Abstract
Galβl, 4GlcNAc a2, 6-sialyltransferase (ST6GalI) mediates the glycosylation of proteins and lipids to form functionally important glycoproteins and glycolipids in the Golgi compartment. Our previous work demonstrated that chronic ethanol feeding in rats caused a marked 59% decrease of ST6GalI activity as well as ST6GalI mRNA level in the liver that were due to decreased stability of the mRNA. Clinical observations show that down-regulation of ST6GalI gene and consequent impaired activity of ST6GalI seems to be the major cause for the appearance of asialoconjugates in the blood of chronic alcoholics. The plasma carbohydrate-deficient transferrin (CDT) and sialic acid index of plasma apolipoprotein J (SIJ) were also altered in the alcoholic group compared to the non drinkers. We have now investigated how alcohol affects the gene regulation of ST6GalI and possible mechanism in post-mortem human liver specimens taken from non drinkers group, moderate-alcohol drinkers and heavy–alcohol drinkers. Real-time PCR analyses of the liver RNA extract showed that ST6GalI mRNA level was progressively decreased by 49% in moderate drinkers (p<0.01) and by 69% in heavy drinkers (p<0.01) compared to those in non drinkers group. Western blot analysis showed that liver ST6GalI protein level was negligibly decreased in moderate drinkers but decreased by 30% (p<0.05) in heavy alcohol drinkers compared to non-drinkers. We further demonstrated a single ST6GalI mRNA binding protein complex in the normal human liver extract, which progressively decreased in the liver extracts of moderate and heavy alcohol drinkers. Thus, it is concluded that the appearance of asialoconjugates in alcoholics is possibly due to the down-regulation of ST6GalI gene expression.
Keywords: Alcoholism, Asialoconjugates, Gene regulation, Real-time PCR, Western Blot
INTRODUCTION
Sialyltransferases catalyze the terminal step in the biosyntheses of various glycoconjugates by mediating the attachment of sialic acids from cytidine-5′-monophosphate- N-acetylneuraminic acid to the end of the carbohydrate chains of glycoproteins and glycolipids. Carbohydrate chains of glycoproteins and glycolipids on cell surface play important roles in biological events, such as cell-cell adhesion and communication (1–3), viral-host recognition (4) and tumor invasiveness (5). The structures of these carbohydrate chains have been clearly established to have sialic acids as the terminal carbohydrate residue in all these glycoconjugates. Therefore, the status of sialyltransferases is critically important in both physiological and pathological conditions. Sialyltransferases may link the sialic acid residues to the glycoconjugate in a number of ways: either through an a2, 3- or an a2, 6-linkage to galactose; or through an a2, 6-bond to N-acetylgalactosamine; or through an a2, 8-bond to another sialic acid to form sialic acid chains. Different sialyl linkages are elaborated by different members in the sialyltransferase family (6, 7).
Galβl, 4GlcNAc a2, 6-sialyltransferase (ST6GalI) mediates the addition of a2, 6-linked sialic acid to glycoproteins in the Golgi compartment. Even though a second a2, 6-sialyltransferase (ST6GalII) has been identified recently (8, 9), its expression was mainly detected in small intestine, colon and fetal brain (8). Moreover, since ST6GalII is an oligosaccharide-specific enzyme that exhibits relatively low and no activities toward some glycoproteins and glycolipids respectively (8–11), ST6GalI remains as the major a2, 6-sialyltransferase responsible for the broad synthesis of glycoproteins and glycolipids.
Clinical observations also show increased appearance of asialoconjugates in the blood of chronic alcoholics (12–14). Since apolipoprotein J (ApoJ) has seven times more sialic acid per mole of ApoJ, we had hypothesized that plasma sialic acid index of plasma ApoJ (moles of sialic acid/mole of ApoJ; SIJ) would be a viable excellent marker for chronic alcohol consumption, Accordingly, we have demonstrated that plasma SIJ is significantly decreased by 50% (p<0.001) in chronic human alcoholics of both genders compared to non-drinkers (14). In our rat alcohol feeding model, ST6GalI mRNA expression is reduced by as much as 59% by chronic alcohol treatment compared to the pair-fed control group in a dose dependent manner (15–16). We have shown that the concomitant decreased hepatic ST6GalI activity is due to its decreased synthetic rate (17) while the down regulation of ST6GalI mRNA is due to its decreased stability (16). In view of the pivotal role played by ST6GalI in alcohol-induced defective glycosylation and consequent appearance of asialoglycoconjugates in the blood, we have now investigated the gene expression of ST6GalI in post-mortem human liver specimens taken from non drinkers group (ND), moderate-alcohol drinkers (MD) and heavy–alcohol drinkers (HD) groups. The present study demonstrates that both liver ST6GalI mRNA and its protein are markedly decreased in liver specimens of heavy-alcohol drinkers compared to non-drinkers, which is consistent with our previous findings in the rat model (15–17). This down regulation of ST6GalI may be due to the gradual loss of a cytosolic binding protein that interacts with ST6GalI mRNA and stabilizes it.
Material and methods
Chemicals & reagents
Tri-Reagent was purchased from Molecular Research Center (Cincinnati, OH). Primers were synthesized by Operon Technologies (Alameda, CA). Molecular biology reagents were procured from Invitrogen Inc (Carlsbad, CA) or Bio-Rad Inc (Hercules, CA).
Ethical guidelines
All human specimens collected conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in the priori and also approved by the Institutional Review Board of Veterans Affairs Medical Center, Washington D.C.
Plasma CDT and SIJ determination
For the measurement of plasma carbohydrate-deficient transferrin (CDT) and sialic acid index of plasma ApoJ (moles of sialic acid/mole of ApoJ; SIJ), 5 ml of whole blood was collected and plasma was prepared from a group of 12 male alcoholics (consuming <60 g ethanol/day) and 12 male non-drinkers, The informed consent was obtained before blood was taken individually. Plasma CDT and SIJ were determined as described by us previously (14).
Liver specimens
Post-mortem human liver specimens (all specimen identities were kept anonymous) were purchased from Tissue Transformation Technologies (Edison, NJ) according to the following criteria:
Non-alcohol drinking group(ND): less than 1 alcoholic beverage/day (<14 g ethanol/day) in the past 10 years before death;
Moderate-alcohol drinkers (MD): 1–3 alcoholic beverage/day (14–42 g ethanol/day) in the past 10 years before death;
Heavy-alcohol drinkers (HD): >6 alcoholic beverage/day (<84 g ethanol/day).
The drinking histories of study subjects were based upon the clinical reports from the donor institution and the clinical classification by the hepatologist.
Patients with history of other drug use were excluded. Sex ratios in each group were biased to male, which is comparable to a similar gender distribution in the real society of alcoholics. Average age for each group is as: Non-alcohol drinking group, 48.08 years; Moderate drinking group, 50.08 years; Heavy drinking group, 50.75 years. Autopsy sampling was randomized for patient’s cause of death (see table 1). Autopsy was performed within 6–8 hours after death and autopsy specimen were promptly frozen in liquid nitrogen and stored at −80°C until analyses.
Table 1.
Patient profiles for post-mortem human liver specimens
| Group | Average age (year) | Gender ratio(M/F) | Cause of death | Daily alcohol consumption |
|---|---|---|---|---|
| Non-drinkers (ND) | 48.08 | 9/4 | Randomized | <14g |
| Moderate-drinkers (MD) | 50.08 | 9/4 | Randomized | 14–42g |
| Heavy-drinkers (HD) | 50.75 | 11/1 | Randomized | <84g |
RNA Isolation
Total RNA was isolated from each liver specimen of all groups using the Tri-Reagent (MRC, Cincinnati, OH) following the manufacturer’s instructions. Adequate measures were undertaken to ensure high quality RNA extraction from all specimens. Briefly, 500 mg of liver were homogenized in 1 ml of Tri-Reagent. The homogenate was left for 5 min at room temperature followed by addition of 0.2 ml of bromochloropropane (MRC, OH) and vigorous shaking for 15 seconds. The mixture was left for 15 min at room temperature. After centrifugation (12,000 × g for 20 min) at 4°C, the upper aqueous phase was carefully transferred into a sterile tube. The RNA was precipitated by addition of 0.5 ml of isopropanol and incubated at room temperature for 5 min. The RNA was pelleted by centrifuging again at 12,000 × g at 4°C for 15 min. The precipitated RNA was washed in 70% ethanol, briefly air-dried, and then solubilized in Formazol (MRC, OH). Total RNA concentrations were measured by absorbance reading at 260 nm using Spectromax 190 (Molecular Devices Co., Sunnyvale, CA). The purity of total RNA specimen was examined by determining the A260/A280 ratio. Isolated RNA was used immediately or stored at −80°C until use.
Golgi fraction preparation
Golgi fractions were prepared according to a method described by Leelavathii et. al (18). All steps were performed on ice with proteinase inhibitors in all the solutions used. Briefly, 2–3 g of livers were homogenized in 5 volume (v/w) of 0.5M sucrose made in 0.1M potassium phosphate buffer (pH6.65) for 30 seconds using a Polytron (Glas-col, Terre Haute, IN ) at 100 rpm. The homogenate was then centrifuged at 600 × g for 10 minutes at 4°C in Beckman model J-6M centrifuge. The supernatant fraction was aspirated and 6 ml was carefully layered on top of 7 ml of 1.3 M sucrose in a 15 ml tube and centrifuged at 105,000 × g for 60 min at 4°C. The thick membrane layer above 1.3M sucrose inter-phase was aspirated and carefully layered on top of 1.1M sucrose and further centrifuged at 105,000 × g for 90 min. Then the Golgi membrane fraction concentrated at the inter-phase of 1.1M sucrose was aspirated and its purity was verified by measuring galactosyltransferase activity (92% of total liver activity in our specimen). 50 μg of the Golgi fraction from each liver was used for SDS-PAGE and subsequent Western Blot analyses.
Liver cytosol extracts preparation
The liver cytosolic fraction was prepared as previously described in our laboratory (19) and was either used directly or stored at −80°C. After determining the total protein concentration of the cytosolic fraction by Bradford method (Bio-Rad, Hercules, CA) it was generally diluted to 5 μg/μl.
Riboprobe template preparation
DNA template for the synthesis of mRNA probe in vitro was prepared by amplifying a 2672 base pair (bp) fragment covering 3′-UTR of hST6GalI by RT-PCR. 5 μg of total RNA extracted from a human liver in the non-alcohol drinking group were used in 20 μl reverse transcription reaction containing 10 pM oligo (dT), 10 μM dNTPs and 1 unit of reverse transcriptase. Typical PCR reaction mixture included 2 μl of cDNA templates from RT, 10 pM of each primer, 10 μM of dNTPs, 3mM of MgCl2, 10X buffer and 2 units of high fidelity Taq DNA polymerase in a reaction volume of 50 μl. The PCR conditions were: 2 min at 94°C followed by 35 cycles at 94°C for 30 seconds, 54°C for 30 seconds and 72 °C for 3 min. The primer pairs used were: Forward primer 5′-TAATACGACTCACTATAGTTGGGAGCTATGGGACATTC-3′ and reverse primer 5′–CTACCCAGTGTCGTCCCAGT-3′. The forward primer included a T7 RNA polymerase promoter sequence, which is underlined.
[32P]-labeled RNA transcripts preparation
32P-labeled and unlabeled RNA probes were transcribed in vitro with T7 RNA polymerase from PCR amplified 3′-UTR of ST6GalI according to the method described by Milligan (20). Briefly, the reactions were performed in the presence of 1 μg of template in a buffer containing 2 nM of a-32P UTP (800Ci/mmol), 2.5 mM of each unlabeled rGTP, rCTP and rATP, 40 units of RNasin, and 2U of T7 polymerase for 1 h at 37°C. 1 unit of RNase-free DNase was then added to the reaction and left at 37°C for another 30 minutes, and unincorporated nucleotides were removed by centricon-30 membrane filtration (Millipore, Billerica, MA). 32P-labeled RNA transcripts were quantified by liquid scintillation counting.
Quantitative Real-Time RT-PCR
cDNA templates for use in quantitative real time PCR were synthesized from 5 μg of total RNA by in vitro transcription in 20 μl reaction containing 0.5 μg Oligo (dT), 10 μM dNTPs and 1 μl of Superscript II reverse transcriptase (Invitrogen, Carlsbad,CA) at 42 °C for 50 min. Typical real time PCR reaction mixture included the same amount of cDNA templates from RT, 10 pM of each primer, 25 ul iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and sterile water in a reaction volume of 50 μl. The PCR conditions were: 3 min at 95°C followed by 40 cycles at 95°C for 30 seconds, 55°C for 30 seconds and 72 °C for 1 min. The primer pairs for human ST6GalI gene were: Forward primer: 5′-GTGGGCACAAAAACTACCAT-3′ and reverse primer 5′-GGCTCTGGGCTCATAAACTG-3. This primer pair was first tested by regular PCR to be highly effective and specific for amplification. β-actin was used as the standard housekeeping gene. Ratios of ST6GalI gene and β-actin gene expression levels were calculated by subtracting the threshold cycle number (Ct) of the target gene from the Ct of β-actin and raising 2 to the power of the negative of this difference. Ct values are defined as the number of PCR cycles at which the fluorescent signal during the PCR reaches a fixed threshold. Target gene expression is expressed relative to β-actin gene expression.
SDS-PAGE and Western Blot analysis
Protein samples were diluted into SDS-PAGE sample buffer [50 mM Tris (pH 6.8), 2% SDS, 10% glycerol, 15 mM 2-mercaptoethanol, and 0.25% bromophenol blue] and boiled for 5 min. The specimen were elctrophoretically resolved on 4–15% SDS-PAGE gels. The gels were transblotted onto PVDF membranes (Bio-Rad, Hercules, CA) and were processed for immuno-quantification using a polyclonal anti ST6GalI antibody (kindly provided by Dr. Karen J. Colley, University of Illinois, Chicago, IL) or anti-β-actin antibody (Santa Cruz biotechnology, CA). The antigen bound primary anti-ST6GalI and anti-β-actin antibodies were detected with horseradish peroxidase conjugated goat anti-rabbit IgG antibodies (Santa Cruz biotechnology, CA). Bound antibodies were visualized by ECL using reagents from Perkin Elmer (Boston, MA) and the intensity of each band was quantified using a Fluorchem 8800 densitometer (Alpha Innotech, San Leandro, CA).
RNA-protein Electrophoretic Gel Mobility Shift Assays (EMSA)
32P-labeled RNA at a concentration of 2 nM was incubated with 20 μg liver cytosol proteins (this amount of protein was determined to be sufficient to visualize a RNA-protein complex in pre-experiments) in a 10 μl solution containing 10 mM HEPES (pH 7.5), 25 mM KCl, 10% glycerol and 1 mM dithiothreitol at 30°C for 30 min followed by incubation with 20 units of RNase T1 (Ambion, Austin, TX) at 37 °C for 10 min in each reaction. For competition experiments, the cytosolic fraction was incubated for 10min with 100-fold molar excess of unlabeled RNA transcripts before incubation with the labeled RNA. Then 2 μl of 6X native gel loading buffer (30% glycerol, 0.025% bromophenol blue and xylene cyanol) was added, and the RNA-protein complexes were resolved on an 8% native polyacrylamide gel in 0.5X TBE buffer (45 mM Tris-HCl pH 8.3, 45 mM borate, 2.5 mM EDTA). Gels were pre-electrophoresed for 30 min at 20 mA followed by electrophoresis at 30 mA for 0.5–1 h at 4°C. Gels were dried and exposed to Kodak XAR-5 film (Sigma-Aldrench, St. Louis, MO) with an intensifying screen at −80°C overnight. The intensity of each band was quantified using a Fluorchem 8800 densitometer (Alpha Innotech, San Leandro, CA). To ensure the comparable measurement of the bands, same numbers of specimen from each group were resolved simultaneously on the same gel; a known sample from non drinkers group that was previously tested to show ST6GalI-protein interaction was used as control for comparable results on separate gels.
Statistical analysis
Data are presented as the Mean ± SEM. Statistical significance was determined by one-way ANOVA test followed by the Tukey’s test (SAS Software, SAS Institute, Cary, NC).
RESULTS
Plasma CDT and SIJ in alcoholics and non drinkers
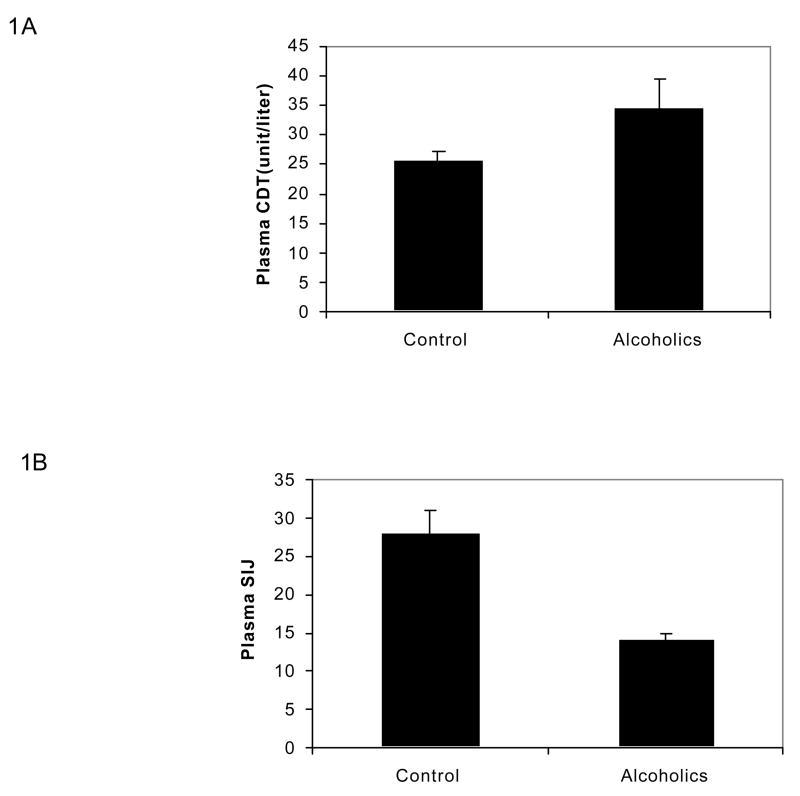
As can be seen in Figure 1, plasma CDT increased by 35% (p<0.05) in alcoholic group compared to non-drinkers (Figure 1A) while plasma SIJ markedly decreased by 50% (p<0.001) in alcoholic group compared to non-drinking group (Figure 1B).
Figure 1. Plasma CDT and SIJ in alcoholics and non drinkers.
A suitable aliquot of each plasma sample was analyzed for CDT and SIJ as described by us previously (14). Plasma CDT is expressed as units/liter based on the Kabi Pharmacia Diagnostics, Uppsala, Sweden. Each value is the Mean ± SEM of 12 specimens from each group.
Liver ST6GalI mRNA level is down-regulated proportionately to the amount of alcohol consumed
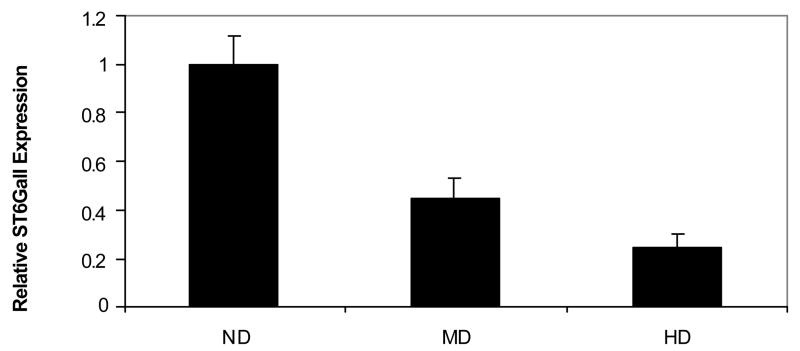
ST6GalI mRNA levels, as determined by quantitative real time RT-PCR, in the post-mortem liver specimens of non drinker, moderate drinker and heavy drinker groups are shown in Figure 2. ST6GalI mRNA level was decreased on an average by 49% (p<0.01) and by 69% (p<0.01) in moderate and heavy-alcohol drinkers, respectively, compared to the non drinkers group.
Figure 2. Real time RT- PCR analyses of ST6GalI in human liver specimens.
Total RNA from the non drinkers group (ND), the moderate drinkers group (MD) and the heavy drinkers group (HD) was reverse transcribed and used in the real time PCR. The RNA levels were normalized to the level of β-actin on experiment to experiment basis. Each sample analysis was performed in triplicate independently and each bar graph represents the Mean ± SEM of 12 specimens in each group with the ND group set at 1 for convenience.
Down-regulation of ST6GalI mRNA by alcohol concomitant with the decrease of ST6GalI protein
Since any change in a given mRNA level does not always reflect in a corresponding change in its protein level or enzyme activity, it is necessary to determine the protein level in order to give a meaningful interpretation of our above observations. Therefore, western blot analyses of the liver Golgi extracts from post-mortem liver specimens of the three experimental groups were performed using anti ST6GalI antibody. The results are shown in Figure 3. The hepatic ST6GalI protein level relative to that of beta actin was not significantly decreased (10%, p=0.4) in the moderate drinkers, whereas it was decreased by 30% (p<0.05) in heavy alcohol drinkers compared to non-drinkers. These results are consistent with our finding of the progressive down regulation of ST6GalI mRNA with the magnitude of alcohol consumption (Figure 1).
Figure 3. Western Blot analysis of ST6GalI in human liver specimens.
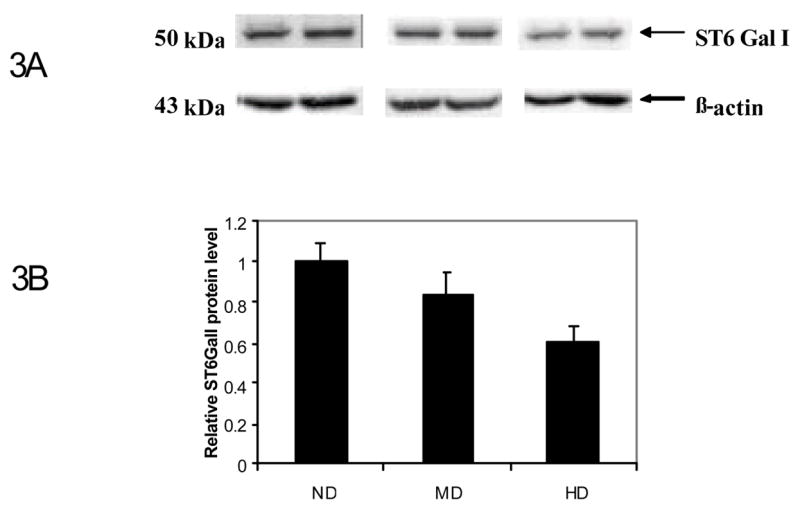
50 μg of liver Golgi fraction extracts from the non drinkers group (ND), the moderate drinkers group (MD) and the heavy drinkers group (HD) was subjected to Western Blot analyses using the polyclonal anti ST6GalI and anti β-actin antibodies. Densitometry analysis of the intensity of each band was then performed. Western blot of two representative specimens from each group are shown in panel A. Bands on the top represent ST6GalI; bands on the bottom represent β-actin. In panel B, the relative intensity of ST6 Gal1 protein level in each specimen was normalized to its β-actin level. The bar graphs represent the relative level of ST6GalI in each group with the ND group set at 1 for convenience. The value in each group is the Mean ± SEM of 12 specimens.
The level of the cytosolic binding protein that protects ST6GalI mRNA stability
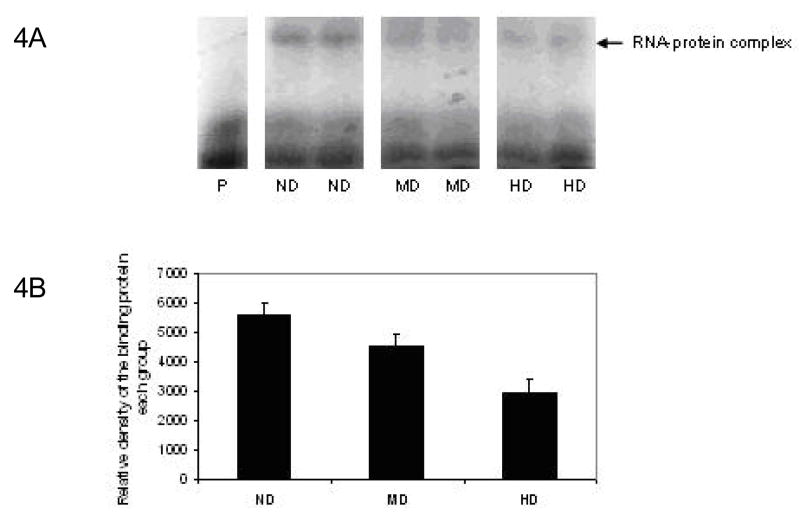
Recently, we clearly characterized a cytosolic protein that stabilizes rat ST6GalI mRNA by specifically interacting with its 3′-UTR domain, but this binding protein was completely depleted after chronic alcohol consumption in rat model (21). In order to find out whether a similar regulatory mechanism operates in the livers of human alcoholics, RNA-Protein Electrophoretic Gel Mobility Shift Assays (EMSA) was performed on all 36 post-mortem liver specimens. Prior experiments were carried out to obtain the optimal amount of cytosol protein to be used and to verify that RNA-protein interaction is specific for ST6GalI. Two representative samples from each ND, MD and HD groups are shown on Figure 4 A. A single cytosolic protein complex with the human ST6GalI mRNA was evident in the liver extracts of non drinkers. When quantitative measurement was performed on all specimens in each group with a known standard, the level of the cytosol protein gradually decreased in the liver extracts from moderate (p<0.05) and heavy alcohol drinkers (p<0.01) (Figure 4B).
Figure 4. RNA-protein Electrophoretic Gel Mobility Shift Assays (EMSA).
32P-labeled RNA at a concentration of 2 nM was incubated with 20 μg of cytosolic proteins from each liver sample and the RNA-protein complexes were resolved on an 8% native polyacrylamide gel in 0.5X TBE buffer. The gel was then dried and exposed to XAR film with an intensifying screen at −80°C overnight. Two representative specimens from each group were shown in 4A. Arrow indicates the RNA-protein complex; Lane P represents RNA probe without the cytosolic protein. 4B: shows the average density of 12 specimens in each group. The value in each group is the Mean ± SEM of 12 specimens. The RNA-protein complex was progressively decreased both in MD and HD groups compared to the ND group.
Discussion
To investigate whether alcohol consumption affects the ST6GalI gene regulation in post-mortem human livers, we determined ST6GalI mRNA levels by quantitative real time RT-PCR in all the liver specimens from the three groups. We show for the first time in humans that moderate drinking down regulates ST6GalI mRNA level by as much as 49% (p<0.01), while heavy drinking further diminishes its level by 69% (p<0.01) (Figure 2.). This is further confirmed by concomitant changes in ST6GalI protein (Figure 3). Similar alterations in plasma CDT and SIJ in the alcoholic group compared to the non drinkers (Figure 1) fully support our concept that the marked down regulation of ST6GalI mRNA in the liver specimens of alcoholics may be responsible for the appearance of these asialoglycoconjugates in alcoholics. Unfortunately, since the blood specimens were not collected in these post-mortem cases, we could not measure plasma CDT and SIJ although we do demonstrate marked decrease in plasma SIJ in alcoholic patients compared to non-drinkers (Figures 1A and 1B). At the same time, the fact that hepatic ST6GalI mRNA has been reported to be unaltered or upregulated in non-alcoholic or neoplastic liver diseases (22–24), liver disease could not be the cause for the down regulation of ST6GalI mRNA in the livers of alcoholics found in the present study. Thus, our data clearly seem to support the concept that alcohol consumption per se, but not the liver disease, may be specifically responsible for ST6GalI mRNA down regulation.
These results are consistent with our earlier findings in animal model in which we previously showed that long term ethanol exposure causes impaired expression of ST6GalI gene (16) accompanied by corresponding changes in plasma CDT and SIJ in chronic alcohol-fed animals (14). In that model, we further demonstrated that chronic alcohol treatment caused decreased ST6GalI protein synthetic rate (17) due to decreased stability of ST6GalI mRNA without affecting its transcription rate (16). We have recently partially characterized a liver cytosolic protein which specifically interacts with ST6GalI mRNA to stabilize it (21). The fact that the level of this binding protein is depleted in the liver specimens of alcoholics (Figure 3) strongly supports our concept that the down regulation of ST6GalI gene may be due to the loss of this mRNA stabilizing protein, as we have shown in the animal system (21). Thus, the possible mechanism/s by which chronic ethanol exposure down-regulates ST6GalI gene in human liver is mediated via the destabilization of its mRNA by depleting a specific cytosol binding protein that interacts with its 3′UTR domain.
Clinical observations also show increased appearance of asialoconjugates in the blood of chronic alcoholics (12–14). Thus, if the specificity of ethanol exposure on this gene is established, it may form the basis for the appearance of asialoconjugates in blood of human alcoholics that can serve as viable biomarkers. Secondly, sialic acids and their derivatives are ubiquitous at the terminal positions of oligosaccharides of glycoproteins and glycolipids in mammalian cells and their roles have been implicated in various biological phenomena, such as cell proliferation, differentiation, signal transduction, cell surface interactions, cell-cell communication and adhesion or even viral-host recognition, or tumor invasiveness (25–27).
Acknowledgments
This work was supported by NIAAA grant R01 AA08149.
Abbreviations
- ST6GalI
Galβ1, 4GlcNAc a2, 6-sialyltransferase
- CDT
carbohydrate-deficient transferrin
- SIJ
sialic acid index of plasma ApoJ
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelm S, Schauer R. Sialic acids in molecular and cellular interactions. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilatte Y, Bignon J, Lambre CR. Sialic acids as important molecules in the regulation of the immune systems: pathophysiological implications of sialidases in immunity. Glycobiology. 1993;3(3):201–218. doi: 10.1093/glycob/3.3.201. [DOI] [PubMed] [Google Scholar]
- 3.Lin S, Kemmner W, Grigull S, Schlag PM. Cell surface alpha 2, 6 sialylation affects adhesion of breast carcinoma cells. Experimental Cell Research. 2002;276(1):101–110. doi: 10.1006/excr.2002.5521. [DOI] [PubMed] [Google Scholar]
- 4.Gagneux P, Cheriyan M, Hurtado-Ziola N, van der Linden EC, Anderson D, McClure H, Varki A, Virki NM. Human-specific regulation of alpha2-6-linked sialic acids. Journal of Biological Chemistry. 2003;278(48):48245–48250. doi: 10.1074/jbc.M309813200. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Srivatana U, Ullah A, Gagneja H, Berenson CS, Lance P. Suppression of a sialyltransferase by antisense DNA reduces invasiveness of human colon cancer cells in vitro. Biochimica et biophysica Acta. 2001;1536(2–3):148–160. doi: 10.1016/s0925-4439(01)00044-8. [DOI] [PubMed] [Google Scholar]
- 6.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83(8):727–37. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 7.Tsuji S. Molecular cloning and functional analysis of sialyltransferases. J Biochem (Tokyo) 1996;120(1):1–13. doi: 10.1093/oxfordjournals.jbchem.a021369. [DOI] [PubMed] [Google Scholar]
- 8.Takashima S, Tsuji S, Tsujimoto M. Characterization of the second type of human β-galactoside a2,6-sialyltransferase (ST6Gal II), which sialylates Galβ1,4GlcNAc structures on oligosaccharides preferentially. Genomic analysis of human sialyltransferase genes. J Biol Chem. 2002;277(48):45719–45728. doi: 10.1074/jbc.M206808200. [DOI] [PubMed] [Google Scholar]
- 9.Krzewinski-Recchi MA, Julien S, Juliant S, Teintenier-Lelievre M, Samyn-Petit B, Montiel MD, Mir AM, Cerutti M, Harduin-Lepers A, Delannoy P. Identification and functional expression of a second human beta-galactoside alpha2, 6-sialyltransferase, ST6Gal II. Eur J Biochem. 2003;270(5):950–961. doi: 10.1046/j.1432-1033.2003.03458.x. [DOI] [PubMed] [Google Scholar]
- 10.Appenheimer MM, Huang RY, Chandrasekaran EV, Dalziel M, Hu YP, Soloway PD, Wuensch SA, Matta KL, Lau JT. Biologic contribution of P1 promoter-mediated expression of ST6GalI sialyltransferase. Glycobiology. 2003;13(8):591–600. doi: 10.1093/glycob/cwg066. [DOI] [PubMed] [Google Scholar]
- 11.Takashima S, Tsuji S, Tsujimoto M. Comparison of the enzymatic properties of mouse beta-galactoside alpha2,6-sialyltransferases, ST6GalI and II. J Biochem (Tokyo) 2003;134(2):287–296. doi: 10.1093/jb/mvg142. [DOI] [PubMed] [Google Scholar]
- 12.Stibler H, Borg S. Evidence of a reduced sialic acid content in serum transferrin in male alcoholics. Alcohol Clin Exp Res. 1981;5(4):545–549. doi: 10.1111/j.1530-0277.1981.tb05358.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsutsumi M, Wang JS, Takada A. Microheterogeneity of serum glycoproteins in alcoholics: is desialo-transferrin the marker of chronic alcohol drinking or alcoholic liver injury? Alcohol Clin Exp Res. 1994;18(2):392–397. doi: 10.1111/j.1530-0277.1994.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh P, Hale EA, Lakshman MR. Plasma sialic acid index of apolipoprotein J (SIJ): a new alcohol intake marker. Alcohol. 2001;25(3):173–179. doi: 10.1016/s0741-8329(01)00187-2. [DOI] [PubMed] [Google Scholar]
- 15.Rao MN, Lakshman MR. Chronic ethanol downregulates 2, 6-sialyltransferase and 2, 3-silayltranferase mRNAs in rat liver. Alcohol Clin Exp Res. 1997;21(2):348–351. [PubMed] [Google Scholar]
- 16.Rao MN, Lakshman MR. Chronic ethanol consumption leads to destabilization of rat liver 2, 6-sialyltransferase mRNA. Metabolism. 1999;48(6):797–803. doi: 10.1016/s0026-0495(99)90182-8. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh P, Liu QH, Lakshman MR. Long-term ethanol exposure impairs glycosylation of both N- and O-glycosylated proteins in rat liver. Metabolism. 1995;44(7):890–898. doi: 10.1016/0026-0495(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 18.Leelavathi DE, Estes LW, Feingold DS, Lombardi B. Isolation of a Golgi-rich fraction from rat liver. Biochim Biophys Acta. 1970;211:124–138. [Google Scholar]
- 19.Marmillot P, Rao MN, Lakshman MR. Chronic ethanol exposure in rats affects rabs-dependent hepatic trafficking of apolipoprotein E and transferrin. Alcohol. 2001;25(3):195–200. doi: 10.1016/s0741-8329(01)00179-3. [DOI] [PubMed] [Google Scholar]
- 20.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA . Nucleic Acids Res. 1987;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garige M, Gong M, Lakshman MR. Ethanol Destabilizes Liver ST6Gal l mRNA by Depleting a 3′-Untranslated Region Specific Binding Protein. J Pharmacol Exp Ther. 2006;318(3):1076–1082. doi: 10.1124/jpet.106.103861. [DOI] [PubMed] [Google Scholar]
- 22.Petretti T, Kemmner W, Schulze B, Schlag PM. Altered mRNA expression of glycosyltransferases in human colorectal carcinomas and liver metastases. Gut. 2000;46(3):359–366. doi: 10.1136/gut.46.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang PH, Lee WL, Lee YR, Juang CM, Chen YJ, Chao HT, Tsai YC, Yuan CC. Enhanced expression of alpha 2,6-sialyltransferase ST6GalI in cervical squamous cell carcinoma. Gynecol Oncol. 2003;89(3):395–401. doi: 10.1016/s0090-8258(03)00127-6. [DOI] [PubMed] [Google Scholar]
- 24.Dall'Olio F, Chiricolo M, D'Errico A, Gruppioni E, Altimari A, Fiorentino M, Grigioni WF. Expression of beta-galactoside alpha2, 6 sialyltransferase and of alpha2, 6-sialylated glycoconjugates in normal human liver, hepatocarcinoma, and cirrhosis. Glycobiology. 2004;14(1):39–49. doi: 10.1093/glycob/cwh002. [DOI] [PubMed] [Google Scholar]
- 25.Schauer R. Sialic acids and their role as biological masks. Trends Biochem Sci. 1985;10:357–360. [Google Scholar]
- 26.Hakomori S. Structure and function of sphingoglycolipids in transmembrane signalling and cell-cell interactions. Biochem Soc Trans. 1993;21 :583–595. doi: 10.1042/bst0210583. [DOI] [PubMed] [Google Scholar]
- 27.Kelm S, Schauer R. Sialic acids in molecular and cellular interactions. Int Rev Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]