Autosomal dominant retinitis pigmentosa results from mutations in 14 known proteins, and at least two further loci have been highlighted by genetic linkage in families (reviewed by the RetNet website; http://www.sph.uth.tmc.edu/Retnet/). The known genes include those encoding components of the phototransduction cascade, retinal transcription factors and retinal structural proteins.1 The list also includes four ubiquitously expressed splicing factors: pre‐mRNA processing factor 8 (PRPF8),2 PRPF31,3 PRPF34 and PAP‐1, also known as RP9.5,6
Splicing is a complex process that involves the precise excision of introns from pre‐mRNA by a macromolecular structure called the spliceosome. Three of the splicing factors implicated in autosomal dominant retinitis pigmentosa (ADRP) are components of the U4/U6‐U5 tri‐snRNP particle, an essential component of the spliceosome.7,8 Mutations in one of these, PRPF31, have been reported to cause between 5 and 20% of ADRP.9,10 In this report, a new mutation in the PRPF31 gene is described, together with the clinical phenotype.
Cases
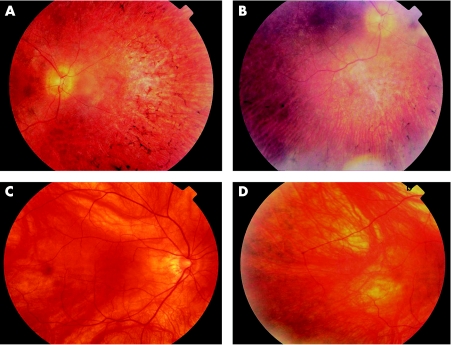
The proband was a 33‐year‐old female with a corrected visual acuity of 58 and 51 ETDRS letters in the right and left eye, respectively (approximate Snellen equivalents of 6/18 and 6/36). She had a myopic refraction with a spherical equivalence of −2 dioptres in each eye. Nyctalopia had been present since the middle of the second decade, and she had noticed a decrease in her central vision since the beginning of the third decade. At the most recent examination, she had early posterior subscapsular cataract, bone spicule formation in all four quadrants (fig 1a,b) attenuated arterioles and pale optic discs in each eye. The maculae appeared normal on clinical examination. On Goldman perimetry, the mean visual field to the V4e target measured 6.5° from fixation. Zeiss OCT 3 examination demonstrated a foveal thickness of 170 and 144 microns, respectively, in the right and left eyes, with absence of the third highly reflective band.11
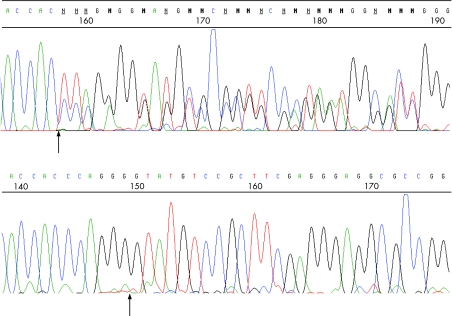
Figure 1 Electropherogram of the mutated (upper) and normal (lower) sequence of PRPF31 in. As the mutation is heterozygous, the upper image shows both the mutated and normal sequences superimposed. The arrow on the mutated sequence denotes the beginning of the deleted sequence, while the arrow on the normal sequence marks the boundary between exon 6 and intron 6. Sequence was generated from PCR‐amplified DNA on a Pharmacia MegaBACE automated DNA sequencer.
Her younger sister had a similar clinical phenotype and age of onset. The 61‐year‐old mother was asymptomatic, with unaided visual acuities of 80 and 81 ETDRS letters (Snellen equivalent of 6/7.5). Fundus examination revealed mild bone spicule attenuation in the peripheral retina (fig 1c,d). Visual field to the V4e target on Goldman perimetry was slightly reduced from normal with a mean of 57.8° from fixation. Foveal thickness on Zeiss OCT 3 examination was 223 and 249 microns in the right and left eyes, respectively. The father of the proband was also asymptomatic with visual acuities of 85 ETDRS letters (Snellen 6/6) in both eyes and normal ocular examinations. No clinical information was available from any other living relative along the maternal line.
Mutation screening
DNA from the proband was included in a large cohort of retinal dystrophy DNAs, which were screened for mutations in a limited set of exons or parts of exons of known retinal degeneration genes. The exons screened were selected from the available literature because they were known mutation hotspots or locations of common founder mutations. Screening was carried out by radioactively labelled single‐strand conformation polymorphism/heteroduplex analysis (SSCP/HA).12
One of the sequences screened was PRPF31 exon 6. Screening of this sequence in the proband revealed a large mobility shift suggestive of a deletion. Sequencing revealed a novel 16 bp deletion present in the three female members of the family but absent from the father and from 120 control Caucasian genomic DNAs (240 chromosomes). This sequence change is denoted c.522–527del&IVS6+1to+10del13 (fig 2). It deletes codons 175 and 176, the last two in exon 6, encoding glutamine and glycine residues. However, it also deletes the first 10 bp of intron 6, including the exon 6/intron 6 boundary and splice donor site, the mutation abolishing the exon 6 splice donor site. This may give rise to an mRNA transcript which includes intron 6, adding seven novel amino‐acids then terminating the encoded protein, or could lead to the skipping of exon 6.
Figure 2 A and B, retinal photographs taken from the proband showing bone spicule attenuation. C and D, retinal photographs from the mother of the proband. The mild bone spicule attenuation was evident only on slit lamp biomicroscopy using a wide angle lens.
Discussion
This novel mutation in the PRPF31 gene causes a severe phenotype in symptomatic cases, with the onset of nyctalopia in the second decade and loss of acuity from the third. Both the age of onset and the phenotype observed are similar to that described by Sato et al14 in Japanese families. In addition, this report is the first to demonstrate variable penetrance of the phenotype in an asymptomatic carrier of the mutation. A high level of non‐penetrance has been described previously, both in families with confirmed PRPF31 mutations and in those linked to the RP11 locus before mutations in PRPF31 were identified.14,15,16,17,18,19 Evans et al15 used the term bimodal expressivity to describe this phenomenon. Sato and colleagues also identified asymptomatic carriers of the mutations in the PRPF31 gene by genetic analysis. One of these was an elderly relative of three generations of symptomatic RP sufferers, though he himself had no ocular abnormalities except for mild cataracts. In our report, the mother of the proband had definite retinal findings and a mildly reduced visual field on Goldman perimetry, though she was totally asymptomatic. This may perhaps imply that the range of phenotypes seen in PRPF31‐RP could be better described as a spectrum of severity, rather than true bimodal expressivity.
The mutation described above is likely to result in a grossly abnormal transcript which may be subject to nonsense mediated decay.20 This brings to 18 the number of published PRPF31 mutations in the literature, comprising six deletions (ranging from one base pair to the whole gene), five splice‐site mutations, two insertion/deletion events, one duplication, one insertion and only three missense mutations.3,14,18,19,21,22,23,24 The lack of missense changes has led others to speculate that mutations in PRPF31 cause RP due to haploinsufficiency and consequent insufficiency of splicing activity.18 Wilkie et al25 concluded that reduced mutant protein solubility in two of the known missense mutations, A194E and A216P, also led to splicing insufficiency.
This hypothesis is further supported by the finding that high‐expressing alleles of PRPF31 from the normal parent compensate for a potentially RP‐causing mutation on the opposing chromosome.26 This phenomenon accounts for the variation in severity described above and predicts that the normal second allele of PRPF31 in the mother from the family described herein is a high‐expressing variant which masks the RP symptoms. However, the alleles inherited by her daughters from their normal father are less well expressed, and so these individuals have a much more severe form of RP. To date, the mechanism controlling this level of expression remains unknown. A bimodal phenotype might be explained by a single diallelic polymorphism in a sequence involved in transcription regulation, whereas a spectrum of severity, as observed herein, might imply a more complex interplay between several such polymorphisms. Understanding the basis of this variation in severity, together with the finding of haploinsufficiency as a cause of disease, could have important implications for the testing of potential new treatments for this relatively common retinal degeneration.
Acknowledgements
We thank Yorkshire Eye Research (grant numbers 009 and 006) and the Leeds Teaching Hospitals Charitable Foundation for funding this research, and Mike Stockton in the medical illustration department, St. James's University Hospital, and of course the patient and her family for enabling us to carry out this lesearch and share the information with the wider medical community.
Footnotes
Competing interests: None declared.
References
- 1.Hims M M, Diager S P, Inglehearn C F. Retinitis pigmentosa: genes, proteins and prospects. Dev Ophthalmol 200337109–125. [DOI] [PubMed] [Google Scholar]
- 2.McKie A B, McHale J C, Keen T J.et al Mutations in the pre‐mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 15 Jul 200110(15)1555–1562. [DOI] [PubMed] [Google Scholar]
- 3.Vithana E N, Abu‐Safieh L, Allen M J.et al A human homolog of yeast pre‐mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001 Aug 8(2)375–381. [DOI] [PubMed] [Google Scholar]
- 4.Chakarova C F, Hims M M, Bolz H.et al Mutations in HPRP3, a third member of pre‐mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet 1 Jan 200211(1)87–92. [DOI] [PubMed] [Google Scholar]
- 5.Keen T J, Hims M M, McKie A B.et al Mutations in a protein target of the Pim‐1 kinase associated with the RP9 form of autosomal dominant retinitis pigmentosa. Eur J Hum Genet 200210245–249. [DOI] [PubMed] [Google Scholar]
- 6.Maita H, Harada Y, Nagakubo D.et al PAP‐1, a novel target protein of phosphorylation by pim‐1 kinase. Eur J Biochem. 2000 Aug 267(16)5168–5178. [DOI] [PubMed] [Google Scholar]
- 7.Makarova O V, Makarov E M, Liu S.et al Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri‐snRNP formation and pre‐mRNA splicing. The EMBO Journal 2002211148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faustino N A, Cooper T A. Pre‐mRNA splicing and human disease. Genes Dev 200315419–437. [DOI] [PubMed] [Google Scholar]
- 9.Inglehearn C F, Tarttelin E E, Plant C.et al A linkage survey of twenty dominant retinitis pigmentosa families: Frequencies of the nine known loci and evidence for further heterogeneity. J Med Genet 1998351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan L S, Bowne S J, Birch D G.et al Prevalence of disease‐causing mutations in families with autosomal dominant retinitis pigmentosa: A screen of known genes in 200 families. Investigative Ophthalmology and Visual Science 2006473052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandberg M A, Brockhurst R J, Gaudio A R.et al The association between visual acuity and central retinal thickness in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2005 Sep 46(9)3349–3354. [DOI] [PubMed] [Google Scholar]
- 12.Toomes C, Marchbank N J, Mackey D A.et al Spectrum, frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet 15 Jun 200110(13)1369–1378. [DOI] [PubMed] [Google Scholar]
- 13.den Dunnen J T, Antonarakis S E. Nomenclature for the description of human sequence variations. Hum Genet. 2001 Jul 109(1)121–124. [DOI] [PubMed] [Google Scholar]
- 14.Sato H, Wada Y, Itabashi T.et al Mutations in the pre‐mRNA splicing gene, PRPF31, in Japanese families with autosomal dominant retinitis pigmentosa. Am J Ophthalmol. 2005 Sep 140(3)537–540. [DOI] [PubMed] [Google Scholar]
- 15.Evans K, al‐Maghtheh M, Fitzke F W.et al Bimodal expressivity in dominant retinitis pigmentosa genetically linked to chromosome 19q. Br J Ophthalmol. 1995 Sep 79(9)841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al‐Maghtheh M, Vithana E, Tarttelin E.et al Evidence for a major retinitis pigmentosa locus on 19q13.4 (RP11) and association with a unique bimodal expressivity phenotype. Am J Hum Genet. 1996 Oct 59(4)864–871. [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez‐Gimeno M, Gamundi M J, Hernan I.et al Mutations in the pre‐mRNA splicing‐factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 200344(5)2171–2177. [DOI] [PubMed] [Google Scholar]
- 18.Abu‐Safieh L, Vithana E N, Mantel I.et al A large deletion in the adRP gene PRPF31: evidence that haploinsufficiency is the cause of disease. Mol Vis 18 Apr 200612384–388. [PubMed] [Google Scholar]
- 19.Rivolta C, McGee T L, Frio T R.et al Variation in retinitis pigmentosa‐11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat 200627(7)644–653. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer P A, Dietz H C. Nonsense‐mediated mRNA decay in health and disease. Hum Mol Genet 19998(10)1893–1900. [DOI] [PubMed] [Google Scholar]
- 21.Xia K, Zheng D, Pan Q.et al A novel PRPF31 splice‐site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol Vis. 2004 20 May 10361–365. [PubMed] [Google Scholar]
- 22.Lu S S, Zhao C, Cui Y.et al Novel splice‐site mutation in the pre‐mRNA splicing gene PRPF31 in a Chinese family with autosomal dominant retinitis pigmentosa] Zhonghua Yan Ke Za Zhi 2005 Apr 41(4)305–311. [PubMed] [Google Scholar]
- 23.Chakarova C F, Cherninkova S, Jordanova A.et al Mutations causing retinitis pigmentosa in gypsy families. Invest Ophth Vis Sci 20064611040 [Google Scholar]
- 24.Wang L, Ribaudo M, Zhao K.et al Novel deletion in the pre‐mRNA splicing gene PRPF31 causes autosomal dominant retinitis pigmentosa in a large Chinese family. Am J Med Genet A 1 Sep 2003121(3)235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkie S E, Morris K J, Bhattacharya S S.et al A study of the nuclear trafficking of the splicing factor protein PRPF31 linked to autosomal dominant retinitis pigmentosa (ADRP). Biochim Biophys Acta. 2006 Mar 1762(3)304–311. [DOI] [PubMed] [Google Scholar]
- 26.Vithana E N, Abu‐Safieh L, Pelosini L.et al Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: A molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci. 2003 Oct 44(10)4204–4209. [DOI] [PubMed] [Google Scholar]