Abstract
It is commonly accepted that the reverse-transcribed cellular RNA molecules, called retroposons, integrate at staggered breaks in mammalian chromosomes. However, unlike what was previously thought, most of the staggered breaks are not generated by random nicking. One of the two nicks involved is primarily associated with the 5′-TTAAAA hexanucleotide and its variants derived by a single base substitution, particularly A → G and T → C. It is probably generated in the antisense strand between the consensus bases 3′-AA and TTTT complementary to 5′-TTAAAA. The sense strand is nicked at variable distances from the TTAAAA consensus site toward the 3′ end, preferably within 15–16 base pairs. The base composition near the second nicking site is also nonrandom at positions preceding the nick. On the basis of the observed sequence patterns it is proposed that integration of mammalian retroposons is mediated by an enzyme with endonucleolytic activity. The best candidate for such enzyme may be the reverse transcriptase encoded by the L1 non-long-terminal-repeat retrotransposon, which contains a freshly reported domain homologous to the apurinic/apyrimidinic (AP) endonuclease family [Martin, F., Olivares, M., Lopez, M. C. & Alonso, C. (1996) Trends Biochem. Sci. 21, 283–285; Feng, Q., Moran, J. V., Kazazian, H. H. & Boeke, J. D. (1996) Cell 87, 905–916] and shows nicking in vitro with preference for targets similar to 5′-TTAAAA/3′-AATTTT consensus sequence [Feng, Q., Moran, J. V., Kazazian, H. H. & Boeke, J. D. (1996) Cell 87, 905–916]. A model for integration of mammalian retroposons based on the presented data is discussed.
The reverse-transcribed cellular RNA molecules, or retroposons, are commonly integrated in mammalian chromosomes (1–3). They are usually flanked by short direct repeats resulting from integration at staggered chromosomal breaks (4). The flanking repeats vary in length and, although rich in adenine (5, 6), they did not seem to contain any specific sequence signals which would indicate enzymatic involvement in retroposon integration (reviewed in ref. 1). This led to a widespread opinion that their origin is attributable to randomly generated staggered breaks. However, sequence analysis of the flanking regions from human and rodent retroposons presented in this paper shows that the staggered breaks are associated with specific, albeit short, sequence signals, consistent with the involvement of an enzyme with endonucleolytic activity. This evidence implies a specific mechanism of retroposon integration in mammals.
MATERIALS AND METHODS
The analysis was performed on sequences flanking human Alu (7) and rodent ID (BC1-like) retroposons (8). GenBank coordinates of the majority of unique Alu elements were obtained from Repbase (ftp http://ncbi.nlm.nih.gov/repository/repbase), and coordinates of ID and additional Alu sequences were directly identified in GenBank (release 97.0) by screening of the corresponding consensus sequences against human and rodent portions of the GenBank database, using the censor program (9). All Alu and ID elements immediately flanked by other repetitive elements were discarded to avoid systematic biases in base compositions of their flanking regions.
Pairs of sequences approximately 50 bp long, flanking each full-length Alu and ID element, were extracted and aligned against each other using an implementation of the Smith–Waterman algorithm (10). Based on the alignment, only identical subsequences at both ends of complete Alu or ID sequences were selected as potential flanking repeats. If the 5′ subsequences in any of the homologous pairs were not immediately adjacent to the 5′ ends of the Alu or ID sequences, the pairs were eliminated from the set by manual editing using the sequence editor mase (11). The finally selected fragments, at least 10 bp long, were considered to be representative flanking repeats of 344 human Alu and 56 rodent ID elements. The remaining 356 Alu and 49 ID flanking repeats 4–9 bp long were put in a separate set for comparative studies, as described in Results.
The 5′ flanking repeats over 9 bp long were adjusted to the left so that they all started at the same position, and the 3′ repeats were adjusted to the right so that they all ended at the same position. A nonadjusted version of the same set was also preserved for comparative purposes. No alignment was attempted to maximize sequence similarity. The adjusted flanking repeats were further extended by additional 15 bp away from the retroposon insertion site, wherever sequence data was available. The extensions are referred to as 5′ and 3′ adjacent sequences.
The second set containing flanking sequences shorter than 10 bp was left unadjusted, since many flanking repeats, particularly on the short end of the distribution, could not be confirmed with any certainty and were likely to represent coincidental matches. Instead, the entire 30-bp regions preceding 5′ ends of Alu and ID retroposons were compared with the analogous regions of the unadjusted set containing longer flanks, in terms of base occurrences at individual positions, as described in the following sections.
Two additional random sets of nonredundant sequence fragments 30 bp long were selected from human sequences deposited in GenBank. One contained 356 human sequence fragments with base composition similar to that of the flanking regions of Alu elements, and another included 913 sequences 30 bp long, chosen irrespectively of their base composition. Both sets were used to illustrate a degree of background fluctuations in base occurrences.
The flanking repeats and the adjacent sequences have been deposited in the repbase/publ directory and are available electronically (see the electronic address above).
RESULTS
Flanking Repeats over 9 bp Long.
The base occurrences at individual positions of 5′ flanking repeats 10 bp long or more, and of the 5′ adjacent regions in Alu and ID retroposons, are listed in Table 1, and analogous data for 3′ flanking repeats and their 3′ adjacent regions are listed in Table 2.
Table 1.
Base occurrences at different positions of flanking repeats and of the 5′ adjacent regions
| N |
Alu
|
ID
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | T | C | A | G | Total | P | T | C | A | G | Total | |
| −15 | T | 110 | 63 | 95 | 76 | 344 | A | 15 | 8 | 22 | 11 | 56 |
| −14 | A | 98 | 66 | 105 | 75 | 344 | C | 16 | 20 | 9 | 11 | 56 |
| −13 | A | 85 | 66 | 129 | 64 | 344 | C | 7 | 29 | 12 | 8 | 56 |
| −12 | A | 95 | 50 | 129 | 70 | 344 | A | 14 | 11 | 23 | 8 | 56 |
| −11 | T | 105 | 65 | 104 | 70 | 344 | A | 10 | 4 | 32 | 10 | 56 |
| −10 | T | 112 | 46 | 107 | 79 | 344 | A | 11 | 10 | 26 | 9 | 56 |
| −9 | T | 107 | 71 | 94 | 72 | 344 | A | 17 | 12 | 18 | 9 | 56 |
| −8 | T | 123 | 64 | 87 | 70 | 344 | T | 17 | 12 | 15 | 12 | 56 |
| −7 | T | 111 | 76 | 90 | 67 | 344 | A | 11 | 15 | 16 | 14 | 56 |
| −6 | T | 123 | 67 | 87 | 67 | 344 | A | 12 | 14 | 16 | 14 | 56 |
| −5 | T | 132 | 60 | 101 | 51 | 344 | T | 19 | 12 | 13 | 12 | 56 |
| −4 | T | 125 | 62 | 97 | 59 | 343 | T | 24 | 13 | 12 | 7 | 56 |
| −3 | T | 131 | 47 | 115 | 51 | 344 | T | 22 | 9 | 15 | 10 | 56 |
| −2 | T | 207 | 34 | 60 | 43 | 344 | T | 43 | 2 | 4 | 7 | 56 |
| −1 | T | 210 | 37 | 71 | 26 | 344 | T | 37 | 10 | 5 | 4 | 56 |
| Comp., % (top) | 36.3 | 16.9 | 28.5 | 18.2 | 32.8 | 21.6 | 28.3 | 17.4 | ||||
| Comp., % (bot.) | 23.4 | 12.8 | 46.1 | 17.8 | 21.0 | 12.5 | 47.7 | 18.9 | ||||
| 1 | A | 30 | 3 | 279 | 32 | 344 | A | 2 | 0 | 51 | 3 | 56 |
| 2 | A | 20 | 4 | 248 | 72 | 344 | A | 1 | 0 | 48 | 7 | 56 |
| 3 | A | 25 | 9 | 238 | 72 | 344 | A | 2 | 1 | 38 | 15 | 56 |
| 4 | A | 43 | 14 | 241 | 46 | 344 | A | 2 | 1 | 46 | 7 | 56 |
| 5 | A | 65 | 38 | 184 | 57 | 344 | A | 6 | 8 | 34 | 8 | 56 |
| 6 | T | 116 | 60 | 114 | 54 | 344 | A | 18 | 6 | 20 | 12 | 56 |
| 7 | A | 97 | 60 | 127 | 60 | 344 | A | 13 | 8 | 20 | 15 | 56 |
| 8 | A | 105 | 49 | 116 | 74 | 344 | A | 14 | 5 | 24 | 13 | 56 |
| 9 | A | 105 | 61 | 107 | 71 | 344 | A | 12 | 6 | 23 | 15 | 56 |
| 10 | A | 90 | 59 | 125 | 70 | 344 | T | 20 | 13 | 12 | 11 | 56 |
| 11 | A | 91 | 53 | 106 | 61 | 311 | A | 15 | 14 | 16 | 7 | 52 |
| 12 | A | 80 | 55 | 96 | 52 | 283 | A | 13 | 13 | 14 | 10 | 50 |
| 13 | A | 80 | 40 | 91 | 37 | 248 | T | 15 | 12 | 14 | 7 | 48 |
| 14 | T | 66 | 38 | 62 | 37 | 203 | T | 17 | 5 | 11 | 9 | 42 |
| 15 | T | 56 | 29 | 44 | 30 | 159 | A | 4 | 8 | 10 | 9 | 31 |
| 16 | T | 34 | 25 | 28 | 20 | 107 | T | 8 | 1 | 4 | 3 | 16 |
| 17 | T | 14 | 12 | 10 | 11 | 47 | T | 5 | 1 | 3 | 0 | 9 |
| 18 | A | 5 | 6 | 12 | 3 | 26 | T | 3 | 0 | 1 | 2 | 6 |
| 19 | T | 7 | 3 | 0 | 1 | 11 | T | 2 | 1 | 1 | 0 | 4 |
| 20 | T | 3 | 1 | 3 | 0 | 7 | A | 0 | 0 | 4 | 0 | 4 |
5′ ends of flanking repeats start at position 1. Regions 5′ adjacent to the flanking repeats are indicated by negative numbers (column 1). Columns 2–13 show the most abundant bases (P), frequencies of T, C, A, G, and the total numbers of bases at the corresponding positions in Alu and ID flanking repeats. Base compositions (Comp.) of flanking repeats (bot.) and of the 5′ adjacent regions (top) are listed in the middle of the table.
Table 2.
Base occurrences at different positions of flanking repeats and of the 3′ adjacent regions
| N |
Alu
|
ID
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | T | C | A | G | Total | P | T | C | A | G | Total | |
| −20 | A | 0 | 0 | 6 | 1 | 7 | A | 0 | 0 | 4 | 0 | 4 |
| −19 | A | 0 | 0 | 11 | 0 | 11 | A | 0 | 0 | 3 | 1 | 4 |
| −18 | A | 2 | 1 | 20 | 3 | 26 | A | 0 | 0 | 6 | 0 | 6 |
| −17 | A | 4 | 0 | 36 | 7 | 47 | A | 0 | 0 | 9 | 0 | 9 |
| −16 | A | 8 | 0 | 82 | 17 | 107 | A | 0 | 1 | 13 | 2 | 16 |
| −15 | A | 8 | 2 | 127 | 22 | 159 | A | 1 | 0 | 26 | 4 | 31 |
| −14 | A | 10 | 1 | 164 | 28 | 203 | A | 2 | 0 | 35 | 5 | 42 |
| −13 | A | 24 | 2 | 186 | 36 | 248 | A | 2 | 0 | 34 | 12 | 48 |
| −12 | A | 45 | 10 | 190 | 38 | 283 | A | 4 | 0 | 42 | 4 | 50 |
| −11 | A | 66 | 20 | 173 | 52 | 311 | A | 8 | 3 | 32 | 9 | 52 |
| −10 | A | 79 | 42 | 162 | 61 | 344 | A | 12 | 9 | 20 | 15 | 56 |
| −9 | A | 72 | 60 | 137 | 75 | 344 | A | 12 | 12 | 21 | 11 | 56 |
| −8 | A | 80 | 58 | 118 | 88 | 344 | A | 12 | 6 | 22 | 16 | 56 |
| −7 | A | 79 | 72 | 135 | 58 | 344 | A | 12 | 5 | 28 | 11 | 56 |
| −6 | A | 87 | 40 | 146 | 71 | 344 | A | 16 | 9 | 21 | 10 | 56 |
| −5 | A | 92 | 61 | 124 | 67 | 344 | A | 14 | 11 | 16 | 15 | 56 |
| −4 | T | 137 | 43 | 119 | 45 | 344 | T | 26 | 8 | 14 | 8 | 56 |
| −3 | T | 118 | 72 | 96 | 58 | 344 | C | 14 | 17 | 11 | 14 | 56 |
| −2 | T | 137 | 90 | 49 | 68 | 344 | T | 21 | 16 | 9 | 10 | 56 |
| −1 | A | 85 | 46 | 149 | 64 | 344 | A | 19 | 7 | 21 | 9 | 56 |
| Comp., % (top) | 23.4 | 12.8 | 46.1 | 17.8 | 21.0 | 12.5 | 47.7 | 18.9 | ||||
| Comp., % (bot.) | 14.3 | 7.8 | 28.2 | 10.9 | 27.4 | 18.4 | 28.6 | 25.7 | ||||
| 1 | A | 48 | 30 | 169 | 94 | 341 | A | 9 | 5 | 21 | 21 | 56 |
| 2 | A | 92 | 53 | 149 | 47 | 341 | A | 14 | 14 | 21 | 7 | 56 |
| 3 | A | 83 | 68 | 104 | 84 | 339 | G | 14 | 14 | 13 | 15 | 56 |
| 4 | A | 98 | 52 | 115 | 71 | 336 | T | 20 | 8 | 14 | 14 | 56 |
| 5 | A | 101 | 50 | 109 | 73 | 333 | A | 13 | 12 | 18 | 13 | 56 |
| 6 | T | 116 | 69 | 81 | 66 | 332 | G | 15 | 8 | 15 | 18 | 56 |
| 7 | A | 94 | 56 | 120 | 62 | 332 | G | 15 | 10 | 12 | 18 | 55 |
| 8 | A | 91 | 79 | 100 | 61 | 331 | A | 16 | 8 | 17 | 14 | 55 |
| 9 | A | 66 | 80 | 102 | 83 | 331 | T | 21 | 6 | 17 | 11 | 55 |
| 10 | A | 91 | 70 | 116 | 54 | 331 | A | 11 | 14 | 15 | 15 | 55 |
| 11 | A | 91 | 71 | 99 | 70 | 331 | T | 19 | 14 | 10 | 11 | 54 |
| 12 | T | 99 | 67 | 99 | 66 | 331 | T | 16 | 14 | 10 | 14 | 54 |
| 13 | T | 104 | 60 | 101 | 65 | 330 | A | 15 | 8 | 18 | 13 | 54 |
| 14 | A | 84 | 71 | 97 | 77 | 329 | T | 16 | 6 | 16 | 16 | 54 |
| 15 | A | 86 | 61 | 111 | 71 | 329 | A | 12 | 11 | 19 | 12 | 54 |
All 3′ ends of the flanking repeats correspond to position −1. Regions 3′ adjacent to the flanking repeats start at position 1 and are indicated by positive numbers (column 1). Columns 2–13 and the base compositions in the middle of the table are analogous to those in table 1.
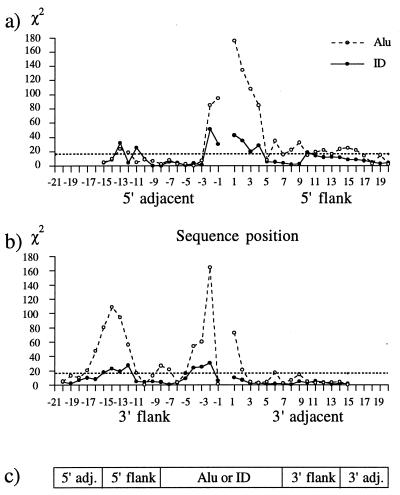
Qualitatively, the most striking pattern emerging from Table 1 is a previously unreported relative abundance of T in the 5′ adjacent regions (columns 3 and 9), particularly at positions −2 and −1 immediately preceding the 5′ flanking repeats of both Alu and ID retroposed elements. In contrast, the 5′ flanking repeats starting at position 1 begin with a very high content of A, particularly at positions 1–4, as observed before (5, 6). It suggests that the preferred 5′ nicking site is either between 5′ TT and AAAA or 3′ AA and TTTT in the complementary strand. This consensus pattern has been verified by a χ2 test that compares observed base occurrences at individual positions listed in Table 1 with the expected ones based on the overall base composition of the 5′ flanking repeats and of the adjacent regions listed in the middle of Table 1, as explained in the legend of Fig. 1. The χ2 values are plotted in Fig. 1a and are compared against the reference value 16.27 indicated by a broken horizontal line, which corresponds to P = 0.001. The χ2 values at positions −2 through +4 discussed above are significant at the 0.001 level in both Alu and ID retroposons.
Figure 1.
χ2 values for individual positions of Alu and ID flanking repeats and of adjacent regions. χ2 = Σi = 14(Oi − Ei)2/Ei; Ei = (Total) × (Compositioni), where individual base occurrences (Oi), total numbers of bases at different positions (Total), and the base compositions (Comp.) for the top and bottom part of the figure are from Tables 1 and 2, respectively. χ2 values above the broken horizontal line correspond to significance levels of P < 0.001 for 3 degrees of freedom. The discrete χ2 values were connected by lines to improve the presentation. (a) A cluster of significant χ2 values occurs at positions −2 through +4 around the 5′ ends of flanking repeats. (b) Significant χ2 values are near the 3′ ends of flanking repeats at positions −4, −3, and −2 immediately preceding the presumed antisense nicking site which is between positions −1 and +1. The significant nonrandomness around positions −11 through −16 corresponds to the 5′ end of flanking repeats presented above. Significant χ2 values not shared by Alu and ID flanking repeats are not considered here. (c) The overall scheme indicating mutual orientation of retroposed elements, flanking repeats, and adjacent regions.
Table 3 shows overall frequencies of different types of hexanucleotides located at statistically significant positions −2 through +4 (see Table 1 and Fig. 1a), in both Alu and ID sequences. The most abundant among them is TTAAAA and its six variants differing by one transition-type mutation (TTAAGA, TTAGAA, TTGAAA, TTAAAG, CTAAAA, and TCAAAA). They represent over 41.5% of all hexamers from Table 3. Over 20% is contributed by hexanucleotides differing from the above TTAAAA consensus and its six variants by an additional base substitution. In general, as the similarity to TTAAAA and its six variants goes down so does the occurrence of different hexanucleotides at the nicking site. The most diverse are hexanucleotides that occur once or twice, and they represent a little over one-third of the total number of hexamers from Table 3.
Table 3.
Overall frequency of hexanucleotides around primary nicking sites associated with 400 Alu and ID sequences
| Frequency | Hexamer(s) |
|---|---|
| 53 | TTAAAA |
| 38 | TTAAGA |
| 32 | TTAGAA |
| 17 | TTGAAA |
| 11 | TTAAAG |
| 10 | CTAAAA |
| 9 | TCAAGA |
| 8 | AAAAAA |
| 7 | TTTAAA |
| 6 | TAAAAA, GTAAGA |
| 5 | TTAAAT, TCAGAA, TCAAAA, GTAAAG |
| 4 | TTAAGT, TCTAAA, GTAGAA, GTAAAA, ATAGAA, AAAAAT |
| 3 | TTAACA, TGAAAA, TAAAAG, GTGAAA, CTAAAG, CCAAAA |
| 2 | TTTAAT, TTATAA, TTAGAT, TTAGAG, TGAGAA, TGAAAT, TGAAAG, TCAAAT, TAAGAA, GGAAAA, GAAAAT, GAAAAG, GAAAAA, CTAAGA, CAAAAA, ATAAGA, ATAAAA, AGAAGT, AGAAAG, ACAAAA, AAAGTT, AAAATC, AAAATA, AAAAAG |
| 1 | TTTTTT, TTTTTC, TTTTAA, TTTGAT, TTTAGA, TTGGAA, TTGAGG, TTATGT, TTAGGA, TTAATT, TTAATC, TTAAAC, TGTTCA, TGTATA, TGGTGA, TGAAGA, TCAGAG, TCAACA, TCAAAG, TCAAAC, TATTAA, TATAAA, TAGAAG, TAAGTG, TAAGGA, TAAGAG, TAAAAC, GTTAAA, GTGAGA, GTAGAC, GTAATG, GGGGGT, GGAGGA, GGAGAA, GGACTG, GGAAAG, GGAAAC, GCAGTT, GCAGAA, GCAAGA, GCAAAG, GCAAAA, GATGCT, GAATGC, GAAGAG, GAAAGT, GAAAAC, CTCAAG, CTATAA, CTAGAA, CTAAGC, CCTGTG, CCATTA, CCATAA, CCAATG, CCAATA, CCAAGA, CATAAA, CAGAAG, CACAGA, CAATTA, CAAGAG, CAAACT, ATTTCA, ATGAGC, ATGAGA, ATGAAA, AGTTTT, AGTAAA, AGCATA, AGAAGA, AGAAAA, ACTAAA, ACAGAA, AATTTT, AATTAC, AAGGGG, AAGCTT, AAGATG, AAGAGT, AAGAAA, AAATGT, AAATCT, AAATAA, AAAGTA, AAAGCT, AAAGAG, AAAGAA, AAACAT, AAACAC, AAAATG, AAAAGT, AAAAGG |
All hexamers include two bases preceding and four bases following the 5′ ends of the flanking repeats.
As illustrated in Table 2 and Fig. 1b, the 3′ ends of flanking repeats also show nonrandom base occurrences at positions −4, −3, and −2 preceding the other nicking site. In particular, all three positions are pyrimidine-enriched, specifically T-enriched, in Alu and ID flanking repeats. The minimum consensus sequence shared by the 3′ ends of flanking repeats in both families of retroposed elements is 5′-TYTN-3′, where Y denotes pyrimidine; T, thymine; and N, any base. The base distribution at position −1 and at the following two positions in the 3′ adjacent regions may also be nonrandom, but it is significant at the 0.001 level only in Alu repeats.
It must be emphasized here that the consensus sequences only summarize the base occurrences from Tables 1 and 2 and should not be viewed as the entire representation of the underlying nonrandomness. For example, the 5′ TYTN consensus described above does not reflect the relative deficiency of A at position −2 (see Table 2) or differences in the relative proportions of T and C at positions −2, −3, and −4. These and similar factors must be included in future comparative analyses of the sequence signals associated with retroposons.
Flanking Repeats 4–9 bp Long.
The distinction between flanking repeats at least 10 bp long and the shorter ones is somewhat arbitrary, but it quite naturally follows the length distribution of Alu and ID flanking repeats as illustrated in Table 4. The distribution is clearly bimodal in the case of ID elements, which have numerous flanking repeats under 8 bp and over 9 bp long, but none exactly 8 or 9 bp long. The distinction between short and long flanking repeats is less sharp in the case of Alu, but it follows a similar pattern with a shallow minimum around the same length range.
Table 4.
Length distribution of Alu and ID flanking repeats
| Length | Frequency
|
Length | Frequency
|
||
|---|---|---|---|---|---|
| Alu | ID | Alu | ID | ||
| 4 | 142 | 30 | 16 | 60 | 7 |
| 5 | 88 | 10 | 17 | 21 | 3 |
| 6 | 45 | 7 | 18 | 15 | 2 |
| 7 | 24 | 3 | 19 | 4 | 0 |
| 8 | 27 | 0 | 20 | 4 | 1 |
| 9 | 30 | 0 | 21 | 1 | 1 |
| 10 | 33 | 4 | 22 | 0 | 0 |
| 11 | 28 | 2 | 23 | 2 | 0 |
| 12 | 35 | 2 | 24 | 0 | 1 |
| 13 | 45 | 6 | 25 | 0 | 0 |
| 14 | 44 | 11 | 26 | 0 | 1 |
| 15 | 52 | 15 | |||
The frequencies of flanking repeats over 9 bp long grow steadily with length and reach a maximum at 15 bp in ID and 16 bp in Alu elements. After that they abruptly decline, indicating some restrictions on flanking repeats over 15–16 bp long. Analogous length limitations over 15 bp were previously reported in non-SINE retroposons (5) (SINE, short interspersed element).
The abundance of flanking repeats under 7 bp may be artifactual, since many flanks may be incomplete, or even entirely missing, at either end of the inserted element. This is compounded by the growing chance of a random match between oligonucleotides as their length goes down. It is estimated from random simulations (data not shown) that ≈56% of 4-bp “flanking repeats” may come from coincidental matches.
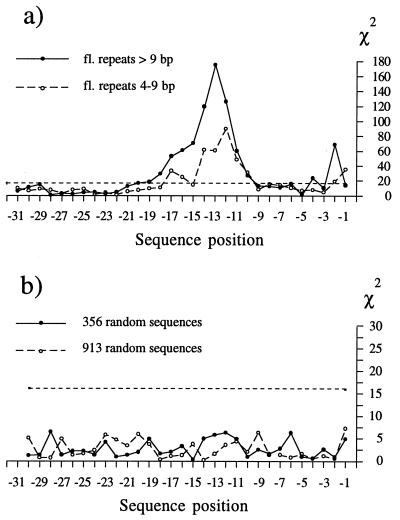
If at least some flanking repeats merely appear to be shorter because their matching copies are either truncated or missing, then the complete copies with the TTAAAA-like nicking signals may still be abundant in the regions preceding or following the retroposons. Fig. 2 compares two groups of 30-bp-long sequence regions immediately preceding 5′ ends of complete Alu elements, using a χ2 test as described for Fig. 1. The first group of 30-bp-long segments includes 344 long flanking repeats discussed in the previous section, and the second one includes 356 shorter flanking repeats. In spite of their various lengths, the 5′ ends of flanking repeats were not adjusted in either group, which contributes to some “blurring” of the χ2 distribution relative to that in Fig. 1a. Nevertheless, the χ2 values go well above the random background in both groups, at positions −10 to −17 corresponding to the TTAAAA consensus signal. Smaller, but significant, χ2 values can also be seen around positions immediately preceding the 5′ ends of Alu retroposons and corresponding to the 3′ signal shown in Fig. 1b. This clearly indicates that many presumed short flanking repeats indeed represent fragments of longer flanks indistinguishable from those described in the previous section.
Figure 2.
χ2 values for individual positions of 30-bp regions preceding 5′ ends of Alu retroposons. The χ2 values were calculated and presented as explained in the legend of Fig. 1, except that flanking repeats and the adjacent regions have not been adjusted and the expected values were calculated using total base composition of the 30-bp sequence segments which include flanking repeats and adjacent regions. All 3′ ends of the 30-bp regions correspond to position −1, immediately preceding the 5′ ends of Alu sequences. (a) χ2 values for the 30-bp regions which include long (>9 bp) and short (4–9 bp) flanking repeats. (b) Analogous χ2 values of two randomly selected sets of 30-bp human sequences as described in Materials and Methods. The χ2 values for these random sets are well below the broken horizontal line, which corresponds to P = 0.001.
Flanking Repeats of Other Retroposed Pseudogenes.
Patterns observed for Alu and ID flanking repeats appear to be shared with B1 and B2 elements from rodents (12). Furthermore, 5′ ends of flanking repeats of recently retroposed rat and human L1 elements (refs. 13–15; see also compilation in ref. 16), and two of the three de novo retroposed mRNA pseudogenes in HeLa cells (17) begin within 5′ TTAAAA sequence or its most common variants listed in Table 3. As illustrated in Fig. 3, similar patterns can be observed in a large variety of processed pseudogenes from mammals. Furthermore, a single example of apoferritin pseudogene from frog, flanked by perfect direct repeats was reported (18); it begins with AAAA and is preceded by tc, thus producing a 5′ tcAAAA pattern at positions −2, +4. This pattern differs from the 5′ TTAAAA consensus signal by a single T → C substitution.
Figure 3.
Examples of direct repeats flanking diverse processed pseudogenes from GenBank 97.0. GenBank accession numbers are listed before each sequence. The flanking repeats are indicated in uppercase letters and the adjacent sequences in lowercase. The omitted portion of each pseudogene is marked by ∼. The pseudogenes listed are as follows: (I) human ferritin H, α-enolase, cytoplasmic 7SL RNA, thiopurine methyltransferase, β-tubulin, γ-actin-like, and chromosomal protein HMG-17; (II) rodent α-tubulin, δ-aminolevulinate dehydratase, cytoplasmic γ-actin, metallothionein 1, cellular tumor antigen p53, small nuclear RNA U3 (rat), small nuclear 4.5S RNA(I), small nuclear RNA U3 (mouse), ribosomal protein L35a-related, thymidine kinase, VH 7183 family for immunoglobulin heavy chain, 7SK RNA, cytochrome oxidase subunit VIa, and acyl-CoA-binding/diazepam binding inhibitor; (III) other mammals, rabbit short interspersed C repeat (SINE) and B2-like repeat from mink; and (IV) frog (Rana catesbeiana) apoferritin pseudogene.
The above examples indicate that at least one of the two patterns observed in Alu and ID flanking repeats can be found in most processed pseudogenes not only in mammals but also in amphibians, although more sequence data will be needed to substantiate the latter. This suggests a universal mechanism of retroposition in mammals and beyond.
DISCUSSION
The nonrandom distribution of bases near both ends of Alu and ID flanking repeats provides a strong argument in support of enzymatic involvement in the generation of the staggered nicks prior to retroposition. Enzymatic nicking is an important first step leading to reverse transcription and integration of R2 retroposed elements in insects (19). It is proposed here that enzymatic nicking also takes place prior to the integration of mammalian retroposons and is guided by short sequence signals described above. Of the two, the 5′ consensus signal (5′-TTAAAA/3′-AATTTT) appears to be more robust statistically and is a likely target for the initial nicking prior to the reverse transcription. The position of the second nick may be determined not only by the sequence signal alone but also by its distance from the first nicking site which, in turn, may depend on the distance between the active sites in the hypothetical nicking enzyme. It can be speculated further that if the best signal is not found within the preferred distance, then a weaker signal is chosen. Conversely, if a strong nicking signal appears closer or further away than the preferred 15- to 16-bp distance it can be accommodated by the nicking enzyme within certain limits. This simple model can explain the variable lengths of flanking repeats and the relative weakness of the sequence patterns associated with the 3′ ends of flanking repeats.
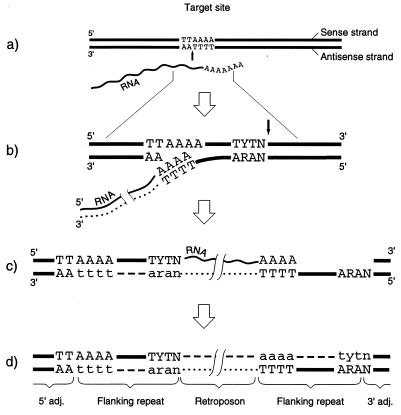
The existing model for RNA-mediated integration of R2 elements (20) does not account for the presence of flanking repeats. In a variant of this model, presented in Fig. 4, it is proposed that the antisense nick originally initiates RNA-dependent DNA polymerization (Fig. 4b), which is followed by the formation of the second nick toward the 3′ end of the sense strand. One possibility is that the formation of the second nick is associated with the ligation of the 5′ end of RNA to the exposed 3′ end of the sense strand. This could help to stabilize the transition from reverse transcription to DNA-dependent DNA polymerization and ligation of the antisense strand (Fig. 4c). However, alternative mechanisms involving specific proteins are also possible. The final stage includes elimination of the RNA and synthesis of the second DNA strand (Fig. 4d).
Figure 4.
Model for retroposon integration in mammals. (a) Enzymatic nicking in the presence of RNA indicated by a vertical black arrow. (b) Synthesis of cDNA, indicated by a dotted line, and formation of the second nick, indicated by a black arrow pointed down. (c) Completion of the reverse transcription and DNA-dependent DNA synthesis, indicated by a dashed line and the lowercase letters, followed by ligation. (d) Elimination of RNA and synthesis of the second DNA strand. Modified after Luan et al. (19).
While this paper was in review, I became aware of new evidence indicating that the reverse transcriptase encoded by the human L1 non-LTR (long terminal repeat) retrotransposon contains a domain homologous to the apurinic/apyrimidinic (AP) endonuclease family (16, 20). It is also capable of nicking DNA in vitro, primarily between runs of pyrimidines and purines in a very A+T-rich region, as shown in experiments with pBS plasmid DNA (16). Furthermore, the authors (16) compiled a list of additional L1 elements with identifiable flanking repeats and concluded that nicking signals in their flanking regions share common patterns with those identified by in vitro cleavage experiments. These conclusions complement independent observations made in this paper that the nicking signals associated with L1 elements follow the 5′-TTAAAA/3′-AATTTT pattern established for Alu and ID elements, which is similar but not identical to the consensus proposed by the authors (16). This adds significant weight to previous hypothesis, based on different premises, that Alu and other mammalian retroposons parasitize on the L1 retroposition machinery (21).
Similarity between AP endonucleases and the RNase H domains of certain reverse transcriptases from insects has also been reported (22), which indicates that the same mechanism for phosphodiester bond cleavage could be used in different steps involved in retroposon integration.
As indicated in Results, some 5′ hexamers from Table 3 do not seem to follow the 5′-TTAAAA consensus or any other significant sequence pattern. They represent about one-third of the studied sample. This leaves the possibility that significant numbers of retroposons still integrate at random chromosomal breaks as envisioned some time ago (23) and substantiated by recent experiments (24, 25). The majority, however, appear to be associated with specific targets, which may explain both frequent clustering and head-to-tail orientation of retroposons (7).
Generation of staggered breaks by an endogenous enzyme at nonrandom targets, combined with homologous recombination (26), can potentially lead to improved targeting of extrachromosomal DNA to predetermined chromosomal sites in mammals.
Acknowledgments
I thank Dr. Thomas Eickbush for helpful suggestions on retroposon integration mechanisms, Drs. Jef Boeke and Haig Kazazian and their coworkers for providing me with their manuscripts prior to submission, Paul Klonowski for skillful computer assistance, and Jolanta Walichiewicz for help with the preparation of this manuscript. This work was supported by the U.S. Department of Energy, Grant DE-FG0395ER62139.
References
- 1.Weiner A M, Deininger P L, Efstratiadis A. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 2.Deininger P L. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 619–636. [Google Scholar]
- 3.Deininger P L, Batzer M A. In: Evolutionary Biology. Hecht M K, MacIntyre R J, Clegg M T, editors. Vol. 27. New York: Plenum; 1993. pp. 157–196. [Google Scholar]
- 4.Van Arsdell S W, Denison R A, Bernstein L B, Weiner A M. Cell. 1981;26:11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- 5.Moos M, Gallwitz D. EMBO J. 1983;2:757–761. doi: 10.1002/j.1460-2075.1983.tb01496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels G R, Deininger P L. Nucleic Acids Res. 1985;13:8939–8954. doi: 10.1093/nar/13.24.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurka J. In: Molecular Biology Intelligence Unit: The Impact of Short Interspersed Elements (SINEs) on the Host Genome. Maraia R, editor. Austin, TX: Landes; 1995. pp. 25–41. [Google Scholar]
- 8.Deininger P L, Batzer M A. In: Molecular Biology Intelligence Unit: The Impact of Short Interspersed Elements (SINEs) on the Host Genome. Maraia R, editor. Austin, TX: Landes; 1995. pp. 43–60. [Google Scholar]
- 9.Jurka J, Klonowski P, Dagman V, Pelton P. Comput Chem. 1996;20:119–122. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 10.Smith T F, Waterman M J. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 11.Faulkner D V, Jurka J. Trends Biochem Sci. 1988;13:321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 12.Jurka J, Klonowski P. J Mol Evol. 1996;43:685–689. doi: 10.1007/BF02202117. [DOI] [PubMed] [Google Scholar]
- 13.Furano A V, Somerville C C, Tsichlis P N, D’Ambrosio E. Nucleic Acids Res. 1986;14:3717–3727. doi: 10.1093/nar/14.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods-Samuels P, Wong C, Mathias S L, Scott A F, Kazazian H H, Jr, Antonorakis S. Genomics. 1989;4:290–296. doi: 10.1016/0888-7543(89)90332-7. [DOI] [PubMed] [Google Scholar]
- 15.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler K W, Vogelstein B, Nakamura Y. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 16.Feng Q, Moran J V, Kazazian H H, Boeke J D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 17.Maestre J, Tchenio T, Dhellin O, Heidmann T. EMBO J. 1995;24:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickey L F, Sreedharan S, Theil E C, Didsbury J R, Wang Y-H, Kaufman R E. J Biol Chem. 1987;262:7901–7907. [PubMed] [Google Scholar]
- 19.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 20.Martin F, Olivares M, Lopez M C, Alonso C. Trends Biochem Sci. 1996;21:283–285. [PubMed] [Google Scholar]
- 21.Smit A F A, Toth G, Riggs A D, Jurka J. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 22.Barzilay G, Hickson I D. BioEssays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 23.Hutchison C A, Hardies S C, Loeb D D, Shehee W R, Edgell M H. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 593–617. [Google Scholar]
- 24.Teng S-C, Kim B, Gabriel A. Nature (London) 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 25.Moore J K, Haber J E. Nature (London) 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 26.Rouet P, Smih F, Jasin M. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]