosphorylated in cells expressing active Src kinase.
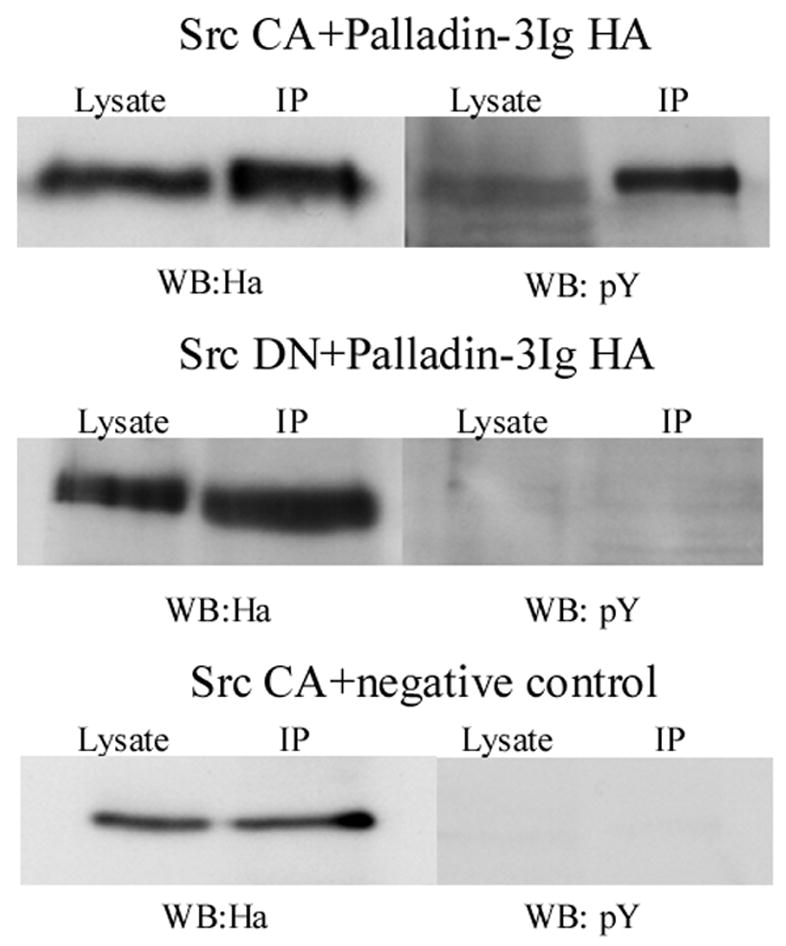
HEK293 cells were co-transfected with HA-tagged palladin together with a constitutively active (CA) or a dominant-negative (DN) Src construct. Palladin was immunoprecipitated with anti-HA antibody and tyrosine phosphorylation analyzed with anti phosphotyrosine (pY) antibody. In cells expressing active Src, palladin is phosphorylated on tyrosine, whereas in cells expressing Src DN no phosphorylation is detected. As a negative control an HA-tagged amino-terminal (amino-acids 1–309/Y145F) ezrin construct was used.
Role of the interaction in subcellular targeting of SPIN90
In order to study whether the interaction with palladin regulates the subcellular distribution of SPIN90 we used two different in vivo targeting assays. We have previously shown that overexpression of palladin in COS-7 cells results in reorganization of the cytoskeleton and formation of thick actin bundles[12, 22]. We analyzed the behavior of HA-tagged SPIN90 (SPIN90-HA) and similarly tagged SH3 domain of PIN90 (SPIN90-SH3-HA) constructs expressed alone or together with GFP-palladin 8–772. When transfected alone, SPIN90 and SPIN90-SH3 constructs were distributed diffusely in the cytoplasm (Fig. 3A and 3D). On the other hand, cotranfrection with palladin resulted in targeting of both SPIN90 constructs to the newly formed actin bundles thus indicating that palladin can target SPIN90 (Fig. 3B–C and 3E–F).
Figure 3. Subcellular targeting of SPIN90 by palladin.
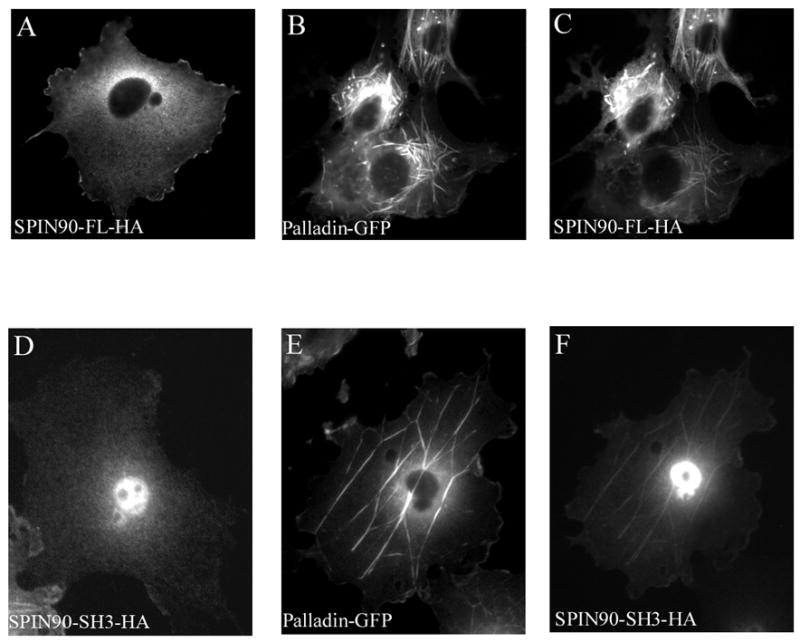
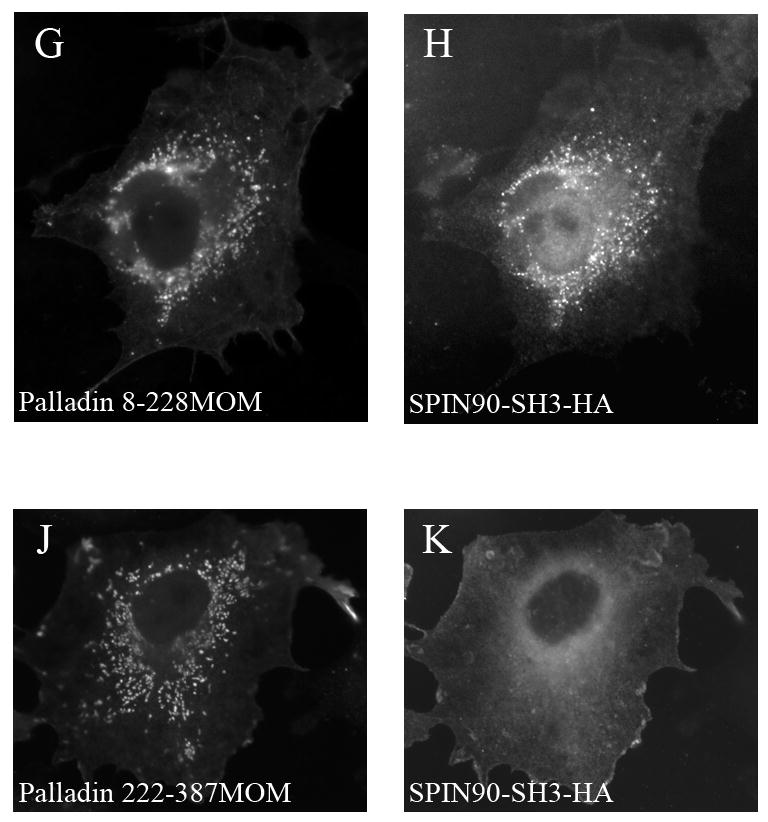
(A–F).COS-7 cells were transfected with GFP-8-772 palladin and HA-tagged full-length (FL) SPIN90 or the SH3 domain of SPIN90 (SPIN90-SH3). SPIN90 was detected with anti-HA antibody. In cotransfected cells, both SPIN90 and SPIN90-SH3 are targeted to palladin-containing actin bundle. (G–K). In a mitochondrial targeting assay, COS-7 cells were cotransfected with palladin 8–228 or 228–387 in a vector containing an outer mitochondrial membrane (MOM) BiPro targeting sequence and with HA-SPIN90-SH3. MOM constructs were detected with a polyclonal palladin antibody and SPIN90-SH3 with HA mAb. When cotransfected with palladin 8-228-MOM construct, SPIN90-SH3 is recruited to the mitochondrial membrane (G,H). Instead, SPIN90-SH3 transfected with the palladin 222–387 MOM is distributed diffusely in the cytoplasm (J,K).
To study, whether the targeting requires direct interaction between the proteins, we used a mitochondrial targeting assay. Palladin 8–228 construct containing the poly-proline sequences was fused with a Mitochondria Outer Membrane (MOM) targeting sequence. This construct was co-transfected with SPIN90-SH3-HA construct to CHO cells. Palladin-8-228-MOM was able to relocalize the SPIN90-SH3 from its normal diffuselocation to the mitochondria (Fig. 3G–H). On the other hand co-expression of palladin 222-387-MOM with SPIN90-SH3 did not result in relocalization of the SPIN90 construct (Fig. 3J–K).
The effect of PDGF treatment and activation of the Src-kinase on palladin and SPIN90 localization
To evaluate, how signaling pathways signaling modulates palladin and SPIN90, we transfected SPIN90 in U251 glioma cells, which express high amounts of endogenous palladin. Under basal conditions co-localization of the two proteins was limited to the lamellipodial extensions of motile cells (not shown). In non-migrating cells palladin localized to the stress fiber dense regions as previously described, while SPIN90 localized rather uniformly through out the cytoplasm (Fig. 4). Both SPIN90 and palladin have been shown to relocate to membrane ruffles after PDGF treatment. In these structures SPIN90 interacts with the Arp2/3 complex and palladin with Eps8 [16, 20]. As expected, stimulation of serum-starved U251 cells with PDGF for 10 minutes resulted in formation of numerous membrane ruffles and wave-like actin rich structures. Both transfected (not shown) and endogenous palladin rapidly re-located to these newly formed structures where it co-localized with SPIN90 and with cortactin (Fig. 4).
Figure 4. Localization of SPIN90 and palladin in PDGF treated U251 cells.
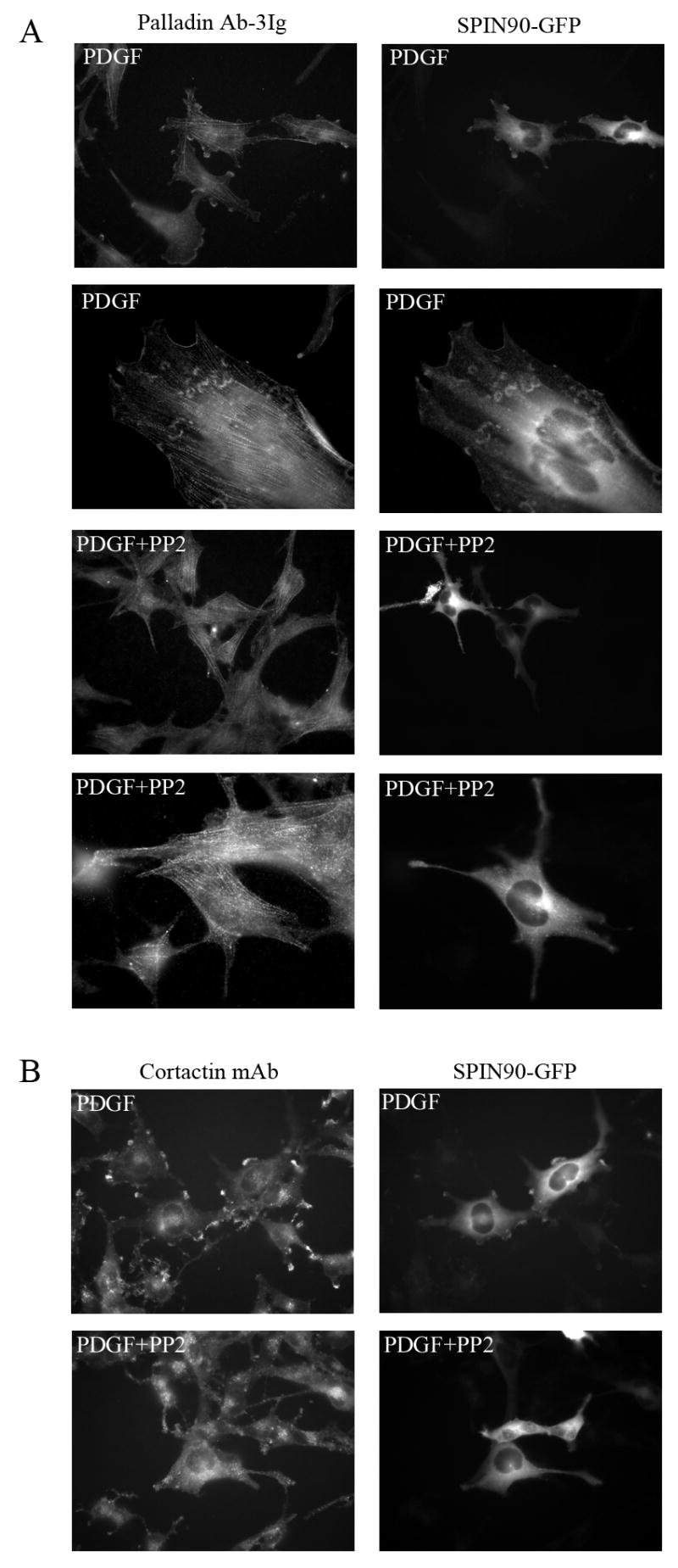
(A) U251 cells transfected with GFP-SPIN90 construct were serum-starved for 16h and stimulated with PDGF for 10 minutes. Cells were fixed and stained for endogenous palladin. Both palladin and SPIN90 are relocated to the PDGF-induced membrane ruffles (top row) and actin containing wave-like structures (second row). Treatment with the Src kinase family inhibitor PP2 prior to PDGF prevents the redistribution and both proteins remain in their normal subcellular positions (third and fourth row). (B) In the same experiment, the co-localization of cortactin, a known Src kinase substrate, and SPIN90 was compared. Cortactin is known to re-locate to the membrane after PDGF treatment and, as expected, cortactin co-localized with SPIN90 in the membrane ruffles in PDGF-treated cells. This re-location was Src-dependent as it was abolished by PP2.
Since the effect of PDGF on actin cytoskeleton has been shown to be, at least partly, mediated by the Src family tyrosine kinases we next studied whether inhibition of kinase activity with Src-selective inhibitor PP2 would have an effect on the re-localization of SPIN90 and palladin in PDGF stimulated cells. As shown in Fig. 4, PP2 completely abolished the formation of the membrane ruffles and thus the re-distribution of SPIN90 and palladin. Similarly, cortactin remained diffusely distributed in the cytoplasm in PP2 treated cells.
To further study the role of Src activity in targeting of SPIN90 and palladin, we expressed either active (CA) or dominant negative (DN) Src constructs in U251 cells. Introduction of active Src in U251 cells resulted in a rapid re-organization of the actin cytoskeleton and drastic changes in cell morphology. Both SPIN90 and palladin re-localized to the Src induced actin rich membrane protrusions. In cells expressing a dominant negative (DN) Src construct both palladin and SPIN90 remained in their normal position (Fig. 5).
Figure 5. SPIN90 and palladin co-localize in cells expressing active Src.
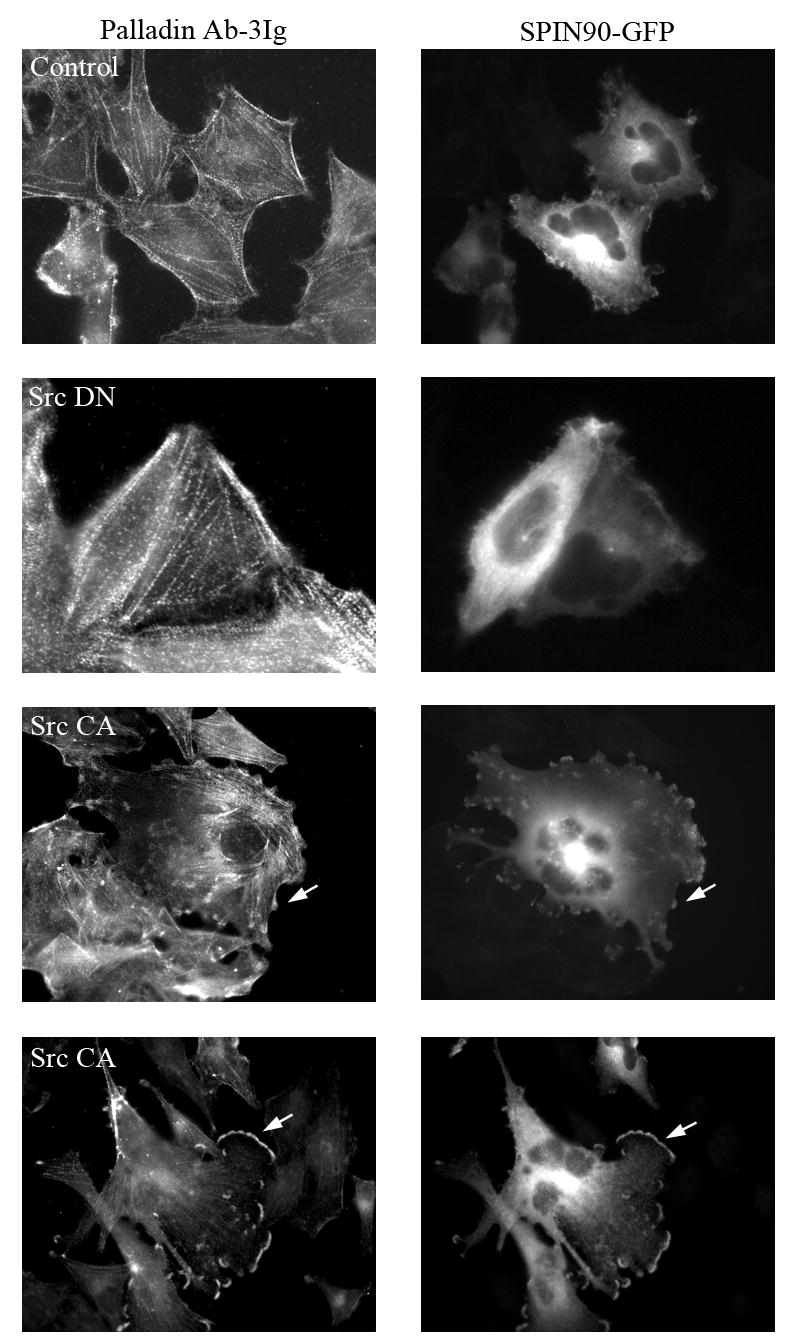
U251 cells were co-transfected with a constitutively active (Src CA) or a dominant negative (Src DN) Src construct together with GFP-SPIN90 and stained for endogenous palladin with palladin Ab-3Ig antibody. Under basal conditions SPIN90 has a predominantly diffuse localization in the cytoplasm whereas palladin localizes to stress fiber dense bodies (Control). In the presence of Src DN, the localization of palladin and SPIN90 is similar to control cells. Expression of Src CA results in formation of numerous membrane projections and ruffles, which contain both palladin and SPIN90 (arrows).
Depletion of palladin leads to cytoskeletal disruption and loss of stress fibers
In order to study the role of palladin in cytoskeletal organization, we designed palladin specific siRNA oligonucleotides and transfected them to U251 cells. As seen in Figure 6, siRNA treatment resulted in a marked down-regulation of palladin as verified by Western Blot analysis and immunofluorescence staining of transfected cells. The remaining weak palladin staining was more diffuse than in control cells, in which palladin was associated with stress fibers. F-actin staining of the treated cells with fluorescent phalloidin demonstrated disruption of stress fibers, disorganization of the actin cytoskeleton and defective cellular spreading. These results are in line with previous studies, in which palladin was down regulated either genetically with an anti-sense cDNA construct[10, 17].
Figure 6. Depletion of palladin results in disruption of actin cytoskeleton and loss of stress fibers.
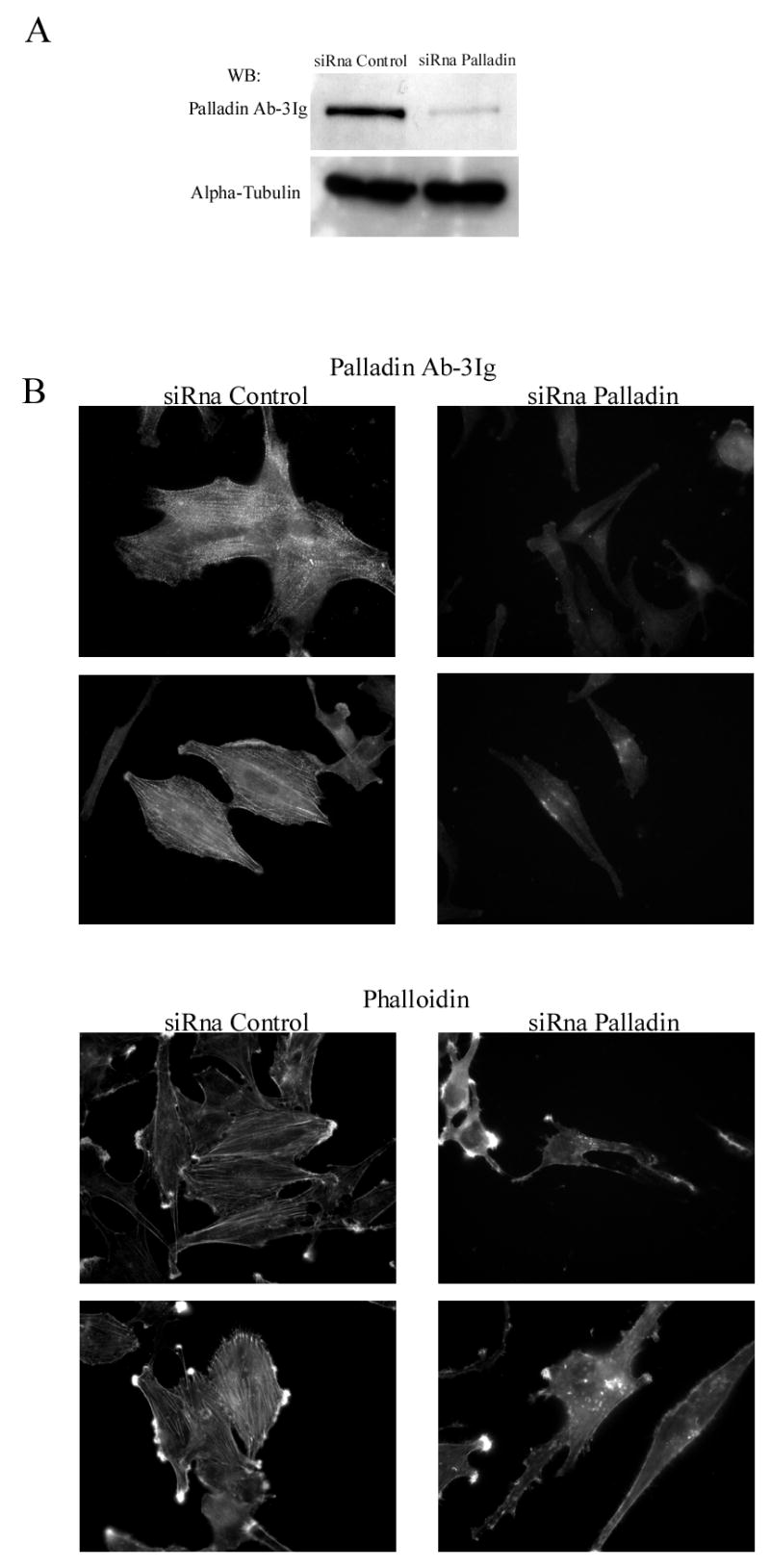
(A). U251 cells were transfected with a palladin specific siRNA oligonucleotide or with a non-targeting control siRNA. The specific siRNA markedly reduced the amount of palladin as compared to control. The alpha-tubulin blot serves as a loading control. (B). Transfected cells were stained with Ab-3Ig palladin antibody and with labeled phalloidin to visualize F-actin. The intensity of palladin staining is markedly reduced and the remaining staining pattern was diffuse compared to the control cells, which showed the typical stress fiber dense region staining. Depletion of palladin resulted in loss of stress fibers and disorganization of the cytoskeleton as visualized by phalloidin staining.
Palladin is required for Src induced actin remodeling
Previous studies have indicated a role for both SPIN90 and palladin in PDGF induced membrane ruffle formation [16, 20]. We wanted to examine whether this is true also for Src-kinase induced actin remodeling. U251 cells were co-transfected with active Src construct and either with SPIN90 specific siRNA construct (siRNA 824-GFP), empty control vector (siVector-GFP) or with a palladin specific siRNA oligonucleotide. Although SPIN90 siRNA efficiently down-regulated SPIN90 expression, we did not detect cytoskeletal differences between cells expressing control siRNA and the specific SPIN90 siRNA together with Src CA. In the presence or absence of SPIN90, Src induced remodeling of actin cytoskeleton and formation of numerous membrane extensions. These newly formed structures contained actin, palladin and also Src (Fig. 7). However, when palladin was knocked-down with a specific siRNA, the effect of Src Ca on the actin cytoskeleton were inhibited. Fewer membrane protrusions were formed in cells co-transfected with palladin siRNA and Src CA than in cells expressing only the Src CA construct. Also, Src kinase, SPIN90 and cortactin were distributed more diffusely in palladin knock-down cells. Taken together, while both palladin and SPIN90 are needed for PDGF induced membrane ruffling, palladin seems to play a more prominent role in Src induced remodeling of actin cytoskeleton.
Figure 7. The role of palladin and SPIN90 in Src induced actin cytoskeleton remodeling.
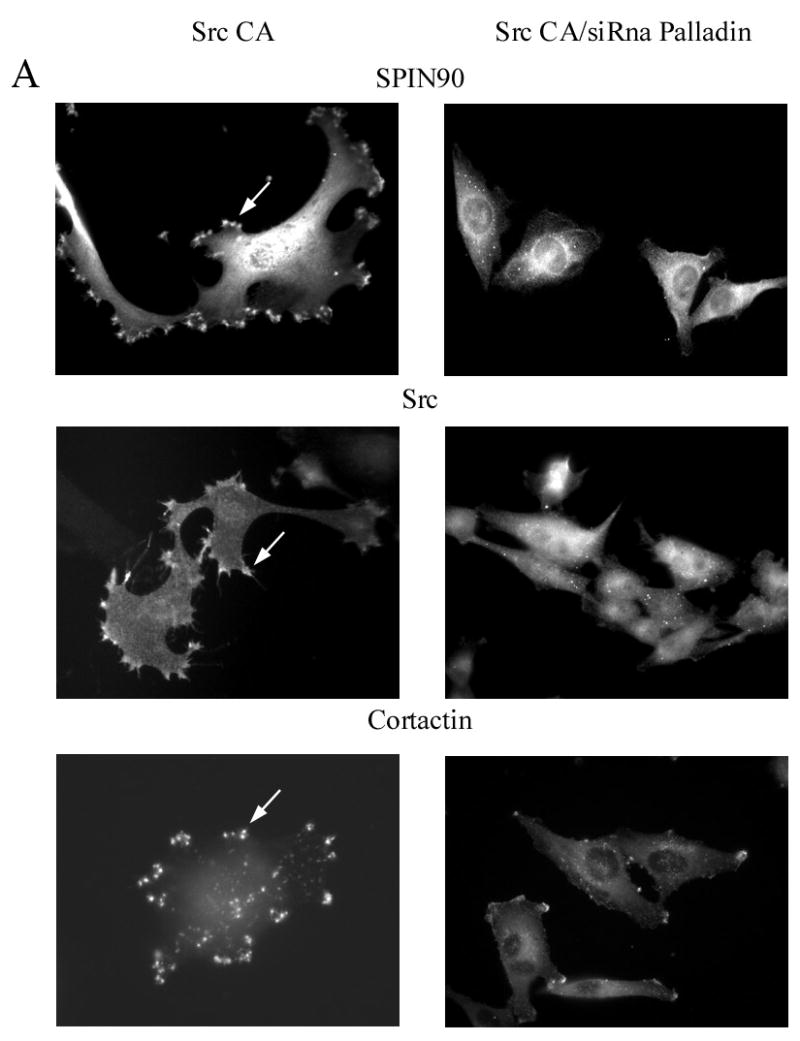
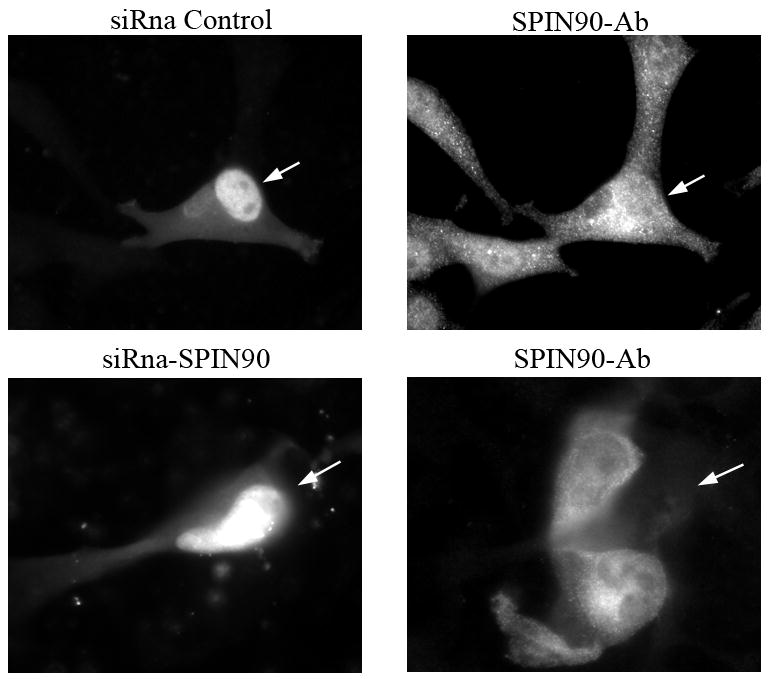
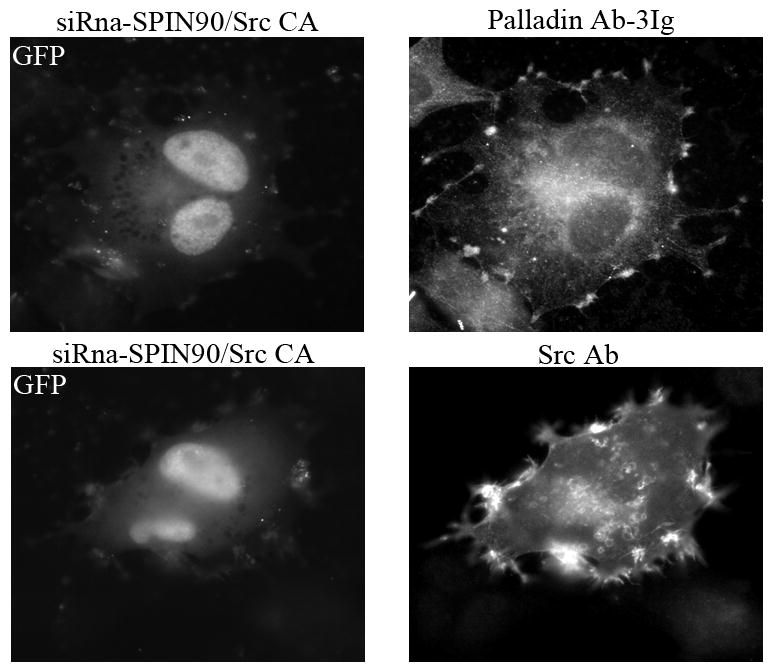
To study the role of palladin and SPIN90 in Src induced actin remodeling, U251 cells were co-transfected with the palladin and SPIN90 siRNA together with constitutively active Src (Src CA). Transfected cells were stained for SPIN90, Src, and cortactin. (A). In cells expressing active Src, all three proteins were located in cell extensions (arrows). However, cells in which palladin was depleted by siRNA exhibited hardly any membrane protrusions and the localization of SPIN90, Src and cortactin was mostly diffuse. (B). U251 cells were transfected with the GFP-conjugated SPIN90 siRNA construct or a control GFP-siRNA. Staining with a SPIN90 antibody shows a clear reduction in staining intensity of the cells transfected with specific siRNA as compared to the non-transfected cells or cells transfected with control GFP-siRNA. Arrows mark transfected cells. (C). Cells co-expressing Src CA and GFP-SPIN90 –siRNA or GFP-control-siRNA demonstrate equal and normal Src-induced actin remodeling. The cells have numerous membrane projections, which contain both palladin and Src.
Discussion
In this work we describe a novel interaction between palladin, SPIN90 and Src. SPIN90 (also known as DIP, mDia interacting protein) contains an SH3 domain, three proline-rich regions, a serine/threonine-rich region and a long C-terminus and shows high sequence similarity to WISH [18]. SPIN90 interacts with several actin-regulating proteins including Nck, N-WASP, βPIX, the Arp2/3 complex, PSD-95 and mDia [19, 20,39,32]. It plays a role in myofibril and sarcomere assembly and depletion of SPIN90 in cardiomyocytes results in sarcomere disruption. Furthermore, it participates in the regulation of filopodia and dendritic spines in neuronal cells [34] and in actin-based motility by promoting the Arp2/3 activity. Finally, SPIN90 can be modulated by ERK1, a kinase activated by cell adhesion or PDGF[21].
Previous reports on palladin and SPIN90 suggest that both proteins act as potent cytoskeletal scaffolds bringing together proteins with different functional modalities into higher-order molecular complexes, whose localization and composition can vary in different cell types and situations. Their role as scaffolds is supported by several observations: (1) the proteins do not have intrinsic enzymatic activity, (2) they contain several domain structures that serve as interaction platforms for actin-associated proteins, (3) the proteins serve as substrates for cytoskeleton modulating kinases, (4) they localize at sites where active actin remodeling takes place, such as lamellipodia and membrane ruffles, and (5) their role in actin remodeling has been demonstrated in knock-down experiments[16, 20]. The present study establishes a molecular link between palladin and SPIN90 but also demonstrates differences in their ability to regulate actin cytoskeleton.
The interaction is mediated by the poly-proline sequences of palladin and the SH3 domain of SPIN90. Palladin differs from the two other protein family members, myotilin and myopalladin, in that its amino-terminus contains proline-rich stretches, which serve as ligands for SH3, EVH and WW domains and also for profilin[26]. One of these proline sequences is conserved between myopalladin and the palladin isoform, 4Ig, and this particular sequence has been shown to mediate direct binding between nebulin and myopalladin[27] and the SH3 domain of LASP-1 and palladin 4Ig[12]. The LASP-1 binding motif is not contained in the major shorter palladin isoform, 3Ig, whose N-terminal poly-prolines bind to the first SH3 domain of ArgBP2 [14], to the EVH domain of VASP [13] and to profilin II[28]. The amino-terminus also binds another SH3 domain protein, Eps8, but this interaction is not mediated by the SH3 domain[16]. While the interactions between SH3 domains and poly-proline sequences are specific, they are usually of low affinity, suggesting that they can be rather short-lived and dynamically regulated by both intra- and extracellular cues. This could also be the case between palladin and SPIN90 because in non-muscle cells the proteins co-localize and thus probably interact in actively remodeled actin rich structures such as lamellipodia.
Our results further demonstrate a bidirectional link between palladin and Src. Src activation results in phosphorylation of palladin in cells and, on the other hand, palladin is involved in subcellular targeting of Src. Src family kinases regulate three main cellular functions that ultimately control the behavior of transformed cells: adhesion, invasion and motility, all of which require active remodeling of the actin cytoskeleton[1, 29]. Src has been previously shown to have several focal adhesion and cytoskeletal substrates, including FAK, vinculin, paxillin, cortactin and ezrin[2, 4]. Activation of Src induces rapid reorganization of the actin cytoskeleton and formation of structures such as lamellipodia and podosomes. Src can be activated by several mechanisms including ligation of receptor tyrosine kinases such as PDGFR and EGFR. When Src is activated by PDGF treatment it rapidly redistributes from central perinuclear location to the plasma membrane. This process requires intact stress fibers and is mediated by the SH3 domain of Src[30, 31,35]. Our results show that knock-down of palladin results in disruption of stress fibers and subsequent inhibition of the relocalization of active Src and SPIN90. Since the formation of membrane ruffles and lamellipodia requires Src activity beneath the plasma membrane, targeting of Src by palladin may be a mechanism to control Src-dependent events.
Similarly to palladin, SPIN90 links directly to Src signaling. Meng et al. showed that SPIN-90 (DIP) is a Src substrate and is involved in targeting effector proteins such as Vav2 and p190RhoGAP [32]. SPIN90 binds to both p190RhoGAP and Vav2 and targets them to the plasma membrane, where all the three components of this complex are phosphorylated by Src. Inhibition of SPIN90 by dominant-negative mutants or siRNA prevented EGF induced actin organization, redistribution of p190RhoGAP and Vav2 and cell movement. SPIN90 inactivates Rho and activates Rac via p190RhoGAP and Vav2, respectively, in a Src dependent manner. Inactivation of Rho leads to loss of stress fibers and activation of Rac to formation of lamellipodia. Our results show that the knock-down of SPIN90 does not affect the targeting of palladin and Src, in line with the results of Meng et al, who showed that SPIN90 (DIP) did not target Src to the plasma membrane [32]. Together these results offer a basis for a model where Src and SPIN90 translocate after stimulation to the membrane via a palladin/stress fiber dependent manner. At the membrane active Src phosphorylates p190RhoGAP and Vav2 via SPIN90 leading to loss of the stress fibers and subsequently formation of lamellipodia and cell movement. Since palladin and SPIN90 have been shown to have several interaction partners localizing to membrane ruffles and lamellipodia it is possible that the interactions can also vary during different stages of lamellipodia organization. In a recent study by Eisenmann et al SPIN90 induced non-apoptotic membrane blebbing, which is known to be associated with amoeboid cell movement and thus cancer-cell migration [25]. This novel finding offers additional support for the assumption that SPIN90 plays an important role in organizing the membrane associated cytoskeleton and cellular movement. Furthermore, fibroblasts derived from palladin negative murine embryos have been to shown to have a greatly diminished motility [17].
Previously, palladin has been shown to be phosphorylated both on tyrosine and serine/threonine residues but kinases involved in this have not been identified[10]. Interestingly, PDGF treatment of A7r5 smooth muscle cells leads to reduced migration of palladin in SDS-PAGE suggesting post-translational modification, possibly phosphorylation of the protein, but this preliminary finding needs still to be experimentally verified[16]. We extend these findings by showing that palladin becomes tyrosine phosphorylated in cells expressing active Src. It remains to be shown, whether Src directly phosphorylates palladin, or whether a kinase downstream of Src would be the actual effector. One of such potential kinases could be FAK.
The role of tyrosine phosphorylation of cytoskeletal proteins is still mostly unclear. One of the best characterized cytoskeletal substrates of Src is cortactin. Cortactin phosphorylation serves a variety of functions, one of which is to provide binding sites for specific signaling proteins with SH2 domains, e.g. Src family kinases and Nck[33]. Tyrosine phosphorylation may also induce other types of novel interactions as shown between ezrin and a kelch-repeat protein family member, KTBDB2[4]. The effects of Src can also be mediated indirectly by other effector proteins, for example by the MAPK family members such as ERK. Src has been shown to directly activate ERK, which is of particular interest since SPIN90 is a substrate of ERK[21]. Phosphorylation of SPIN90 by ERK1 promotes the interaction between SPIN90-βPIX-WASP complex and the adaptor protein Nck[21]. These findings offer some clues for the consequences of palladin phosphorylation, but further experiments are needed to elucidate its specific function.
In conclusion our results show that palladin interacts directly with both SPIN90 and Src and that palladin plays a role in maintaining stress fiber integrity and targeting of SPIN90 and Src to sites where active actin remodeling takes place. Thus palladin can act as a true protein scaffold bringing together effector proteins and their cyroskeletal targets.
Supplementary Material
Supplementary figure 1. A schematic illustration of the palladin’s poly-proline sequences
Above is the amino-terminal (amino-acids 1–228) protein sequence of the major palladin 3Ig isoform (protein accession KIAA0992). Below are the sequences of the three GST-constructs used in the pull-down assays. Proline residues are in capital and the two especially proline-rich sequences located between amino-acids 65–85 and 185–220 are in italic.
Acknowledgments
We thank Helena Ahola for expert technical assistance. This work was supported by grants from the Academy of Finland, Sigrid Juselius Foundation, and Finnish Heart Association, by NIH grants GM61743 and NS43253 to Carol Otey and grants from the National Research Laboratory (The Ministry of Science & Technology) and the Brain Korea 21 Program to Woo Keun Song.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brunton VG, MacPherson IR, Frame MC. Cell adhesion receptors, tyrosine kinases and actin modulators: A complex three-way circuitry. Biochim Biophys Acta. 2004;1692:121–144. doi: 10.1016/j.bbamcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Frame MC. Newest findings on the oldest oncogene; how activated Src does it. J Cell Sci. 2004;117:989–998. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- 3.Brown MT, Cooper JA. Regulation, substrates and functions of Src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 4.Heiska L, Carpen O. Src phosphorylates ezrin at tyrosine 477 and induces a phosphospecific association between ezrin and a kelch-repeat protein family member. J Biol Chem. 2005;280:10244–10252. doi: 10.1074/jbc.M411353200. [DOI] [PubMed] [Google Scholar]
- 5.Frame MC. Src in cancer: Deregulation and consequences for cell behaviour. Biochim Biophys Acta. 2002;1602:114–130. doi: 10.1016/s0304-419x(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 6.Otey CA, Rachlin A, Moza M, Arneman D, Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int Rev Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- 7.Hauser MA, Horrigan SK, Salmikangas P, Torian UM, Viles KD, Dancel R, Tim RW, Taivainen A, Bartoloni L, Gilchrist JM, Stajich JM, Gaskell PC, Gilbert JR, Vance JM, Pericak-Vance MA, Carpen O, Westbrook CA, Speer MC. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum Mol Genet. 2000;9:2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 8.Selcen D, Engel AG. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- 9.Otey CA, Carpen O. Alpha-actinin revisited: A fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 10.Parast MM, Otey CA. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mykkänen OM, Gronholm M, Rönty M, Lalowski M, Salmikangas P, Suila H, Carpen O. Characterization of human palladin, a microfilament-associated protein. Mol Biol Cell. 2001;12:3060–3073. doi: 10.1091/mbc.12.10.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rachlin AS, Otey CA. Identification of palladin isoforms and characterization of an isoform-specific interaction between lasp-1 and palladin. J Cell Sci. 2006;119:995–1004. doi: 10.1242/jcs.02825. [DOI] [PubMed] [Google Scholar]
- 13.Boukhelifa M, Parast MM, Bear JE, Gertler FB, Otey CA. Palladin is a novel binding partner for Ena/VASP family members. Cell Motil Cytoskeleton. 2004;58:17–29. doi: 10.1002/cm.10173. [DOI] [PubMed] [Google Scholar]
- 14.Rönty M, Taivainen A, Moza M, Kruh GD, Ehler E, Carpen O. Involvement of palladin and alpha-actinin in targeting of the Abl/Arg kinase adaptor ArgBP2 to the actin cytoskeleton. Exp Cell Res. 2005;310:88–98. doi: 10.1016/j.yexcr.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Boukhelifa M, Moza M, Johansson T, Rachlin A, Parast M, Huttelmaier S, Roy P, Jockusch BM, Carpen O, Karlsson R, Otey CA. The proline-rich protein palladin is a binding partner for profilin. FEBS J. 2006;273:26–33. doi: 10.1111/j.1742-4658.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 16.Goicoechea S, Arneman D, Disanza A, Garcia-Mata R, Scita G, Otey CA. Palladin binds to Eps8 and enhances the formation of dorsal ruffles and podosomes in vascular smooth muscle cells. J Cell Sci. 2006;119:3316–3324. doi: 10.1242/jcs.03076. [DOI] [PubMed] [Google Scholar]
- 17.Luo H, Liu X, Wang F, Huang Q, Shen S, Wang L, Xu G, Sun X, Kong H, Gu M, Chen S, Chen Z, Wang Z. Disruption of palladin results in neural tube closure defects in mice. Mol Cell Neurosci. 2005;29:507–515. doi: 10.1016/j.mcn.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Lim CS, Park ES, Kim DJ, Song YH, Eom SH, Chun JS, Kim JH, Kim JK, Park D, Song WK. SPIN90 (SH3 protein interacting with nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem. 2001;276:12871–12878. doi: 10.1074/jbc.M009411200. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Kim S, Lee S, Kim SH, Kim Y, Park ZY, Song WK, Chang S. Interaction of SPIN90 with dynamin I and its participation in synaptic vesicle endocytosis. J Neurosci. 2005;25:9515–9523. doi: 10.1523/JNEUROSCI.1643-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim JK, Song WK. Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem. 2006;281:617–625. doi: 10.1074/jbc.M504450200. [DOI] [PubMed] [Google Scholar]
- 21.Lim CS, Kim SH, Jung JG, Kim JK, Song WK. Regulation of SPIN90 phosphorylation and interaction with nck by ERK and cell adhesion. J Biol Chem. 2003;278:52116–52123. doi: 10.1074/jbc.M310974200. [DOI] [PubMed] [Google Scholar]
- 22.Rönty M, Taivainen A, Moza M, Otey CA, Carpen O. Molecular analysis of the interaction between palladin and alpha-actinin. FEBS Lett. 2004;566:30–34. doi: 10.1016/j.febslet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann U, Zuppinger C, Waibler Z, Rudiger M, Urbich C, Martin B, Jockusch BM, Eppenberger H, Starzinski-Powitz A. The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J Cell Sci. 2000;113:4121–4135. doi: 10.1242/jcs.113.22.4121. [DOI] [PubMed] [Google Scholar]
- 24.Rönty M, Leivonen S, Hinz B, Rachlin A, Otey CA, Kahari VM, Carpen O. Isoform-specific regulation of the actin-organizing protein palladin during TGF-beta1-induced myofibroblast differentiation. J Invest Dermatol. 2006;126:2387–2396. doi: 10.1038/sj.jid.5700427. [DOI] [PubMed] [Google Scholar]
- 25.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-Interacting protein modulates Formin-mediated actin assembly at the cell cortex. Current Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Kay BK, Williamson MP, Sudol M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 27.Bang ML, Mudry RE, McElhinny AS, Trombitas K, Geach AJ, Yamasaki R, Sorimachi H, Granzier H, Gregorio CC, Labeit S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol. 2001;153:413–427. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boukhelifa M, Moza M, Johansson T, Rachlin A, Parast M, Huttelmaier S, Roy P, Jockusch BM, Carpen O, Karlsson R, Otey CA. The proline-rich protein palladin is a binding partner for profilin. FEBS J. 2006;273:26–33. doi: 10.1111/j.1742-4658.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 29.Frame MC, Fincham VJ, Carragher NO, Wyke JA. v-src’s hold over actin and cell adhesions. Nat Rev Mol Cell Biol. 2002;3:233–245. doi: 10.1038/nrm779. [DOI] [PubMed] [Google Scholar]
- 30.Fincham VJ, Brunton VG, Frame MC. The SH3 domain directs acto-myosin-dependent targeting of v-src to focal adhesions via phosphatidylinositol 3-kinase. Mol Cell Biol. 2000;20:6518–6536. doi: 10.1128/mcb.20.17.6518-6536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fincham VJ, Unlu M, Brunton VG, Pitts JD, Wyke JA, Frame MC. Translocation of src kinase to the cell periphery is mediated by the actin cytoskeleton under the control of the rho family of small G proteins. J Cell Biol. 1996;135:1551–1564. doi: 10.1083/jcb.135.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng W, Numazaki M, Takeuchi K, Uchibori Y, Ando-Akatsuka Y, Tominaga M, Tominaga T. DIP (mDia interacting protein) is a key molecule regulating Rho and Rac in a Src-dependent manner. EMBO J. 2004;23:760–771. doi: 10.1038/sj.emboj.7600095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S, Lee K, Hwang S, Kim S, Song WK, Park ZY, Chang S. SPIN90/WISH interacts with PSD-95 and regulates dendritic spinogenesis via an N-WASP-independent mechanism. EMBO J. 2006;25:4983–4995. doi: 10.1038/sj.emboj.7601349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timpson P, Jones GE, Frame MC, Brunton VG. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Current Biol. 2001;11:1836–1846. doi: 10.1016/s0960-9822(01)00583-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. A schematic illustration of the palladin’s poly-proline sequences
Above is the amino-terminal (amino-acids 1–228) protein sequence of the major palladin 3Ig isoform (protein accession KIAA0992). Below are the sequences of the three GST-constructs used in the pull-down assays. Proline residues are in capital and the two especially proline-rich sequences located between amino-acids 65–85 and 185–220 are in italic.


