Abstract
Aim
In an effort to identify patients with uveal melanoma at high risk of metastasis, the authors undertook correlation of gene expression profiles with histopathology data and tumour‐related mortality.
Methods
The RNA was isolated from 27 samples of uveal melanoma from patients who had consented to undergo enucleation, and transcripts profiled using a cDNA array comprised of sequence‐verified cDNA clones representing approximately 4000 genes implicated in cancer development. Two multivariate data mining techniques—hierarchical cluster analysis and multidimensional scaling—were used to investigate the grouping structure in the gene expression data. Cluster analysis was performed with a subset of 10 000 randomly selected genes and the cumulative contribution of all the genes in making the correct grouping was recorded.
Results
Hierarchical cluster analysis and multidimensional scaling revealed two distinct classes. When correlated with the data on metastasis, the two molecular classes corresponded very well to the survival data for the 27 patients. Thirty two discrete genes (corresponding to 44 probe sets) that correctly defined the molecular classes were selected. A single gene (ectonucleotide pyrophosphatase/phosphodiesterase 2; autotaxin) could classify the molecular types. The expression pattern was confirmed using real‐time quantitative PCR.
Conclusions
Gene expression profiling identifies two distinct prognostic classes of uveal melanoma. Underexpression of autotaxin in class 2 uveal melanoma with a poor prognosis needs to be explored further.
About 5% of all melanomas arise from the ocular and adnexal structures and uveal melanoma is the most common primary intraocular malignant tumour with an incidence of approximately 4.3 cases per million per year in the USA.1,2 Although, cutaneous and uveal melanocytes share a common embryologic origin, uveal melanoma and cutaneous melanoma have many differing clinical, epidemiological and prognostic features.3,4
The five‐year survival rates following enucleation, brachytherapy and other methods of treatment of primary uveal melanoma range from 6%–53% depending upon the size of the tumour.5,6,7,8,9,10 Despite achieving great accuracy in diagnosing uveal melanoma,11 mortality from uveal melanoma in the USA has remained unchanged over a period of 25 years from 1973 to 1997.12
Recent studies have identified specific chromosomal changes of prognostic significance such as monosomy 3, alteration of 8q, and 6p13,14,15,16 but the genetic mechanisms involved in the pathogenesis of uveal melanoma remain unknown. Microarray gene profiling of uveal melanoma tumour samples17 and cell cultures18 has revealed two subtypes of uveal melanoma. One previous study has suggested a correlation between molecular classes of uveal melanoma and survival.19 To get a better insight into genes that are involved in uveal melanoma tumorigenesis and metastasis, we analysed gene expression profiles in 27 patients with uveal melanoma with known survival outcome.
Materials and methods
Tumour collection
The tumour specimens were collected from the eyes of patients undergoing enucleation. The tumour samples were snap frozen in liquid nitrogen at the point of collection, and subsequently transferred to −180°C for long‐term storage.
Clinical data
Twenty seven patients with medium to large sized uveal melanoma were included in the study following approval by the institutional review board. The clinical details (age, sex), tumour features (location, largest basal diameter, height), histopathological features (per cent epithelioid component, presence or absence of matrix patterns), and outcome (follow‐up duration, survival status (alive, dead due to metastasis, dead due to other causes)) were recorded for each patient. On the largest tumour face, a periodic acid‐Schiff stain was carried out, without counterstain, to assess the tumour extracellular matrix patterns. Nine morphological patterns of extracellular matrix deposition have been defined for ciliary body or choroidal melanomas.20,21 The presence of extracellular closed loops and networks (a network is defined as at least three back‐to‐back closed loops), is the feature strongly associated with death from metastatic disease.20,21
RNA isolation and target RNA preparation
RNA was isolated from uveal melanoma tumour samples using the RNAqueous kit (Ambion) as described by the manufacturer. Following isolation, the RNA was DNAse treated using the reagents from the DNA‐free kit (Ambion, Foster City, USA), again as described by the manufacturer. The quality of the isolated RNA was verified using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, USA). Target RNA was generated in a T7 polymerase‐based linear amplification reaction using a modified version of a published protocol.22 Two micrograms of total RNA and 5 pmol of T7‐(dT)24 primer [5′‐GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG‐(dT)24‐3′] in a total volume of 5.5 μl was incubated at 70°C for 10 min and chilled on ice. For first‐strand cDNA synthesis, the annealed RNA template was incubated for 1 h at 42°C in a 10 μl reaction mixture containing first‐strand buffer (Invitrogen, Carlsbad, CA, USA), 10 mM dithiothreitol, 1 unit of anti‐RNase (Ambion) per μl, 500 μM deoxynucleoside triphosphates, and 2 units of Superscript II (Invitrogen) per μl. Second‐strand synthesis was for 2 h at 16°C in a total reaction volume of 50 μl containing first‐strand reaction products, second‐strand buffer (Invitrogen), 250 μM deoxynucleoside triphosphates, 0.06 unit of DNA ligase (Ambion) per μl, 0.26 unit of DNA polymerase I (New England Biolabs, Ipswich, USA) per μl, and 0.012 U of RNase H (Ambion) per µl followed by the addition of 3.3 units of T4 DNA polymerase (3 units per µl; New England Biolabs) and a further 15 min of incubation at 16°C. Second‐strand reaction products were purified by phenol‐chloroform‐isoamyl alcohol extraction in Phaselock microcentrifuge tubes (Eppendorf, Westbury, NY, USA) according to the manufacturer's instructions and ethanol precipitated. In vitro transcription was performed by using the T7 megascript kit (Ambion) according to a modified protocol in which purified cDNA was combined with 1 μl (each) of 10× ATP, GTP, CTP, and UTP and 1 μl of T7 enzyme mix in a 10 μl reaction volume and incubated for 9 h at 37°C. Amplified RNA was purified by using the RNAeasy RNA purification kit (QIAGEN, Valencia, USA) according to the manufacturer's instructions.
cDNA microarray construction
The tumour cDNA array used in this study comprised a subset of sequence‐verified cDNA clones from Research Genetics Inc (Carlsbad, USA), a 40 000‐clone set representing approximately 4000 genes involved in tumorigenesis.23,24,25 The 4000 cDNAs were selected based on their implication in metastasis and cancer development in general from the literature and from the Affymetrix cancer G110 array, the presence of AU‐rich elements in their 3′‐ and/or 5′‐untranslated region25 as well as encoding proteins responsive to cytokines, containing a zinc‐finger or being implicated in apoptosis. The complete list of spotted cDNAs on this tumour array can be downloaded from the site http://geacf.cwru.edu/geacf/geaaspotteddescriptions.shtml. DNA preparation and slide printing were as previously described, except for the use of 40% dimethyl sulfoxide in place of 1.5× SSC as the printing solution.25
RNA labelling
Cy3‐ or Cy5‐labeled cDNA was prepared by indirect incorporation. Two micrograms of amplified RNA, 1 µl of dT12‐18 primer (1 µg per µl; Invitrogen), 2.6 µl of random hexanucleotides (3 µg per µl; Invitrogen), and 1 µl of anti‐RNase (Ambion) were combined in a reaction volume of 15.5 µl and incubated for 10 min at 70°C. Reverse transcription was for 2 h at 42°C in a 30 µl reaction mixture containing annealed RNA template, first‐strand buffer, 500 µM (each) dATP, dCTP, and dGTP, 300 µM dTTP, 200 µM aminoallyl‐dUTP (Sigma), 10 mM dithiothreitol, and 12.7 units of Superscript II per µl. For template hydrolysis, 10 µl of 0.1 M NaOH was added to the reverse transcription reaction mixture and the mixture was incubated for 10 min at 70°C, allowed to cool at room temperature for 5 min, and neutralised by the addition of 10 µl of 0.1 M HCl. cDNA was precipitated at −20°C for 30 min after the addition of 1 µl of linear acrylamide (Ambion), 4 µl of 3 M sodium acetate (pH 5.2), and 100 µl of absolute ethanol, and then resuspended in 5 µl of 0.1 M NaHCO3. For dye coupling, the contents of one tube of N‐hydroxysuccinimide ester containing Cy3 or Cy5 dye (product numbers PA25001 and PA25002; GE Healthcare) was dissolved in 45 µl of dimethyl sulfoxide. Five µl of dye solution was mixed with the cDNA and incubated for 1 h in darkness at room temperature. Labelled cDNA was purified on a QIAquick PCR purification column (QIAGEN) according to the manufacturer's instructions. Eluted cDNA was dried under a vacuum and resuspended in 30 µl of Slidehyb II hybridisation buffer (Ambion). After 2 min of denaturation at 95°C, the hybridisation mixture was applied to the microarray slide under a coverslip. Hybridisation proceeded overnight in a sealed moist chamber in a 55°C water bath. After hybridisation, slides were washed successively for 5 min each in 2× SSC–0.1% SDS at 55°C, then in 2× SSC at 55°C, and finally in 0.2× SSC at room temperature.
Acquisition of data
Data were acquired with a GenePix 4000B laser scanner and GenePix Pro software, version 5.0, as previously described.22
Real‐time quantitative PCR
One μg of total RNA was reverse transcribed using random hexamers (Superscript first‐strand synthesis system; Invitrogen). Real‐time quantitative RTQ‐PCR was performed on the cDNA using a Taqman gene expression assay for autotaxin (Hs00196470 µm1) normalised with a eukaryotic 18S rRNA endogenous control assay (Applied Biosystems, Foster City, CA, USA). Reactions were performed in triplicate on a 7900HT Sequence Detection System instrument (Applied Biosystems) using Real Master Mix Probe ROX according to the manufacturers instructions (Eppendorf). Relative quantification was determined by comparison of each sample to a reference cDNA (Universal human reference RNA, Stratagene, La Jolla, CA).
Data analysis
Two multivariate data mining techniques, Hierarchical cluster analysis performed with 10 000 randomly selected subsets and multidimensional scaling,26 were used to investigate the grouping structure in the gene expression data of the 27 tumour samples. Pearson correlation coefficient was used as the proximity matrix27 in the hierarchical cluster analysis and in multidimensional scaling. Hierarchical cluster analysis with average linkage method26 was used to generate the dendrograms. Multidimensional scaling was also applied to investigate the grouping structure. Tumour metastatic survival data within molecular classes was correlated with Kaplan–Meier survival curves.
Results
Clinical aspects
Twenty seven patients were included in the study. The average age of the patients was 61 years, of whom 13 were female; 16 patients were dead by the follow‐up date. The clinical details, tumour features, histopathological features and outcome are summarised in table 1.
Table 1 Clinical and pathologic profile of 27 patients with uveal melanoma.
| No | Clinical | Tumour features | Outcome | |||||
|---|---|---|---|---|---|---|---|---|
| Age (years) | Sex | Location | LBD (mm) | Height (mm) | E (%) | Networks | ||
| 202 | 45 | Male | Choroid | 16 | 11 | 0 | Absent | Alive |
| 205 | 73 | Male | CBD + choroid | 14 | 8 | 70 | Present | Dead (Other) |
| 209 | 64 | Female | Choroid | 15 | 7 | 30 | Present | Dead (Mets) |
| 212 | 45 | Male | Choroid | 17 | 1 | 30 | Present | Dead (Mets) |
| 213 | 71 | Female | CBD + choroid | 17 | 13 | 40 | Absent | Dead (Mets) |
| 218 | 42 | Male | Choroid | 13 | 6 | 30 | Present | Dead (Mets) |
| 222 | 36 | Female | Choroid | 12 | 7 | 5 | Absent | Alive |
| 232 | 67 | Male | CBD + choroid | 12 | 12 | 40 | Present | Dead (Mets) |
| 234 | 69 | Male | CBD + choroid | 19 | 12 | 30 | Present | Dead (Mets) |
| 235 | 71 | Female | CBD + choroid | 14 | 14 | 10 | Present | Dead (Mets) |
| 236 | 63 | Male | CBD + choroid | 21 | 9 | 10 | Present | Dead (Mets) |
| 237 | 49 | Male | Choroid | 22 | 8 | 0 | Present | Dead (Mets) |
| 239 | 43 | Female | Choroid | 16 | 8 | 10 | Present | Dead (Mets) |
| 240 | 35 | Female | Choroid | 17 | 8 | 0 | Absent | Alive |
| 242 | 83 | Male | CBD + choroid | 11 | 10 | 30 | Present | Dead (Mets) |
| 245 | 44 | Male | Choroid | 12 | 12 | 0 | Absent | Alive |
| 249 | 66 | Male | Choroid | 9 | 10 | 0 | Absent | Alive |
| 250 | 60 | Female | Choroid | 19 | 13 | 10 | Absent | Dead (Mets) |
| 253 | 50 | Male | CBD + choroid | 16 | 12 | 0 | Absent | Alive |
| 255 | 63 | Female | CBD + choroid | 17 | 8 | 1 | Present | Dead (Mets) |
| 256 | 80 | Female | Choroid | 10 | 5 | 1 | Absent | Alive |
| 263 | 70 | Male | Choroid | 13 | 8 | 10 | Absent | Alive |
| 266 | 61 | Female | Choroid | 16 | 9 | 0 | Present | Alive |
| 267 | 49 | Female | Choroid | 17 | 9 | 0 | Absent | Alive |
| 268 | 78 | Female | Choroid | 17 | 9 | 40 | Present | Dead (Mets) |
| 280 | 85 | Male | Choroid | 18 | 15 | 20 | Absent | Dead (Mets) |
| 289 | 88 | Female | Choroid | 19 | 10 | 40 | Present | Dead (Mets) |
LBD, largest basal diameter; E%, percent epithelioid component; Networks, presence or absence of aggressive matrix patterns; CBD, ciliary body; Mets, metastasis.
Gene expression profile analysis
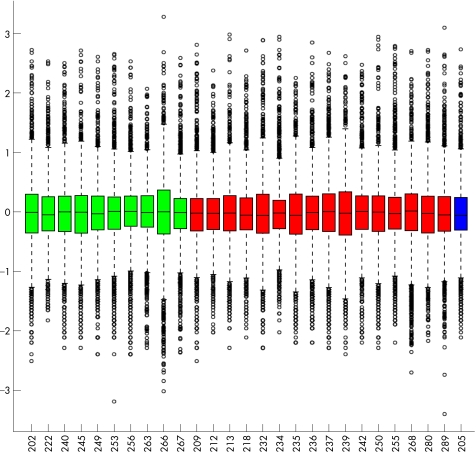
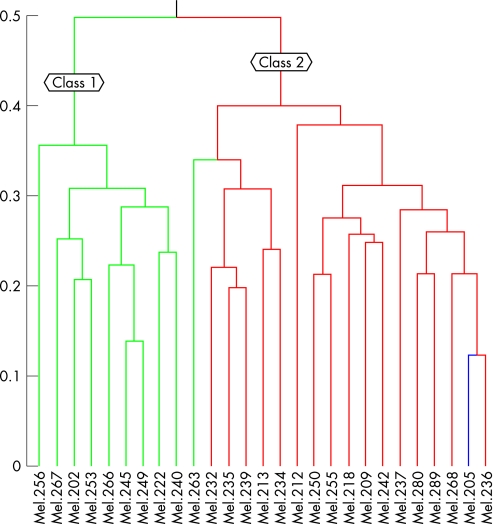
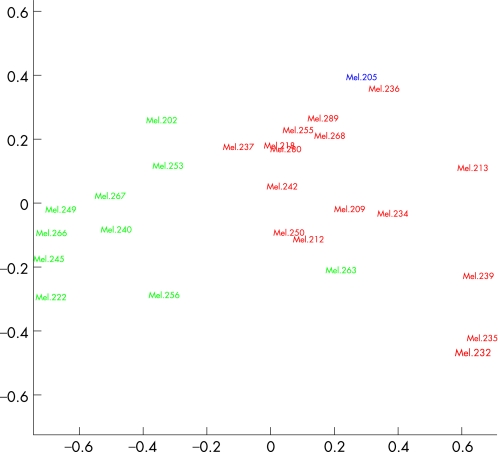
No obvious pitfalls were found when we looked at the overall gene expression profiles across the tumour samples, so we analysed all 27 samples provided (fig 1). Hierarchical cluster analysis revealed two distinct molecular classes (class 1 and class 2). Hierarchical cluster analysis with average linkage method was used to generate the dendrogram (fig 2). Multidimensional scaling analysis also revealed two molecular classes identical to those found with the hierarchical cluster analysis (fig 3).
Figure 1 Box plot of the 27 tumour samples. The colour indicates the metastatic death status (green for alive, red for dead, and blue for death from other cause).
Figure 2 Dendrogram from the hierarchical cluster analysis using Pearson correlation as distance matrix. Two molecular classes were found, the one in the green box corresponds to class 1 that contains all the patients alive and the other one in the red box corresponds to class 2 with all people dead except for one alive.
Figure 3 Plot of the multidimentional scaling with Pearson correlation as distance matrix. Two molecular classes, class 1 and class 2, are obvious and the colour indicates the metastatic death (green, alive; red, metastatic death; blue, one death of other cause).
Key genes that are in favour of the grouping structure
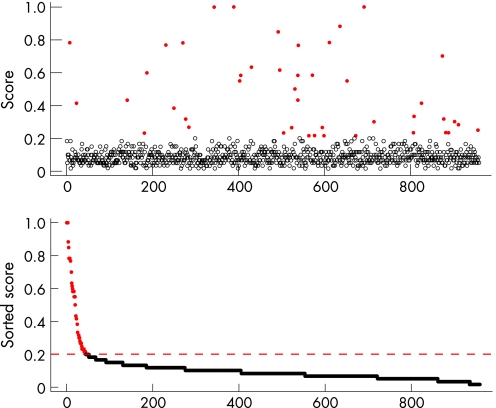
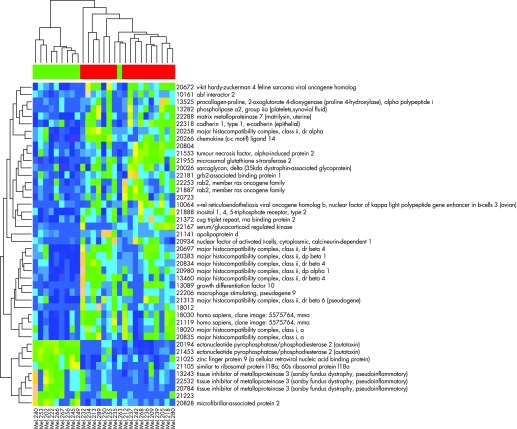
We next identified the genes that distinguished the two molecular classes. First, all genes were pre‐filtered and those with data missing were deleted. Wilcoxon rank sum test was performed on the remaining 3514 genes. Based on the p values from the Wilcoxon test, the adjusted p values for controlling the false discovery rate were calculated. We filtered out all the genes with false discovery rate adjusted p values greater than 0.1, leaving 972 genes. Second, cluster analysis was repeatedly performed with 10 000 randomly selected subset of genes, and the cumulative contributions of all the genes in making the correct grouping were recorded. The cumulative contribution (score) was plotted for all 972 genes and we selected the top 32 discrete genes (corresponding to 44 probe sets) with the highest classification contribution (fig 4; indicated as red dots). The 44 probe sets selected from the above algorithm are listed in table 2. The 44 probe sets are clustered into two major groups, with either high expression of molecular class 1 and low expression of molecular class 2, or low expression of class 1 and high expression of class 2 (fig 5).
Figure 4 Top key genes selected from the selection algorithm. Gene weights was the average gene selection rate, and 20% was used as the cutoff to select the key genes that were correctly defined the molecular class 1 and class 2.
Table 2 Summary of the properties of the 32 key genes (44 probe sets).
| Gene* | Score† | Mean ratio‡ | p Value§ | Common | Genbank | UniGene |
|---|---|---|---|---|---|---|
| 20026 | 1 | 1.6 | 9.40E‐03 | Sarcoglycan, delta (35 kda dystrophin‐associated glycoprotein) | AA234982 | Hs.387207 |
| 20194 | 1 | 0.22 | 4.30E‐07 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | T80232 | Hs.190977 |
| 21453 | 1 | 0.26 | 4.30E‐07 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | AA476508 | Hs.190977 |
| 21223 | 0.88 | 0.28 | 3.50E‐03 | AA775616 | Data not found | |
| 20672 | 0.85 | 1.5 | 1.10E‐02 | v‐kit hardy‐zuckerman 4 feline sarcoma viral oncogene homolog | N24824 | Hs.479754 |
| 10064 | 0.78 | 2 | 2.20E‐02 | v‐rel reticuloendotheliosis viral oncogene homolog b, nuclear factor of kappa light polypeptide gene enhancer in b‐cells 3 (avian) | AA258001 | Hs.307905 |
| 18012 | 0.78 | 2.2 | 2.10E‐04 | T63324 | Data not found | |
| 21141 | 0.78 | 2.4 | 6.40E‐03 | Apolipoprotein d | AA456975 | Hs.522555 |
| 13460 | 0.77 | 2 | 3.50E‐04 | Major histocompatibility complex, class ii, dr beta 4 | AA630549 | Hs.534322 |
| 20835 | 0.77 | 2.2 | 1.20E‐03 | Major histocompatibility complex, class i, a | AA644657 | Hs.181244 |
| 22167 | 0.7 | 3.6 | 2.40E‐03 | Serum/glucocorticoid regulated kinase | AA486082 | Hs.296323 |
| 20383 | 0.63 | 2 | 4.70E‐04 | Major histocompatibility complex, class ii, dp beta 1 | AA486627 | Hs.485130 |
| 20697 | 0.62 | 2 | 1.50E‐03 | Major histocompatibility complex, class ii, dr beta 4 | W88967 | Hs.534322 |
| 13282 | 0.6 | 2.1 | 2.20E‐03 | Phospholipase a2, group iia (platelets, synovial fluid) | T61323 | Hs.466804 |
| 20266 | 0.58 | 1.9 | 2.40E‐03 | Chemokine (c‐c motif) ligand 14 | R96668 | Hs.272493 |
| 20834 | 0.58 | 2.1 | 9.80E‐04 | Major histocompatibility complex, class ii, dr beta 4 | AA664195 | Hs.520049 |
| 20980 | 0.58 | 2.1 | 1.50E‐04 | Major histocompatibility complex, class ii, dp alpha 1 | AA634028 | Hs.347270 |
| 20258 | 0.55 | 1.8 | 5.10E‐03 | Major histocompatibility complex, class ii, dr alpha | R47979 | Hs.520048 |
| 21313 | 0.55 | 2 | 2.40E‐03 | Major histocompatibility complex, class ii, dr beta 6 (pseudogene) | H50623 | Hs.545653 |
| 20804 | 0.5 | 1.5 | 1.70E‐02 | H26426 | Data not found | |
| 13089 | 0.43 | 2 | 6.00E‐04 | Growth differentiation factor 10 | R52085 | Hs.2171 |
| 20828 | 0.43 | 0.51 | 1.20E‐02 | Microfibrillar‐associated protein 2 | N67487 | Hs.389137 |
| 10161 | 0.42 | 1.8 | 2.00E‐02 | abl interactor 2 | N21334 | Hs.471156 |
| 21955 | 0.42 | 1.9 | 4.70E‐04 | Microsomal glutathione s‐transferase 2 | W73474 | Hs.81874 |
| 13525 | 0.38 | 1.8 | 9.40E‐03 | Procollagen‐proline, 2‐oxoglutarate 4‐dioxygenase (proline 4‐hydroxylase), alpha polypeptide i | AA457671 | Hs.500047 |
| 21888 | 0.33 | 2.6 | 3.60E‐04 | Inositol 1,4,5‐triphosphate receptor, type 2 | R68021 | Hs.512235 |
| 18020 | 0.32 | 1.8 | 8.90E‐03 | Major histocompatibility complex, class i, a | AA644657 | Hs.181244 |
| 22181 | 0.32 | 1.4 | 1.60E‐02 | grb2‐associated binding protein 1 | R96595 | Hs.80720 |
| 21553 | 0.3 | 1.4 | 2.40E‐02 | Tumour necrosis factor, alpha‐induced protein 2 | W33183 | Hs.525607 |
| 22288 | 0.3 | 1.5 | 1.20E‐02 | Matrix metalloproteinase 7 (matrilysin, uterine) | AA031513 | Hs.2256 |
| 22318 | 0.28 | 1.6 | 1.50E‐02 | Cadherin 1, type 1, e‐cadherin (epithelial) | H97778 | Hs.461086 |
| 18030 | 0.27 | 2 | 6.20E‐03 | Homo sapiens, clone image: 5575764, mRNA | W60701 | Hs.519978 |
| 20784 | 0.27 | 0.51 | 1.20E‐02 | Tissue inhibitor of metalloproteinase 3 (sorsby fundus dystrophy, pseudoinflammatory) | AA099153 | Hs.297324 |
| 21105 | 0.27 | 0.42 | 1.30E‐05 | Similar to ribosomal protein l18a; 60s ribosomal protein l18a | W81118 | Hs.517776 |
| 22532 | 0.25 | 0.51 | 9.40E‐03 | Tissue inhibitor of metalloproteinase 3 (sorsby fundus dystrophy, pseudoinflammatory) | AA479202 | Hs.297324 |
| 13243 | 0.23 | 0.52 | 1.50E‐02 | Tissue inhibitor of metalloproteinase 3 (sorsby fundus dystrophy, pseudoinflammatory) | AA479202 | Hs.297324 |
| 20723 | 0.23 | 1.4 | 1.70E‐02 | AA455270 | Data not found | |
| 21887 | 0.23 | 1.3 | 2.10E‐02 | rab2, member ras oncogene family | T82415 | Hs.369017 |
| 22206 | 0.23 | 2.6 | 1.20E‐03 | Macrophage stimulating, pseudogene 9 | T51539 | Hs.475654 |
| 22253 | 0.23 | 1.4 | 2.90E‐03 | rab2, member ras oncogene family | R38260 | Hs.369017 |
| 20934 | 0.22 | 1.4 | 2.70E‐02 | Nuclear factor of activated t‐cells, cytoplasmic, calcineurin‐dependent 1 | AA679278 | Hs.534074 |
| 21025 | 0.22 | 0.44 | 5.40E‐05 | Zinc finger protein 9 (a cellular retroviral nucleic acid binding protein) | AA625995 | Hs.518249 |
| 21119 | 0.22 | 2.2 | 4.00E‐03 | Homo sapiens, clone image: 5575764, mRNA | W60701 | Hs.519978 |
| 21372 | 0.22 | 1.9 | 3.50E‐04 | cug triplet repeat, RNA binding protein 2 | T84491 | Hs.309288 |
*Gene number used in the array data.
†The average selection rate.
‡The ratio between the means of the two molecular classes.
§p Value from the Wilcoxon rank sum test on the two molecular classes.
All other columns are from the original data provided. Genes 20194 and 21453, which correspond to one unique gene alone, can classify the molecular classes detected using all the data.
Figure 5 Gene profiles of the 44 key genes across the 27 samples with the dendrogram from the samples on the top and the dendrogram from the 44 key genes on the side. Pink indicates the high expression and cerulean blue indicates low expression. The colour bar on the top indicates the metastatic death status—green for alive and red for dead. The 44 key genes are clustered into two major groups shown in the site dendrogram: one group has high expressions in molecular class 1 and low expressions in molecular class 2 and the other low in molecular class 1 and high in molecular class 2.
Correlation with survival data
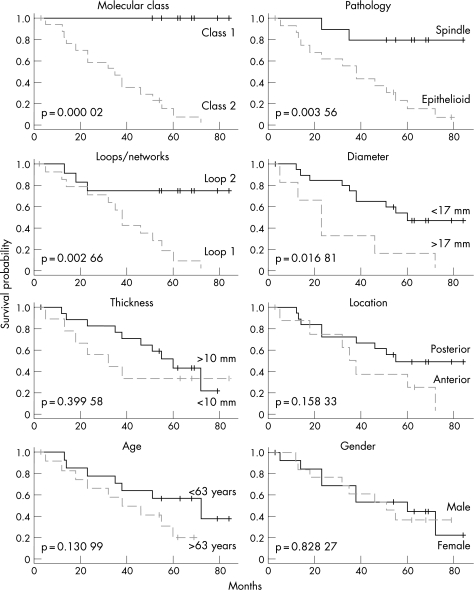
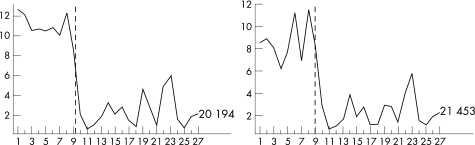
The two molecular classes corresponded very well to the Kaplan–Meier survival curves of the two classes. Class 1 had all nine patients alive and class 2 had 17 dead and one alive patient. The prognosis obtained with molecular classification was superior to other well known prognosticators such as tumour size, histopathology, location and extracellular matrix patterns (fig 6).28 Moreover, among the 32 key genes, gene expression pattern of autotaxin (ectonucleotide pyrophosphatase‐phosphodiesterase 2, ENPP2) alone was sufficient to distinguish molecular class 1 and class 2, where class 2 represents poor prognosis uveal melanoma (fig 7). The expression pattern of autotaxin was confirmed using real‐time quantitative PCR (in triplicate) and was highly concordant.
Figure 6 Kaplan–Meier survival analysis on molecular classes and seven histopathological and clinical measures. All mortality was melanoma metastasis. Statistical significance is based on log‐rank test and indicated in the plot for each parameter.
Figure 7 Individual profile for ENPP2 expression (autotaxin, 20194). The x axis corresponds to 27 samples that are grouped onto molecular class 1 and class 2, in that order, and the dashed vertical lines indicates the boundary between the two classes. Note lower expression in molecular class 2 sample.
Discussion
Uveal melanoma cells are shed into circulation at initial presentation and even in adequately treated cases.29 The fate of the uveal melanoma cells in the circulation and hence the features that influence their ability to establish metastasis determines the survival of patients with uveal melanoma. Microarray gene profiling is a powerful tool that can reveal such genetic attributes. In our study, cluster analysis performed with a subset of 10 000 randomly selected genes revealed 44 genes with the highest classification contribution and cumulative contribution (score). Overexpression of E‐cadherin was noted in class 2 uveal melanoma associated with poor prognosis. E‐cadherin is an important transmembrane protein which is indicative of epithelial differentiation.30 This attribute correlates with epithelial cell‐like (epithelioid) morphology of aggressive uveal melanoma.31 Although overexpression of E‐cadherin in class 2 uveal melanoma associated with poor prognosis initially appears to be paradoxical, E‐cadherin is also upregulated in other cancers that spread haematogenously, such as advanced cutaneous melanoma and hepatocellular carcinoma.32
In the circulation, uveal melanoma cells with high HLA class I expression enable the cells to evade natural killer cell‐mediated tumour surveillance.33 Therefore, it is not unexpected that several genes of the histocompatibility complex class I and class II were overexpressed in class 2 uveal melanoma associated with a poor prognosis. It is also likely that the presence of tumour‐infiltrating macrophages and lymphocytes, features of a poor prognosis for uveal melanoma,34 may be the reason for the observed overexpression of immune response genes.
Tissue inhibitor of metalloproteinase 3 (TIMP3) inhibits cell invasion and metastasis by inhibiting degradation of the extracellular matrix.35 TIMP3 also induces apoptosis36 and inhibits angiogenesis.37 Comparative expression profiling of metastasis in uveal melanoma cell lines and primary uveal melanoma cell lines revealed a fivefold lower expression of TIMP3 in metastasis cell lines.38 The lower expression of TIMP3 in class 2 uveal melanoma with a poor prognosis, as observed in our study, indicates pro‐angiogenic and anti‐apoptotic attributes of aggressive melanoma cells that favour establishment of tumour metastatic sites.
Results of gene expression profiling do not always generate reproducible or identical genetic profiles due to several factors including biological and technical variability (table 3). Previous studies have identified sets of 201 genes,17 7 genes,39 and 3 genes19 sufficient for accurate class prediction in uveal melanoma. It is remarkable that autotaxin was also one of the discriminating genes in two previous studies.19,39 In any event, it is prudent not to focus on one gene; the expression pattern of several genes should be considered for determining the expression profile.40 What is remarkable is the fact that non‐metastasising (class 1) and metastasising (class 2) uveal melanoma can be readily discriminated by their genetic profiles as determined by microarray techniques.
Table 3 Comparison of top discriminating genes from published studies that are underexpressed in class 2 (poor prognosis) uveal melanoma.
| Present study | Tschentscher17 | Onken19 | Worley39 |
|---|---|---|---|
| ENPP2 | CHL1 | ENPP2 | LPIN1 |
| MAP2 | HIFX | PHLDA1 | ENPP2 |
| TIMP3 | CLONE 24421 | FZD6 | LMCD1 |
| RIB118a | PDE3A | SLC6A15 | |
| ZFP9 | fls485 | ||
| ROBO1 | |||
| HCGIV.9 | |||
| IL12RB2 | |||
| SPP1 | |||
| PDE4B | |||
| MSC | |||
| TIMP3 | |||
| ATIP1 | |||
| KIAA0828 | |||
| PHLDA1 | |||
| PLXNB1 | |||
| COX8A2 | |||
| KIAA0977 | |||
| 8ETMAR | |||
| PTRF | |||
| FZD6 | |||
| PPP13RC | |||
| ET81 | |||
| IL1RAP | |||
| ID2 | |||
| NR1D2 | |||
| 36b3cDNA | |||
| ALDH5A1 | |||
| RAF1 | |||
| TFAP2A | |||
| NTT73 | |||
| ADFP | |||
| LPIN1 |
Genes found to be discriminating in more than one study are shown in bold.
Autotaxin is synthesised as a pre‐pro‐enzyme and after proteolytic cleavage the protein is secreted.41 Autotaxin is identical to lysophospolipase D and generates lysophosphatidic acid by hydrolysing lysophosphatidyl choline.42 Lysophosphatidyl choline enhances cell motility, cell proliferation, and angiogenesis41,43,44 through G protein‐coupled receptors.45 Autotaxin is upregulated in various malignancies, including breast,46 lung,47 thyroid carcinoma48 and glioblastoma multiforme.49
As autotaxin is a tumour cell motility‐stimulating factor, originally isolated from melanoma cell supernatants,50 it is possible that autotaxin may be detected in serum and serve as a biomarker of metastasis. Underexpression of autotaxin in class 2 uveal melanoma with a poor prognosis needs to be explored further.
Footnotes
Grant support: American Cancer Society.
Competing interests: None declared.
References
- 1.Singh A D, Topham A. Incidence of uveal melanoma in the United States: 1973–1997. Ophthalmology 2003110956–961. [DOI] [PubMed] [Google Scholar]
- 2.Singh A D, Bergman L, Seregard S. Uveal malignant melanoma: epidemiologic aspects. In: Singh AD, Damato BE, Pe'er J, et al eds. Clinical ophthalmic oncology. Philadelphia: Saunders‐Elsevier, 2007198–204.
- 3.Egan K M, Seddon J M, Glynn R J.et al Epidemiologic aspects of uveal melanoma. Surv Ophthalmol 198832239–251. [DOI] [PubMed] [Google Scholar]
- 4.Singh A D, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18: 75–84, viii, [DOI] [PubMed]
- 5.Diener‐West M, Hawkins B S, Markowitz J A.et al A review of mortality from choroidal melanoma. Arch Ophthalmol 1992110245–250. [DOI] [PubMed] [Google Scholar]
- 6.Anonymous The COMS randomized trial of iodine 125 brachytherapy for choroidal melanoma, III: initial mortality findings. COMS report no. 18. Arch Ophthalmol 2001119969–982. [DOI] [PubMed] [Google Scholar]
- 7.Anonymous The Collaborative Ocular Melanoma Study (COMS) randomized trial of pre‐enucleation radiation of large choroidal melanoma II: initial mortality findings. COMS report no. 10. Am J Ophthalmol 1998125779–796. [DOI] [PubMed] [Google Scholar]
- 8.Anonymous Mortality in patients with small choroidal melanoma. COMS report no. 4. The Collaborative Ocular Melanoma Study Group. Arch Ophthalmol 1997115886–893. [PubMed] [Google Scholar]
- 9.Kujala E, Makitie T, Kivela T. Very long‐term prognosis of patients with malignant uveal melanoma. Investigative Ophthalmology & Visual Science 2003444651–4659. [DOI] [PubMed] [Google Scholar]
- 10.Singh A D, Borden E C. Metastatic uveal melanoma. Ophthalmol Clin North Am. 2005;18: 143–50, ix, [DOI] [PubMed]
- 11.Anonymous Accuracy of diagnosis of choroidal melanomas in the Collaborative Ocular Melanoma Study. COMS report no. 1. Arch Ophthalmol 19901081268–1273. [DOI] [PubMed] [Google Scholar]
- 12.Singh A D, Topham A. Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology 2003110962–965. [DOI] [PubMed] [Google Scholar]
- 13.Prescher G, Bornfeld N, Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J Natl Cancer Inst 1990821765–1769. [DOI] [PubMed] [Google Scholar]
- 14.Prescher G, Bornfeld N, Hirche H.et al Prognostic implications of monosomy 3 in uveal melanoma. Lancet 19963471222–1225. [DOI] [PubMed] [Google Scholar]
- 15.Singh A D, Boghosian‐Sell L, Wary K K.et al Cytogenetic findings in primary uveal melanoma. Cancer Genetics & Cytogenetics 199472109–115. [DOI] [PubMed] [Google Scholar]
- 16.Sisley K, Rennie I G, Parsons M A.et al Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes, Chromosomes & Cancer 19971922–28. [DOI] [PubMed] [Google Scholar]
- 17.Tschentscher F, Husing J, Holter T.et al Tumor classification based on gene expression profiling shows that uveal melanomas with and without monosomy 3 represent two distinct entities. Cancer Res 2003632578–2584. [PubMed] [Google Scholar]
- 18.Zuidervaart W, van der Velden P A, Hurks M H.et al Gene expression profiling identifies tumour markers potentially playing a role in uveal melanoma development. Br J Cancer 2003891914–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onken M D, Worley L A, Ehlers J P.et al Novel molecular classification of uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res 2004647205–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folberg R, Pe'er J, Gruman L M.et al The morphologic characteristics of tumor blood vessels as a marker of tumor progression in primary human uveal melanoma: a matched case‐control study. Hum Pathol 1992231298–1305. [DOI] [PubMed] [Google Scholar]
- 21.Folberg R, Rummelt V, Parys‐Van Ginderdeuren R.et al The prognostic value of tumor blood vessel morphology in primary uveal melanoma. Ophthalmology 19931001389–1398. [DOI] [PubMed] [Google Scholar]
- 22.Wang E, Miller L D, Ohnmacht G A.et al High‐fidelity mRNA amplification for gene profiling. Nat Biotechnol 200018457–459. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Kessler P, Williams B R. Transcript profiling of Wilms tumors reveals connections to kidney morphogenesis and expression patterns associated with anaplasia. Oncogene 200524457–468. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Kessler P, Yeger H.et al A gene expression signature for relapse of primary wilms tumors. Cancer Res 2005652592–2601. [DOI] [PubMed] [Google Scholar]
- 25.Frevel M A, Bakheet T, Silva A M.et al p38 Mitogen‐activated protein kinase‐dependent and ‐independent signaling of mRNA stability of AU‐rich element‐containing transcripts. Mol Cell Biol 200323425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Friedman J.The elements of statistical learning. New York: Springer Verlag, 2001
- 27.Eisen M B, Spellman P T, Brown P O.et al Cluster analysis and display of genome‐wide expression patterns. Proc Natl Acad Sci U S A 19989514863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A D, Shields C L, Shields J A. Prognostic factors in uveal melanoma. Melanoma Research 2001111–9. [DOI] [PubMed] [Google Scholar]
- 29.Callejo S A, Antecka E, Blanco P L.et al Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye 2006 [DOI] [PubMed]
- 30.Haass N K, Smalley K S, Li L.et al Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res 200518150–159. [DOI] [PubMed] [Google Scholar]
- 31.Hendrix M J, Seftor E A, Seftor R E.et al Biologic determinants of uveal melanoma metastatic phenotype: role of intermediate filaments as predictive markers. Lab Invest 199878153–163. [PubMed] [Google Scholar]
- 32.Onken M D, Ehlers J P, Worley L A.et al Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res 2006664602–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jager M J, Hurks H M, Levitskaya J.et al HLA expression in uveal melanoma: there is no rule without some exception. Hum Immunol 200263444–451. [DOI] [PubMed] [Google Scholar]
- 34.Makitie T, Summanen P, Tarkkanen A.et al Tumor‐infiltrating macrophages (CD68(+) cells) and prognosis in malignant uveal melanoma. Invest Ophthalmol Vis Sci 2001421414–1421. [PubMed] [Google Scholar]
- 35.Hojilla C V, Mohammed F F, Khokha R. Matrix metalloproteinases and their tissue inhibitors direct cell fate during cancer development. Br J Cancer 2003891817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahonen M, Baker A H, Kahari V M. Adenovirus‐mediated gene delivery of tissue inhibitor of metalloproteinases‐3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res 1998582310–2315. [PubMed] [Google Scholar]
- 37.Handsley M M, Edwards D R. Metalloproteinases and their inhibitors in tumor angiogenesis. Int J Cancer 2005115849–860. [DOI] [PubMed] [Google Scholar]
- 38.van der Velden P A, Zuidervaart W, Hurks M H.et al Expression profiling reveals that methylation of TIMP3 is involved in uveal melanoma development. Int J Cancer 2003106472–479. [DOI] [PubMed] [Google Scholar]
- 39.Worley L A, Onken M D, Person E.et al Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res 2007131466–1471. [DOI] [PubMed] [Google Scholar]
- 40.Conlon M R, Albert D M. Uveal melanoma. International Ophthalmology Clinics 19933367–76. [DOI] [PubMed] [Google Scholar]
- 41.Jansen S, Stefan C, Creemers J W.et al Proteolytic maturation and activation of autotaxin (NPP2), a secreted metastasis‐enhancing lysophospholipase D. J Cell Sci 20051183081–3089. [DOI] [PubMed] [Google Scholar]
- 42.Umezu‐Goto M, Kishi Y, Taira A.et al Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol 2002158227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam S W, Clair T, Kim Y S.et al Autotaxin (NPP‐2), a metastasis‐enhancing motogen, is an angiogenic factor. Cancer Res 2001616938–6944. [PubMed] [Google Scholar]
- 44.Umezu‐Goto M, Tanyi J, Lahad J.et al Lysophosphatidic acid production and action: validated targets in cancer? J Cell Biochem 2004921115–1140. [DOI] [PubMed] [Google Scholar]
- 45.Moolenaar W H, van Meeteren L A, Giepmans B N. The ins and outs of lysophosphatidic acid signaling. Bioessays 200426870–881. [DOI] [PubMed] [Google Scholar]
- 46.Yang S Y, Lee J, Park C G.et al Expression of autotaxin (NPP‐2) is closely linked to invasiveness of breast cancer cells. Clin Exp Metastasis 200219603–608. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Mou L, Liu N.et al Autotaxin expression in non‐small‐cell lung cancer. Am J Respir Cell Mol Biol 199921216–222. [DOI] [PubMed] [Google Scholar]
- 48.Kehlen A, Englert N, Seifert A.et al Expression, regulation and function of autotaxin in thyroid carcinomas. Int J Cancer 2004109833–838. [DOI] [PubMed] [Google Scholar]
- 49.Kishi Y, Okudaira S, Tanaka M.et al Autotaxin is overexpressed in glioblastoma multiforme and contributes to cell motility of glioblastoma by converting lysophosphatidylcholine to lysophosphatidic acid. J Biol Chem 200628117492–17500. [DOI] [PubMed] [Google Scholar]
- 50.Stracke M L, Krutzsch H C, Unsworth E J.et al Identification, purification, and partial sequence analysis of autotaxin, a novel motility‐stimulating protein. J Biol Chem 19922672524–2529. [PubMed] [Google Scholar]