Abstract
Cytoplasmic mislocalization of p27 (cyclin-dependent kinase inhibitor-1B, CDKN1B/KIP1) is caused by activated AKT1, and has been associated with poor prognosis in various cancers. CpG island methylator phenotype (CIMP) in colorectal cancer is characterized by extensive promoter methylation, and is associated with microsatellite instability-high (MSI-H) and BRAF mutations. We have recently shown a positive correlation between MSI/CIMP and loss of nuclear p27. However, no study has examined cytoplasmic p27 mislocalization in relation to CIMP and MSI in colorectal cancer. Using MethyLight assays, we quantified DNA methylation in eight CIMP-specific gene promoters {CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1} in 853 colorectal cancer samples obtained from two large prospective cohorts. We assessed expressions of nuclear and cytoplasmic p27 and nuclear p53 by immunohistochemistry. Cytoplasmic p27 expression was inversely associated with loss of nuclear p27 (p<0.0001), CIMP-high (p<0.0001), MSI-H (p<0.0001) and BRAF mutations (p<0.0001). The inverse association of cytoplasmic p27 with CIMP-high (or MSI-H) was independent of MSI (or CIMP) status. In addition, the inverse association of cytoplasmic p27 with CIMP-high was independent of KRAS/BRAF status. BRAF and CDKN2A (p16) methylation were not correlated with cytoplasmic p27 after stratification by CIMP status. The inverse associations of cytoplasmic p27 with MSI-H and CIMP-high were much more pronounced in p53-negative tumors than p53-positive tumors. In conclusion, cytoplasmic p27 expression is inversely associated with MSI-H and CIMP-high, particularly in p53-negative tumors, suggesting interplay of functional losses of p27 and p53 in the development of various molecular subtypes of colorectal cancer.
Keywords: colon cancer, CIMP, p27, cyclin-dependent kinase inhibitor, MSI
Introduction
Progression through the cell cycle involves sequential activation and inactivation of cyclin-dependent kinases (CDKs) [1]. CDKs are activated through association with positive regulators (cyclins) and inactivated by cyclin-CDK inhibitors. p27 {CDKN1B (cyclin-dependent kinase inhibitor 1B) / KIP1} is one of the cyclin-CDK inhibitors and plays a key role in preventing progression into S phase of the cell cycle [1]. Regulation of p27 levels is achieved post-translationally through ubiquitin-mediated protein degradation [2]. The F-box protein SKP2 has been identified as the substrate recognition component that binds and targets p27 for ubiquitination and subsequent degradation [3]. Low levels of p27 have been associated with tumor progression and poor prognosis in various cancers including colon cancer [4, 5]. Although mutations or deletions of p27 allele rarely occur, down-regulation of p27 in colorectal cancer mainly result from abnormal activation of ubiquitin-mediated proteolysis [4, 5]. CDKN1B (p27) promoter has not been shown to be methylated in colorectal cancer [6]. In addition to loss of nuclear p27 expression, cytoplasmic mislocalization of p27 has been observed in colon cancer [7]. In various cancers, cytoplasmic p27 mislocalization has been associated with activated AKT1 (protein kinase B) [8, 9], overexpression of cyclin D3 [10], and poor prognosis [1, 11]. However, biological implications and differences between p27 loss and p27 mislocalization in colorectal cancer, particularly in relation to other molecular alterations, have not been comprehensively evaluated.
Transcriptional inactivation by cytosine methylation at promoter CpG islands of tumor suppressor genes is thought to be an important mechanism in human carcinogenesis [12]. A number of tumor suppressor genes are silenced by promoter methylation in colorectal cancer [12]. Promoter CpG island methylation has been shown to occur early in colorectal carcinogenesis [13]. A subset of colorectal cancers exhibit promoter methylation in multiple genes, referred to as the CpG island methylator phenotype (CIMP) [12, 14]. CIMP-positive colorectal tumors appear to have a distinct clinical, pathologic and molecular profile, including associations with female sex, proximal tumor location, mucinous and poor differentiations, microsatellite instability (MSI) and BRAF mutations [14–17]. We have recently demonstrated that both MSI and CIMP are correlated positively with loss of nuclear p27 [18], and inversely with down-regulation of p21 (CDKN1B / KIP1) [19], another cyclin-dependent kinase inhibitor. However, no study to date has examined relationship between cytoplasmic p27 expression, MSI and CIMP in colorectal cancer. In this study, using quantitative real-time PCR (MethyLight) assays [16, 20, 21], and relatively unbiased samples of colorectal cancer from two large prospective cohort studies, we show that loss of p27 expression and cytoplasmic p27 localization have opposite molecular features in terms of MSI and CIMP. MethyLight assays can reliably distinguish high from low levels of DNA methylation, the latter of which likely have little or no biological significance [22].
Materials and methods
Study group
We utilized the databases of two large prospective cohort studies; the Nurses’ Health Study (N = 121,700 women followed since 1976) [23], and the Health Professional Follow-up Study (N = 51,500 men followed since 1986) [24]. Informed consent was obtained from all participants prior to inclusion in the cohorts. All cohort participants were free of cancer at the time of study entry. A subset of the cohort participants developed colorectal cancers during prospective follow-up. We included cases only if there was adequate paraffin-embedded tumor tissue and results were available for MSI status and p27 at the time of this study. As a result, a total of 855 colorectal cancer cases (364 from the men’s cohort and 491 from the women’s cohort) were included in this study. Many tumors have been previously characterized for status of CIMP, MSI, KRAS, BRAF and nuclear p27 [18, 19, 25]. However, no tumor has previously been studied for cytoplasmic p27 expression. Tissue collection and analyses were approved by the Institutional Review Boards.
Microsatellite instability (MSI) analysis
Genomic DNA was extracted as previously described [16]. Whole genome amplification (WGA) of genomic DNA was performed by PCR using random 15-mer primers [26]. Methods for MSI analysis were previously described [16]. In addition to D2S123, D5S346, D17S250, BAT25 and BAT26 (the NCI panel), we used BAT40, D18S55, D18S56, D18S67 and D18S487 (i.e., a 10-marker panel). “MSI-high (MSI-H)” was defined as instability in 30% or more of the markers, “MSI-low (MSI-L)” as instability in less than 30% of the markers, and “microsatellite stability (MSS)” as no unstable marker.
Real-time PCR (MethyLight) for quantitative DNA methylation analysis
Sodium bisulfite treatment on genomic DNA was performed as previously described [22]. For DNA methylation analysis, we typically used 1 to 2 tissue sections (10 μm thick) when large tumor sections were available. Real-time PCR to measure DNA methylation (MethyLight) was performed as previously described [20, 21, 27]. We used ABI 7300 (Applied Biosystems, Foster City, CA USA) for quantitative real-time PCR. Using nine sets of primers and probes, we amplified eight CIMP-specific promoters (CACNA1G, CDKN2A (p16/INK4A), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1) [16, 17] and COL2A1. The use of these eight markers in a CIMP-specific panel has been validated by examining 920 colorectal cancers, and all of the eight markers showed good sensitivity and specificity for the prediction of overall CIMP status (manuscript submitted). COL2A1 (the collagen 2A1 gene) was used to normalize for the amount of input bisulfite-converted DNA [21, 22]. Primers and probes were previously described as follows: CACNA1G, CRABP1 and NEUROG1 [16, 17]; CDKN2A and COL2A1 [21]; MLH1 [22]; and IGF2, RUNX3 and SOCS1 [17]. The percentage of methylated reference (PMR, i.e., degree of methylation) at a specific locus was calculated by dividing the GENE:COL2A1 ratio of template amounts in a sample by the GENE:COL2A1 ratio of template amounts in SssI-treated human genomic DNA (presumably fully methylated) and multiplying this value by 100 [28]. A PMR cutoff value of 4 was based on previously validated data [21, 22, 28]. Based on the distribution of PMR values at the CRABP1 and IGF2 loci, we raised PMR cutoff to 6 for CRABP1 and IGF2. Precision and performance characteristics of bisulfite conversion and subsequent MethyLight assays have been previously evaluated and the assays have been validated [22].
The CpG island methylator phenotype-high (CIMP-high) was defined as the presence of ≥6/8 methylated promoters, CIMP-low as the presence of 1/8 to 5/8 methylated promoters CIMP-0 as the absence (0/8) of methylated promoters, based on the fact that CIMP-high and CIMP-low are associated with high BRAF and KRAS mutation rates, respectively [25].
Sequencing of KRAS and BRAF
Methods of KRAS and BRAF Pyrosequencing have been previously described.[25, 26] Pyrosequecing was performed using the PSQ96 HS System (Biotage AB and Biosystems, Uppsala, Sweden) according to the manufacturer’s instructions.
Tissue microarray (TMA) and immunohistochemistry for p27 and p53
Methods for p27 immunohistochemistry were previously described [29]. The extent of nuclear and cytoplasmic p27 expressions were visually estimated using whole tissue sections. Nuclear p27 expression was interpreted as “loss” (no staining, only weakly staining, or <10% of tumor cells positive for moderate/strong staining) (Figure 1A), positive in 10–49% of cells (1+), or positive in ≥ 50% of cells (2+) (Figure 1B). Cytoplasmic p27 expression was interpreted as negative (no staining or <10% of tumor cells staining) or positive (≥ 10% of tumor cells staining). Inflammatory cells and normal colonic epithelial cells served as internal positive controls.
Figure 1. Cytoplasmic or nuclear p27 expression in colorectal cancer.
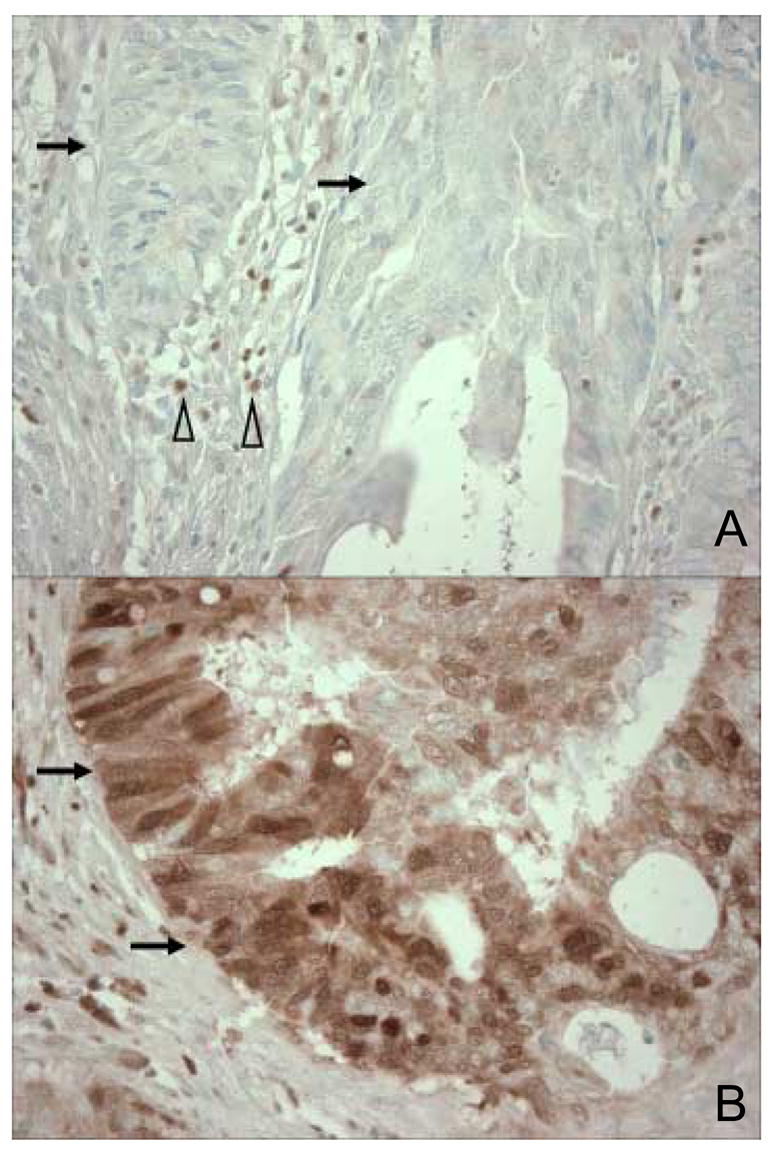
A. Tumor cells with no cytoplasmic or nuclear p27 expression are indicated by arrows. Inflammatory cells serve as internal positive controls (empty arrowheads). B. Tumor cells with cytoplasmic and nuclear p27 expression are indicated by arrows.
Tissue microarrays (TMAs) were constructed for p53 analysis as previously described [30], using the Automated Arrayer (Beecher Instruments, Sun Prairie, WI USA). For p53 immunohistochemistry, deparaffinized tissue sections in a citrate buffer (BioGenex, San Ramon, CA) were microwaved in a pressure cooker at high power for 15 min. Tissue sections were incubated with 3% H2O2 (10 min) to block endogenous peroxidase, and then incubated with protein block (Vector Laboratories, Burlingame, CA) (10 min). Primary anti-p53 mouse monoclonal antibody (clone DO-1, Calbiochem, San Diego, CA) (dilution 1:50) was applied for 30 min at room temperature. Then, biotinylated secondary Multi-Link antibody (Biogenex) was applied (20 min), horse radish peroxidase avidin complex (Biogenex) was added (20 min) and sections were visualized by DAB (5 min) and methyl-green counterstain. We visually estimated the fraction of tumor cells with unequivocal strong nuclear staining for p53, by examining at least two tissue cores in TMAs, or the whole tissue section in each case for which there was not enough tissue for TMAs or results were equivocal in TMAs. p53 positivity was defined as 50% or more of tumor cells with unequivocal strong nuclear staining.
Appropriate positive and negative controls were included in each run of p27 and p53 immunohistochemistry. All immunohistochemically-stained slides were interpreted by a pathologist (S.O.) blinded from any other laboratory data.
Statistical analysis
In statistical analysis, chi-square test (or Fisher’s exact test when any category was less than 10) was performed for categorical data, using the SAS program (version 9.1, SAS Institute, Cary, NC). All p values were two-sided, and statistical significance was set at p ≤ 0.05.
RESULTS
Cytoplasmic p27 Localization Is Inversely Correlated with Loss of Nuclear p27
We examined nuclear and cytoplasmic p27 expressions in 855 colorectal cancer specimens by immunohistochemistry; 408 (48%) and 447 (52%) were positive and negative, respectively, for cytoplasmic p27 expression. Among the 855 tumors, 160 (19%) were extensively positive (2+) for nuclear p27, 427 (50%) were focally positive (1+) for nuclear p27, and the remaining 268 (31%) showed loss of nuclear p27. Figure 2 illustrates relationship between cytoplasmic and nuclear p27 expressions. Cytoplasmic p27 positivity was significantly more frequent in tumors with nuclear p27 1+ or 2+ expression (63–64%, p < 0.0001) than in tumors with nuclear p27 loss (12%), raising the possibility that cytoplasmic sequestration of p27 and loss of nuclear p27 expression tend to be mutually exclusive events in colorectal cancer. We used inflammatory cells and normal colonic epithelial cells as internal positive controls. Therefore, it is unlikely that this positive correlation was due to unstainable tumors with no or low overall antigenicity.
Figure 2. Relationship between nuclear and cytoplasmic expressions of p27 in 855 colorectal cancers.
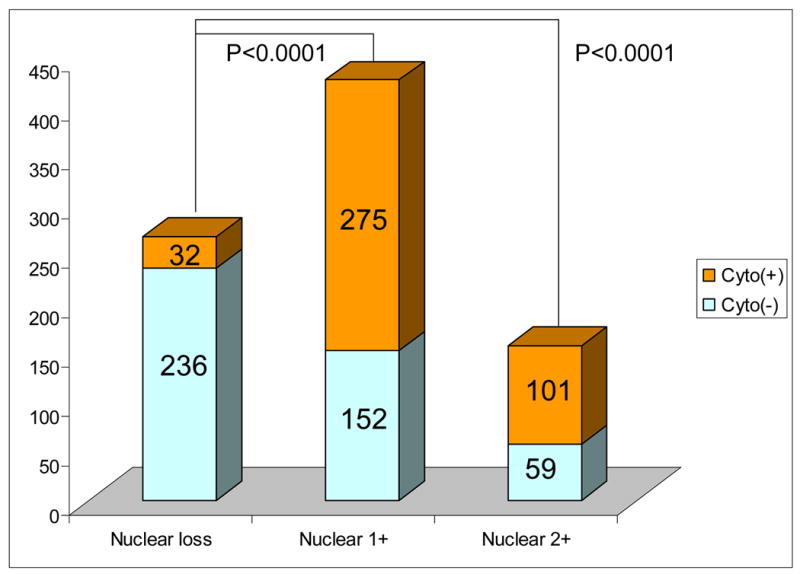
The Y axis indicates the number of tumors. P values indicate statistical significance levels when comparing the fractions of tumors expressing cytoplasmic p27. Cyto(+), cytoplasmic p27 positive; Cyto(−), cytoplasmic p27 negative.
Cytoplasmic p27 expression is inversely associated with microsatellite instability (MSI) and CpG island methylator phenotype (CIMP)
We quantified DNA methylation in a panel of the eight CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1) [16, 17] by MethyLight to determine CIMP status in tumors. The selection and use of these eight markers for the determination of CIMP status have been validated [16, 17]. Among 833 tumors with DNA methylation data, 132 tumors (16%) were classified as CIMP-high (≥ 6/8 methylated promoters).
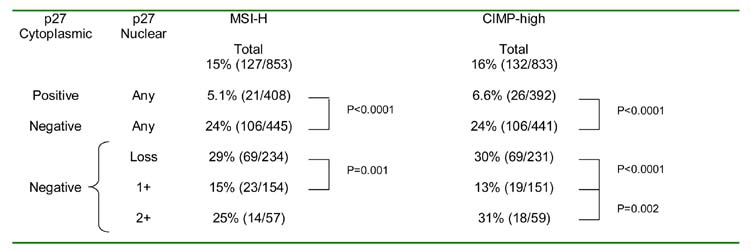
Tumors with cytoplasmic p27 expression showed significantly lower frequencies of MSI-H (5.1%, p < 0.0001) and CIMP-high (6.6%, p < 0.0001) than tumors negative for cytoplasmic p27 (MSI-H 24%, and CIMP-high 24%) (Table 1). Among cytoplasmic p27-negative tumors, nuclear p27-negativity was associated with higher frequencies of MSI-H (29%, p = 0.001) and CIMP-high (30%, p < 0.0001) compared to nuclear p27 1+ positivity (MSI-H 15%, and CIMP-high 13%) (Table 1). Interestingly, among cytoplasmic p27-negative tumors, nuclear p27 2+ positive tumors also showed higher frequencies of MSI-H (25%, p = 0.10) and CIMP-high (31%, p = 0.002) than nuclear p27 1+ tumors (MSI-H 15%, and CIMP-high 13%). In contrast, among cytoplasmic p27-positive tumors, there were no significant differences in the MSI-H or CIMP-high frequencies between nuclear p27-negative, 1+ positive and 2+ positive tumors (data not shown).
Table 1.
Frequencies of MSI-H and CIMP-high in Tumors with Various Cytoplasmic and Nuclear p27 Expressions
Abbreviations: CIMP, CpG island methylator phenotype; MSI, microsatellite instability.
In order to determine whether the inverse association of cytoplasmic p27 positivity with CIMP-high was due to a correlation between p27 and methylation of CDKN2A (p16, another important cell cycle regulator), we examined the frequencies of cytoplasmic p27 positivity among CIMP-high and CIMP-low/0 tumors stratified by CDKN2A methylation status (Supplemental Material 1, after Figure 5). For this analysis, CDKN2A was excluded from the CIMP panel to avoid its confounding effect on diagnosing CIMP status, and CIMP-high was defined as the presence of ≥5/7 methylated promoters. After stratification by CIMP status, CDKN2A methylation was not correlated with cytoplasmic p27 expression. In contrast, the inverse association between cytoplasmic p27 and CIMP-high was still present even after tumors were stratified by CDKN2A methylation status.
Figure 5. Frequency of MSI-H and CIMP-high in colorectal cancer with various combined p53/p27 status.
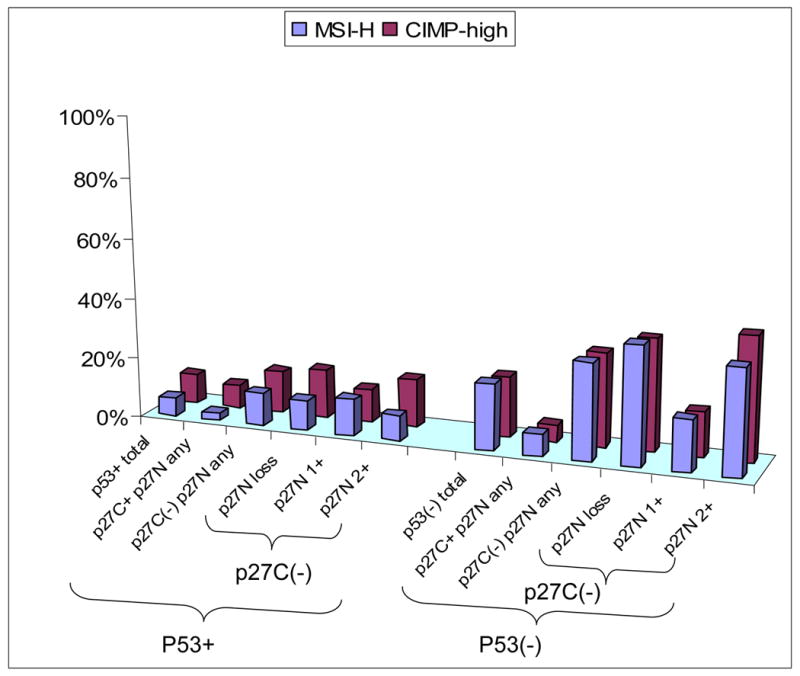
The inverse association between cytoplasmic p27 and MSI-H (or CIMP-high) is much more pronounced in p53 negative tumors than in p53 positive tumors. For raw data, see Supplemental material 2. CIMP, CpG island methylator phenotype; MSI-H, microsatellite instability-high; p27C, p27 cytoplasmic; p27N, p27 nuclear.
Cytoplasmic/nuclear p27 expression, and mutations of KRAS and BRAF
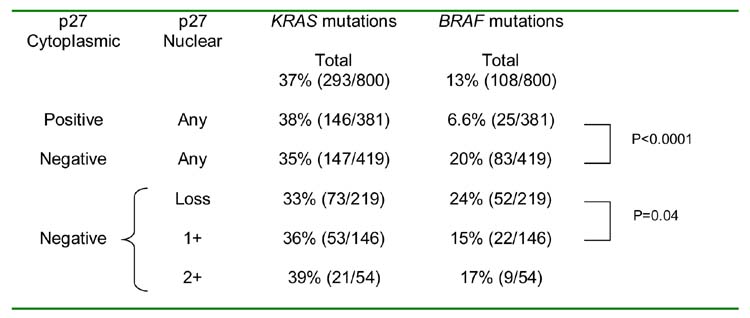
We examined relations between cytoplasmic/nuclear p27 expression and mutations of KRAS and BRAF. Tumors with cytoplasmic p27 expression showed a significantly lower frequency of BRAF mutations (6.6%, p < 0.0001) than tumors negative for cytoplasmic p27 (20%) (Table 2). Among cytoplasmic p27-negative tumors, nuclear p27-negativity was associated with a higher frequency of BRAF mutations (24%, p = 0.04) compared to nuclear p27 1+ positivity (BRAF mutations 15%). Cytoplasmic/nuclear p27 expression was not correlated with KRAS mutations.
Table 2.
Frequencies of KRAS and BRAF Mutations in Tumors with Various Cytoplasmic and Nuclear p27 Expressions
Cytoplasmic p27 expression in various CIMP/KRAS/BRAF subtypes of colorectal cancer
In order to examine an association between cytoplasmic p27 and KRAS/BRAF mutations independent of CIMP status, we examined the frequencies of cytoplasmic p27 expression in six subtypes of colorectal cancer according to combined status of CIMP/KRAS/BRAF (Figure 3). There was no significant difference in the frequencies of cytoplasmic p27 expression between BRAF-mutated (KRAS wild-type) tumors and BRAF/KRAS wild-type tumors after tumors were stratified by CIMP status. Thus, BRAF mutations did not seem to be directly associated with cytoplasmic p27 expression.
Figure 3. Frequency of cytoplasmic p27+ in various CIMP/KRAS/BRAF subtypes of colorectal cancer.
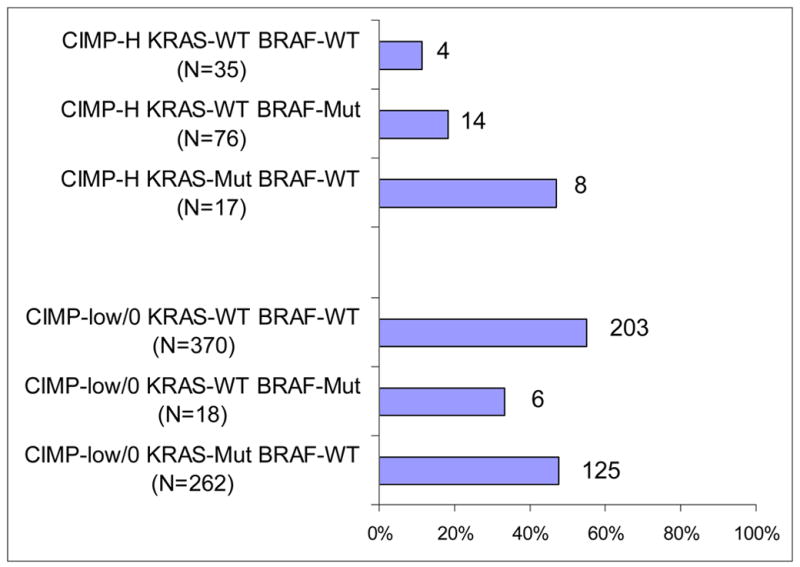
BRAF or KRAS mutations do not appear to be consistently correlated with cytoplasmic p27 expression after stratification by CIMP status. CIMP, CpG island methylator phenotype; Mut, mutated; WT, wild-type.
Cytoplasmic p27 Expression in various MSI/CIMP subtypes of colorectal cancer
In order to examine an association between cytoplasmic p27 expression and CIMP-high (or MSI-H) independent of MSI (or CIMP) status, we examined the frequencies of cytoplasmic p27 expression in four subtypes of colorectal cancer stratified by both MSI and CIMP status (Figure 4). Overall, both MSI-H and CIMP-high appeared to be independently correlated (inversely) with cytoplasmic p27 expression. Among MSI-H tumors, MSI-H CIMP-high tumors showed a significantly lower frequency of cytoplasmic p27 expression (9.1%) than MSI-H CIMP-low/0 tumors (32%, p = 0.002). Among MSI-L/MSS tumors, MSI-L/MSS CIMP-high tumors showed a lower frequency of cytoplasmic p27 expression (39%) than MSI-L/MSS CIMP-low/0 tumors (52%, p = 0.09) though statistical significance was not reached. Thus, cytoplasmic p27 expression appeared to be inversely associated with CIMP-high independent of MSI status.
Figure 4. Frequency of cytoplasmic p27+ in various MSI/CIMP subtypes of colorectal cancer.
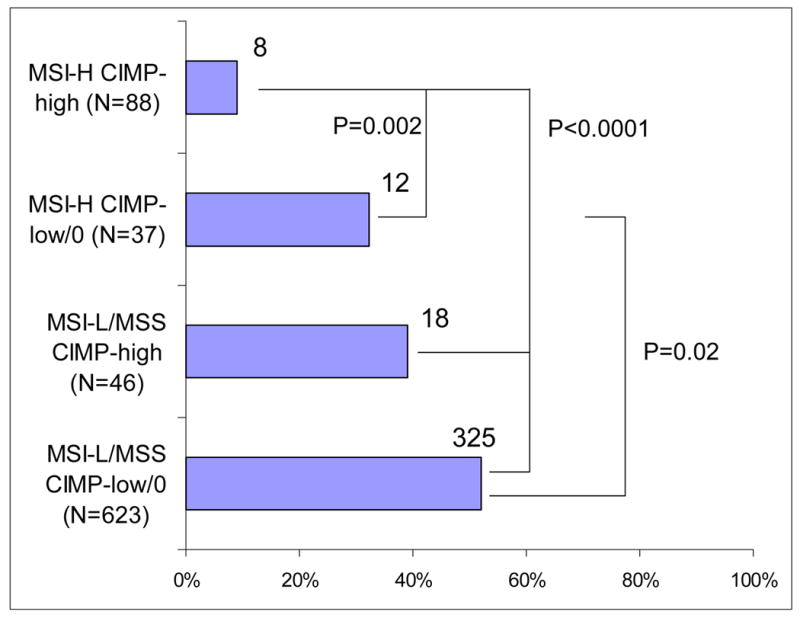
MSI-H and CIMP-high are synergistically inversely associated with cytoplasmic p27 expression. CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable.
We also stratified tumors by CIMP status. Among CIMP-high tumors, MSI-H CIMP-high tumors showed a significantly lower frequency of cytoplasmic p27 expression (9.1%) than MSI-L/MSS CIMP-high tumors (39%, p < 0.0001). Among CIMP-low/0 tumors, MSI-H CIMP-low/0 tumors showed a lower frequency of cytoplasmic p27 expression (32%) than MSI-L/MSS CIMP-low/0 tumors (52%, p = 0.02). Therefore, cytoplasmic p27 expression was inversely associated with MSI-H independent of CIMP status.
Combined p27/p53 status, CIMP and MSI
Because of important roles of both p27 and p53 in regulating the cell cycle, we correlated combined status of p27 and p53 with CIMP and MSI. The inverse associations of cytoplasmic p27 expression with CIMP-high and MSI-H were much more pronounced among p53-negative tumors than p53-positive tumors (Figure 5, raw data in Supplemental material 2). For instance, p53-negative/cytoplasmic p27-negative tumors showed much higher frequencies of MSI-H (32%) and CIMP-high (31%) than p53-negative/cytoplasmic p27-positive tumors (MSI-H 7.2% and CIMP-high 5.6%) (Figure 5). These results suggest that the functional status of p27 is much more relevant in tumors with wild-type p53.
DISCUSSION
We conducted this study to examine significance of cytoplasmic localization of p27. We evaluated relationship between nuclear and cytoplasmic expression of p27 and other molecular features of colorectal cancer, including CpG island methylator phenotype (CIMP), microsatellite instability (MSI) and mutations in the KRAS and BRAF oncogenes. In the current study, the use of quantitative DNA methylation assays (MethyLight) as well as population-based samples of colorectal cancer from two large prospective cohorts has enabled us to precisely estimate the frequency of colorectal cancers with specific molecular features (such as CIMP-high, MSI-H, cytoplasmic p27 positive, etc.) at a population level. We have demonstrated that cytoplasmic p27 expression is inversely correlated with CIMP-high and MSI-high (MSI-H) in colorectal cancer. In addition, we have shown that cytoplasmic p27 is correlated with nuclear p27 expression (retention), raising the possibility that cytoplasmic sequestration of p27 and loss of nuclear p27 tend to be mutually exclusive events in colorectal carcinogenesis. Taken together with our previous data of positive correlations for nuclear p27 loss with MSI-H and CIMP-high in colorectal cancer [18], our current data imply that MSI-H CIMP-high tumors preferentially lose nuclear p27 whereas non-MSI-H non-CIMP-high tumors preferentially sequester p27 in cytoplasm.
Because CIMP-high and MSI-H (as well as CIMP-high and BRAF mutations) are tightly associated with each other [15–17], we stratified tumors according to CIMP and MSI status, and according to CIMP and KRAS/BRAF gene status. The inverse association of cytoplasmic p27 expression with CIMP-high (or MSI-H) appeared to be independent of MSI (or CIMP) status. In addition, the inverse association of cytoplasmic p27 expression with CIMP-high also appeared to be independent of BRAF status. However, the inverse correlation of cytoplasmic p27 expression with BRAF mutations failed to persist when tumors were stratified by both CIMP and KRAS/BRAF status. Thus, we conclude that cytoplasmic p27 expression is inversely associated with CIMP-high and MSI-H, but not directly with BRAF mutations.
Because of important roles of both p27 and p53 in regulating the cell cycle, we examined whether p53 status in colorectal cancer might modify molecular characteristics of tumors with various cytoplasmic/nuclear p27 status. Interestingly, the inverse associations of cytoplasmic p27 expression with CIMP-high and MSI-H were much more pronounced among p53-negative (mostly wild-type) tumors than p53-positive (mostly mutated) tumors. Our results imply that functional status of p27 may be much more important in p53 wild-type tumors than p53-mutated tumors. Although p53 immunohistochemistry has been shown to have both false positives and false negatives for the assessment of TP53 gene mutations, when higher threshold of p53 positivity is used (as in the current study), p53 immunohistochemistry can generally predict the presence or absence of mutations of TP53 [31].
A cause and biological significance of cytoplasmic mislocalization of p27 in cancer have been investigated. Liang et al. [8] have demonstrated that activated AKT1 (protein kinase B) phosphorylates p27, impairing nuclear import of p27 in breast cancer cells, and that cytoplasmic mislocalization of p27 is associated with poor prognosis in breast cancer. An association between cytoplasmic p27 mislocalization and phosphorylation of AKT1 has also been observed in acute myelogenous leukemia [9] and thyroid carcinoma [32]. Cytoplasmic mislocalization of p27 has been shown to be associated with poor prognosis in various cancers [1, 11]. Cytoplasmic mislocalization of p27 has also been reported in colon cancer [7]; however, its biological significance in colon cancer has not been studied.
The prognostic significance of nuclear p27 loss in colorectal cancer has been examined in previous studies, which have shown that nuclear p27 loss was a significant predictor of worse survival by multivariate analysis [4, 33, 34]. Manne et al. [35] examined p27 expression in colorectal cancer stratified by stage and found that nuclear p27 loss was associated with local recurrence and poor survival only for stage III colorectal cancer. The authors also found that p27 loss was associated with poor differentiation (in stage II tumors) and distal location [35]. In contrast, Palmqvist et al. [33] showed that p27 loss was associated with proximal location. These discrepant results may be attributable to differences in patient populations and criteria of p27 interpretation. We have previously examined nuclear p27 expression in relation to response to combination chemotherapy against advanced colorectal cancer, and shown that p27 positivity might confer a better response rate although statistical significance was not reached [29].
In conclusion, cytoplasmic localization of p27 in colorectal cancer is inversely associated with nuclear p27 loss, CIMP-high and MSI-H. Our results imply that there are mutually exclusive pathways of functional inactivation of p27, namely loss of nuclear p27 and cytoplasmic sequestration. Exact biological significance of cytoplasmic p27 mislocalization and influence of CIMP and MSI status on biological effect of p27 need to be further elucidated.
Supplementary Material
Frequency of Cytoplasmic p27+ in Colorectal Cancer with Various CIMP and CDNK2A (p16) Methylation Status. CDKN2A methylation is not correlated with cytoplasmic p27 expression after tumors are stratified by CIMP status. CIMP, CpG island methylator phenotype; M, methylated; UM, unmethylated.
Raw Data for Figure 5 (Frequencies of MSI-H and CIMP-high in Tumors with Various Combined p53/p27 Status).
Acknowledgments
This work was supported by National Institute of Health (NIH) grants P01 CA87969-03 and P01 CA55075-13. We deeply thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires. We thank Graham Colditz, Walter Willett, and many other staff members who implemented and have maintained the cohort studies. We thank Peter Laird, Daniel Weisenberger, and Mihaela Campan for assisting in the development of the MethyLight assay. No conflict of interest is present.
This work was supported by National Institute of Health (NIH) grants P01 CA87969-03 and P01 CA55075-13.
Abbreviations and HUGO Gene Nomenclature Committee (HGNC)-approved official gene symbols used
- CACNA1G
calcium channel, voltage-dependent, T type alpha-1G subunit
- CDKN1B
cyclin-dependent kinase inhibitor 1B (p27/KIP1)
- CDKN2A
cyclin-dependent kinase inhibitor 2A (p16/INK4A)
- CIMP
CpG island methylator phenotype
- CRABP1
cellular retinoic acid binding protein 1
- IGF2
insulin-like growth factor 2
- MSI
microsatellite instability
- MSI-H
microsatellite instability-high
- MSI-L
microsatellite instability-low
- MSS
microsatellite stable
- NEUROG1
neurogenin 1
- PMR
percentage of methylated reference (degree of DNA methylation)
- RUNX3
runt-related transcription factor 3
- SOCS1
suppressor of cytokine signaling 1
Footnotes
No conflict of interest is present.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Macaluso M, Montanari M, Cinti C, et al. Modulation of cell cycle components by epigenetic and genetic events. Semin Oncol. 2005;32:452–7. doi: 10.1053/j.seminoncol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Pagano M, Tam SW, Theodoras AM, et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–5. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 3.Carrano AC, Eytan E, Hershko A, et al. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 4.Loda M, Cukor B, Tam SW, et al. Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat Med. 1997;3:231–4. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- 5.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 6.Xu XL, Yu J, Zhang HY, et al. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–54. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sgambato A, Ratto C, Faraglia B, et al. Reduced expression and altered subcellular localization of the cyclin-dependent kinase inhibitor p27(Kip1) in human colon cancer. Mol Carcinog. 1999;26:172–9. doi: 10.1002/(sici)1098-2744(199911)26:3<172::aid-mc6>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Liang J, Zubovitz J, Petrocelli T, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med. 2002;8:1153–60. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- 9.Min YH, Cheong JW, Kim JY, et al. Cytoplasmic mislocalization of p27Kip1 protein is associated with constitutive phosphorylation of Akt or protein kinase B and poor prognosis in acute myelogenous leukemia. Cancer Res. 2004;64:5225–31. doi: 10.1158/0008-5472.CAN-04-0174. [DOI] [PubMed] [Google Scholar]
- 10.Baldassarre G, Belletti B, Bruni P, et al. Overexpressed cyclin D3 contributes to retaining the growth inhibitor p27 in the cytoplasm of thyroid tumor cells. J Clin Invest. 1999;104:865–74. doi: 10.1172/JCI6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SP, Lipman J, Goldman H, et al. Loss or altered subcellular localization of p27 in Barrett’s associated adenocarcinoma. Cancer Res. 1998;58:1730–5. [PubMed] [Google Scholar]
- 12.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 13.Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- 14.Toyota M, Ohe-Toyota M, Ahuja N, et al. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–5. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samowitz W, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 18.Ogino S, kawasaki T, Kirkner GJ, et al. Loss of nuclear p27 (CDKN1B / KIP1) in colorectal cancer is correlated with microsatellite instability and CpG island methylator phenotype (CIMP) Mod Pathol. doi: 10.1038/modpathol.3800709. In press. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, kawasaki T, Kirkner GJ, et al. Down-regulation of p21 (CDKN1A / CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol. 2006;210:147–54. doi: 10.1002/path.2030. [DOI] [PubMed] [Google Scholar]
- 20.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widschwendter M, Siegmund KD, Muller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–13. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 22.Ogino S, kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–17. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–96. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 24.Wei EK, Giovannucci E, Fuchs CS, et al. Low Plasma Adiponectin Levels and Risk of Colorectal Cancer in Men: A Prospective Study. J Natl Cancer Inst. 2005;97:1688–94. doi: 10.1093/jnci/dji376. [DOI] [PubMed] [Google Scholar]
- 25.Ogino S, kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006 doi: 10.2353/jmoldx.2006.060082. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eads CA, Danenberg KD, Kawakami K, et al. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–6. [PubMed] [Google Scholar]
- 28.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–8. [PubMed] [Google Scholar]
- 29.Ogino S, Meyerhardt JA, Cantor M, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 30.Ogino S, Brahmandam M, kawasaki T, et al. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–64. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall PA, McCluggage WG. Assessing p53 in clinical contexts: unlearned lessons and new perspectives. J Pathol. 2006;208:1–6. doi: 10.1002/path.1913. [DOI] [PubMed] [Google Scholar]
- 32.Motti ML, Califano D, Troncone G, et al. Complex regulation of the cyclin-dependent kinase inhibitor p27kip1 in thyroid cancer cells by the PI3K/AKT pathway: regulation of p27kip1 expression and localization. Am J Pathol. 2005;166:737–49. doi: 10.1016/S0002-9440(10)62295-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmqvist R, Stenling R, Oberg A, et al. Prognostic significance of p27(Kip1) expression in colorectal cancer: a clinico-pathological characterization. J Pathol. 1999;188:18–23. doi: 10.1002/(SICI)1096-9896(199905)188:1<18::AID-PATH311>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Noguchi T, Kikuchi R, Ono K, et al. Prognostic significance of p27/kip1 and apoptosis in patients with colorectal carcinoma. Oncol Rep. 2003;10:827–31. [PubMed] [Google Scholar]
- 35.Manne U, Jhala NC, Jones J, et al. Prognostic significance of p27(kip-1) expression in colorectal adenocarcinomas is associated with tumor stage. Clin Cancer Res. 2004;10:1743–52. doi: 10.1158/1078-0432.ccr-03-0037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency of Cytoplasmic p27+ in Colorectal Cancer with Various CIMP and CDNK2A (p16) Methylation Status. CDKN2A methylation is not correlated with cytoplasmic p27 expression after tumors are stratified by CIMP status. CIMP, CpG island methylator phenotype; M, methylated; UM, unmethylated.
Raw Data for Figure 5 (Frequencies of MSI-H and CIMP-high in Tumors with Various Combined p53/p27 Status).