Abstract
The folding and assembly of nascent proteins in the endoplasmic reticulum (ER) is assisted by molecular chaperones that are themselves retained within the ER. We now report that a number of different ER proteins, including molecular chaperones, are selectively expressed on the surface of immature thymocytes, but their surface expression is extinguished upon further differentiation. Escape from the ER is only possible for newly synthesized ER proteins before they become permanently retained. Thus, the cellular process of ER retention is incomplete in immature thymocytes and provides an explanation for surface expression of partial receptor complexes that transduce differentiative signals during thymic development.
The functions performed in intracellular organelles of eukaryotic cells are essential for survival of the host cell. One of the most central is performed in the endoplasmic reticulum (ER), which is the site of synthesis, folding, and assembly of protein components of the vesicular transport system (i.e., ER, Golgi complex, endosomes, lysozomes, plasma membrane, etc.). The performance of this task in the ER is facilitated by a class of proteins referred to as molecular chaperones (1). Molecular chaperones assist the folding and assembly of nascent proteins, while retaining proteins that persist in a malfolded or incompletely assembled state (2, 3). The chaperones themselves are thought to be retained in the ER by cognate receptors, which constantly retrieve escaped chaperones from a dynamic intermediate compartment between the ER and Golgi complex (4–10). The retention receptors recognize motifs encoded in the primary amino acid sequences of chaperones: the C-terminal lys-asp-glu-leu (KDEL) tetrapeptide for lumenal chaperones and the C-terminal dilysine (lys-lys-X-X) motif for membrane bound chaperones (11, 12). Recently, another mode of ER retention has been described that involves ill-defined sequences that anchor proteins within the ER, preventing even transient escape (13). The basis for this mode of retention is unclear, but may involve lateral associations with other proteins as has been reported for resident Golgi proteins (14–16).
In the thymus, immature CD4−CD8− T cell precursors are normally signaled to differentiate into CD4+CD8+ cells by a surface pre-T cell receptor complex consisting of clonotypic T cell receptor β chains assembled with invariant pre-Tα and CD3 proteins (17–20). However, even CD4−CD8− thymocytes, which do not express surface pre-T cell receptor complexes (because they lack T cell receptor β) can be induced to differentiate into CD4+CD8+ cells by administration of anti-CD3 mAb (21–23). Indeed, immature CD4−CD8− thymocytes were recently found to express surface receptor complexes comprised of CD3γɛ and CD3δɛ heterodimers complexed with the molecular chaperone calnexin (24–28). This finding was remarkable because calnexin had never previously been found on the cell surface and because calnexin, CD3γ, and CD3δ chains all have ER retention signals near their C termini (29, 30). The calnexin–CD3 complexes that escape to the cell surface appear to do so because interactions between the cytoplasmic domains of calnexin and CD3 sterically mask their retention sequences, as has been reported for subunits of the immunoglobulin E receptor (24, 31). This study was undertaken to evaluate whether escape of calnexin–CD3 complexes from the ER to the surface of immature thymocytes was unique to these particular protein complexes or alternatively whether multiple ER proteins were able to escape ER retention in these developmentally immature cells.
We report here that calnexin–CD3 complexes are not unique and that immature thymocytes allow many, but not all, resident ER proteins to escape from ER retention and reach the cell surface, suggesting that ER retention in immature thymocytes is incomplete.
MATERIALS AND METHODS
Cell Lines and Antibodies.
VL3–3M2, a thymic lymphoma line that closely approximates the phenotype of an immature CD4+CD8+ thymocyte (32), was provided by Cynthia Guidos (Hospital for Sick Children, Toronto). VL3–3M2 cells, as well as the BW5147 thymic lymphoma cell line (33), were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). The rabbit antibodies (Ab) used in this study were raised against the following immunogens: (i) anti-cal-N, fusion protein encompassing the N-terminal 374 aa of mouse calnexin (24); (ii) anti-cal-C, C-terminal 12 aa of mouse calnexin (34); (iii) anti-rI, aa 563–583 of rat ribophorin I (35); (iv) anti-rII, aa 1–22 of ribophorin II (35); (v) anti-SSR, aa 266–286 of signal sequence receptor α subunit (36); and (vi) anti-CRT, recombinant human calreticulin (CRT) (Affinity BioReagents, Golden, CO). The following mAb were used: (i) anti-CD3ɛ, 145–2C11 (37) and (ii) anti-KDEL (StressGen Biotechnologies, Victoria, BC).
Surface Reexpression Assay.
VL3–3M2 cells were washed two times in Hanks’ balanced salts solution (HBSS), resuspended at 5 × 106/ml either in HBSS (mock) or in HBSS containing 2 mg/ml pronase (Calbiochem), and incubated at 37°C for 15 min. Pronase treatment was quenched with an equal volume of ice-cold HBSS containing 5% FCS and 100 μg/ml DNase. After three more washes in HBSS containing 5% FCS, any remaining pronase was inactivated with 0.5 mM phenylmethylsulfonyl fluoride (PMSF)/0.1 mM 7-amino-1-chloro-3-tosylamido-2-heptanone (TLCK) for 10 min on ice. Pronase treated cells were resuspended at 106/ml in RPMI 1640 medium containing 10% FCS and cultured for 4 hr: (i) on ice, (ii) at 37°C, (iii) at 37°C with 1 μg/ml Brefeldin A (BFA; Epicentre Technologies, Madison, WI), or (iv) at 37°C with 10 μg/ml cycloheximide (CHX) (Sigma). After culture, the cells were washed three times with HBSS, resuspended at 20 × 106/ml, and cooled on ice for 15 min. Sulfo-NHS-biotin (Pierce) was added to 1 mg/ml and the biotin labeling was allowed to proceed for 30 min on ice, then terminated with 25 mM lysine/HBSS. After three more washes, any dead cells were removed by pelleting through a cushion of lympholyte M (Cedarlane Laboratories). Viabilities were consistently greater than 98%.
Cell Lysis, Immunoprecipitation, and Immunoblotting.
Cells to be immunoprecipitated with anti-CRT Ab were lysed by boiling 5 min in 1% SDS, diluted 10-fold with lysis buffer (50 mM Tris, pH 7.4/150 mM NaCl/1.8 mg/ml iodoacetamide/0.5 mM PMSF/0.1 mM TLCK/40 μg/ml aprotinin/20 μg/ml leupeptin) containing 1% Nonidet P-40 (Calbiochem), and clarified by microcentrifugation for 10 min at 4°C. Otherwise, cells were extracted with buffer containing 1% digitonin (Wako Biochemicals, Kyoto) for 20 min on ice and clarified by microcentrifugation for 10 min at 4°C. Clarified extracts were immunoprecipitated for 2 hr with Ab preadsorbed to protein A-Sepharose (Pharmacia) except for the anti-KDEL mAb, which was adsorbed to protein G-Sepharose (Sigma) (24). The resultant immune complexes were eluted by boiling in sample buffer, resolved on SDS/PAGE gels, and transferred onto Immobilon poly(vinylidene difluoride) membranes (Millipore). Surface biotinylated proteins were visualized with horseradish peroxidase-conjugated streptavidin (Southern Biotechnology Associates) and then the same membranes were reprobed with Ab to assess total protein levels as described (24). Rabbit Ab were used for blots at the indicated dilution: (i) anti-cal-C, 1:500; (ii) anti-rI, 1:800; (iii) anti-rII, 1:800; (iv) anti-SSRα, 1:50; (v) anti-CRT, 1:1000. The anti-KDEL mAb was used at 1:500 (StressGen Biotechnologies).
Production of Calnexin Fusion Protein.
The bacterial extracts were produced as follows. A 1.1-kb fragment encoding the N terminus of mouse calnexin (34) was subcloned directionally into the BamHI and NdeI sites of the pET-21a vector and transformed into BL21 cells (Novagen), which were used to inoculate 0.5 liter of Luria broth containing 100 μg/ml ampicillin. The cultures were shaken at 37°C until the OD600 was between 0.8–1.0, induced for 2 hr in 1 mM isopropyl β-d-thiogalactoside (Calbiochem) and then a final hour in 200 μg/ml rifampicin (Calbiochem). Cells were harvested by centrifugation (9,000 rpm for 10 min in a GSA-3 rotor; Dupont/Sorvall), resuspended in 25 ml of 50 mM Tris/5 mM EDTA (pH 8.0), and pelleted for 5 min at 15,000 rpm in an SA-600 rotor (Sorvall). Each gram of bacteria was resuspended in 10 ml of 50 mM Tris/5 mM EDTA (pH 8.0) containing 0.1 mg/ml lysozyme (Calbiochem) and incubated 1 hr at 37°C. The cell extract was then sonicated until the viscosity disappeared and then clarified for 5 min at 15,000 rpm in an SA-600 rotor. The clarified extract was found to be between 70 and 90% recombinant calnexin.
Cell Preparation and Flow Cytometry.
Fresh thymic and lymph node explants from adult C57BL/6 mice (National Cancer Institute, Fredrick, MD) were gently teased to produce single-cell suspensions that were analyzed by flow cytometry immediately, or after removal of immature thymocytes/enriching for lymph node T cells using anti-HSA mAb and rabbit complement. For each sample, 106 cells were stained for 30 min with anti-cal-N diluted 1:50 in staining buffer (0.1%BSA/0.1% sodium azide/HBSS), alone or after a 30-min preincubation in staining buffer containing 30 ng of bacterial extract from control cells or those expressing a calnexin fusion protein. Bound Ab was visualized by washing the cells 3 times in staining buffer and incubating for an additional 30 min with 0.5 μg of fluorescein-conjugated goat anti-rabbit Ab (Caltag, South San Francisco, CA). The stained cells were analyzed in a FACSscan cytometer using cellquest software (Becton Dickinson). Dead cells were excluded from analysis by propidium iodide gating.
Concanavalin A (ConA) Chromatography and Glycosidase Digestion.
Surface biotin-labeled BW5147 thymic lymphoma cells were lysed in digitonin lysis buffer supplemented to 1 mM with CaCl2 and MgCl2 and the extracts halved. One-half was held on ice (unfractionated), while the other half was adsorbed to ConA-Sepharose (EY Laboratories) for 2 hr with agitation at 4°C. After pelleting the ConA-Sepharose, the unbound material (Con-A unbound fraction) was held on ice, while the Sepharose was washed three times with lysis buffer containing 0.2% digitonin. Bound glycoproteins were eluted two times for 30 min at 4°C (ConA bound fraction) with lysis buffer containing 0.5 M α-methyl mannoside. The “unfractionated,” “bound,” and “unbound” material was immunoprecipitated with specific Ab as indicated. Immune complexes adsorbed to Sepharose beads were divided into three equal parts, resuspended in the appropriate stock digestion buffer, and treated as follows: (i) Mock, resuspended in 50 μl of stock digestion buffer for endoglycosidase H (Endo H; New England Biolabs); (ii) Endo H, resuspended in 50 μl of Endo H buffer and supplemented with 5000 units of Endo Hf (New England Biolabs) as described (12); and (iii) jackbean mannosidase, resuspended in 50 μl of commercially prepared stock buffer for jackbean mannosidase and 0.5 unit of jackbean mannosidase (Oxford Glycosystems, Rosedale, NY). All samples were incubated for 18 hr at 37°C and then quenched with 25 μl of 3 × SDS/PAGE sample buffer.
RESULTS
It is difficult by current biochemical surface labeling techniques to confidently distinguish intentionally labeled proteins that are present on the surface of viable cells from artifactually labeled proteins present in the cytosol of dead cells. Consequently, we derived a strategy to do so, which we refer to as the surface reexpression assay (Fig. 1A). Cells are treated with extracellular proteases to digest surface proteins on viable cells (as well as accessible internal proteins in dead cells), and placed into culture at 37°C to allow ongoing protein transport to reestablish surface expression of the digested proteins, which can then be visualized by the biotin surface-labeling reaction. Importantly, intracellular proteins might become accessible to labeling if cells die during the culture period. To evaluate the extent of internal labeling, parallel cultures of protease-treated cells are supplemented with an inhibitor of vesicular transport, BFA, which interferes with reexpression of surface proteins by viable cells, but not with internal labeling of cytosolic proteins in dying cells (38). Indeed, as predicted from our previous results (24), CD3-associated calnexin molecules were detected on mock-treated cells (Fig. 1B, lane 1), were largely destroyed by exogenously added proteases (lane 2), were reexpressed following culture for 4 hr at 37°C (lane 3), and were not reexpressed when the cells were treated with BFA (lane 4). Reprobing the same membranes with anti-calnexin Ab revealed that the overall cellular abundance of CD3-associated calnexin molecules was unaffected by BFA treatment, demonstrating that only a small fraction (≈1%) of the calnexin–CD3 complexes assembled in these cells were actually escaping to the cell surface.
Figure 1.
Surface reexpression assay. (A) Schematic of the surface reexpression assay. Proteins expressed on the cell surface are removed by treating cells with extracellular proteases. Protease-treated cells are then placed into culture at 37°C to allow ongoing protein transport to reestablish surface expression, which is evaluated biochemically by biotin labeling. It should be appreciated that proteins might also become accessible to labeling if there is cell death during culture. To distinguish legitimate surface reexpression from artefactual internal labeling, protease-treated cells are cultured in parallel with the protein transport inhibitor BFA, which blocks surface reexpression by viable cells, but not internal labeling of cytosolic proteins in dying cells. (B) An immature thymocyte cell line was either mock treated (HBSS, 15 min at 37°C) or protease treated (2 mg/ml pronase/HBSS for 15 min at 37°C), following which cells were cultured in complete medium at 4°C. In addition, proteolyzed cells were cultured at 37°C in complete medium alone or supplemented with 1 μg/ml BFA following which the cells were surface labeled with biotin, lysed in digitonin lysis buffer, and immunoprecipitated with anti-CD3ɛ mAb (145–2C11). The immune complexes were resolved on SDS/PAGE gels, blotted to Immobilon poly(vinylidene difluoride) membranes and visualized with HRP-Av and chemiluminescent detection. The same membranes were then washed in PBS-Tween, reblocked in 5% milk/PBS, and immunoblotted with the immunoprecipitating Ab to determine the effect of treatment on total protein levels. Bound Ab was visualized with 125I protein A and autoradiography. The immature thymocyte cell line used in these experiments was the VL3–3M2 thymic lymphoma. Similar results were obtained with primary thymocytes from tissue explants. (C) The surface reexpression assay was performed as above except that a parallel culture supplemented with the protein synthesis inhibitor, CHX, was added and the resultant detergent extracts were immunoprecipitated with rabbit anti-calnexin Ab reactive with calnexin’s lumenal domain (anti-cal-N). (D) The surface reexpression assay was performed as above using rabbit anti-calnexin Ab reactive with calnexin’s C-terminal 12 aa (anti-cal-C, Left) and calnexin’s lumenal domain (anti-cal N, Right). It should be noted that the surface-labeled species migrating beneath calnexin (Right) may represent breakdown products possibly generated during turnover of surface calnexin. Consistent with this explanation, breakdown products are not seen among newly reexpressed calnexin molecules (lane 9) suggesting that they result from degradation of calnexin molecules after they have reached the cell surface.
To determine if the escaped calnexin molecules represented a steady loss of preformed proteins from the ER, we performed the surface reexpression assay but, in addition, cultured pronase stripped cells in the presence of the protein synthesis inhibitor CHX. Direct immunoprecipitation of the detergent extracts with anti-calnexin Ab revealed that reexpression of calnexin on the cell surface was effectively blocked by not only by BFA but by CHX as well (Fig. 1C), demonstrating that escape of calnexin from the ER to the cell surface requires ongoing protein synthesis. Thus, these results indicate that preformed calnexin molecules are efficiently retained within the ER of immature thymocytes and that it is only newly synthesized calnexin molecules that can escape to the cell surface.
To document that calnexin molecules were able to reach the cell surface while still bearing ER retention motifs, we performed immunoprecipitations on surface biotin-labeled thymocytes using an Ab reactive with calnexin’s C-terminal 12 aa (Fig. 1D), which includes calnexin’s ER retention signal. Indeed, surface-labeled calnexin molecules were detected using Ab to calnexin’s C terminus, demonstrating that calnexin molecules were present on the cell surface despite bearing ER retention signals (Fig. 1D). In agreement, we have previously demonstrated that surface calnexin molecules associated with CD3 comigrated precisely with respect to both charge and size with internal calnexin molecules bearing ER retention signals, as demonstrated by blotting with Ab to calnexin’s C terminus (24).
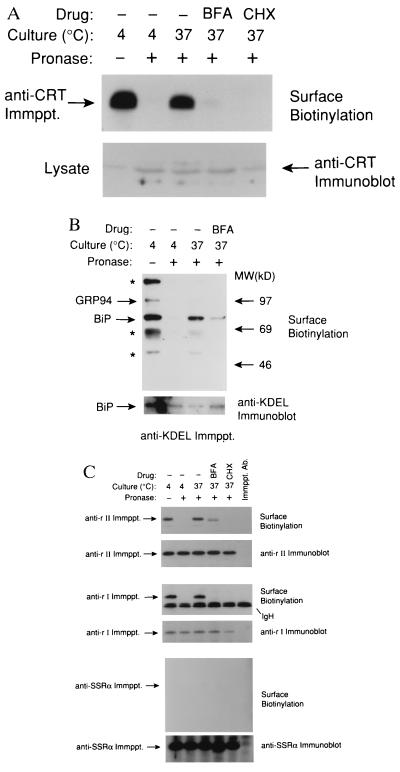
The escape of nascent calnexin molecules from ER retention suggested that in these developmentally immature thymocytes ER retention might be incomplete. Consequently, we asked if other ER proteins (both soluble and integral membrane proteins) were also able to escape from the ER and reach the cell surface. CRT is a multifunctional, peptide and Ca2+-binding molecular chaperone that is highly homologous to calnexin’s lumenal domain (39, 40). However, unlike calnexin, CRT is a soluble protein that is found in the ER lumen and that is retained by a distinct motif (KDEL) and a distinct retention receptor (KDEL receptor). Using the surface reexpression assay we found that CRT was destroyed by proteolysis, reexpressed during culture at 37°C, and the reexpression was blocked by both BFA and CHX (Fig. 2A). Escape of CRT was not unique among lumenal ER proteins as immunoprecipitation with anti-KDEL mAb revealed at least five different surface-labeled proteins, two of which we identified conclusively using monospecific Ab as BiP and GRP94 (Fig. 2B, arrows). BiP (or GRP78) and GRP94 are lumenal ER chaperones that are members of the glucose regulated protein (GRP) family, which are upregulated in response to glucose starvation (41–43). Whereas reexpression of BiP is readily apparent following culture at 37°C, reexpression of GRP94 and other proteins precipitated with the anti-KDEL Ab require longer exposure times to be visible, possibly because the biosynthetic rates of these proteins differ. The identity of the remaining proteins (Fig. 2B, asterisks) is currently unclear. These may be KDEL-containing ER proteins themselves or may be proteins bound to GRP94 and BiP. Indeed, since GRP94 and BiP are soluble proteins their presence on the cell surface must reflect interaction with other membrane bound proteins.
Figure 2.
Immature thymocytes express ER-resident proteins, including molecular chaperones, on the cell surface. Thymic lymphoma cells were proteolyzed and then cultured in medium alone, medium containing 1 μg/ml BFA, or medium containing 10 μg/ml of the protein synthesis inhibitor CHX. After biotin surface-labeling of cells, digitonin cell extracts were immunoprecipitated with Ab specific for (A) CRT, (B) KDEL, (C) rI, rII, and signal sequence receptor α. It should be appreciated that the anti-KDEL Ab was originally generated by immunization with peptides encompassing the retention signal of BiP and does not recognize all KDEL-containing ER proteins equally.
To determine if integral membrane proteins in addition to calnexin are expressed on immature thymocytes, we examined immature thymoma cells for surface expression of ribophorin I, ribophorin II, and the signal sequence receptor α (also referred to as translocon-associated protein or TRAP) subunit (Fig. 2C), all of which are associated with a macromolecular complex (translocon) involved in the translocation of nascent proteins from the cytosol to the ER lumen or their cotranslational modification (44–46). We could clearly detect surface expression of both ribophorin I and ribophorin II, although detection of ribophorin I expression required longer exposure times than that for ribophorin II. In contrast, we could not detect signal sequence receptor α on the cell surface at all (Fig. 2C). Thus, many, but not all, ER proteins are able to escape from ER retention in immature thymocytes. However, escape from the ER appears to be possible only for newly synthesized proteins, since surface reexpression is always blocked by CHX, suggesting that preformed ER proteins are retained efficiently.
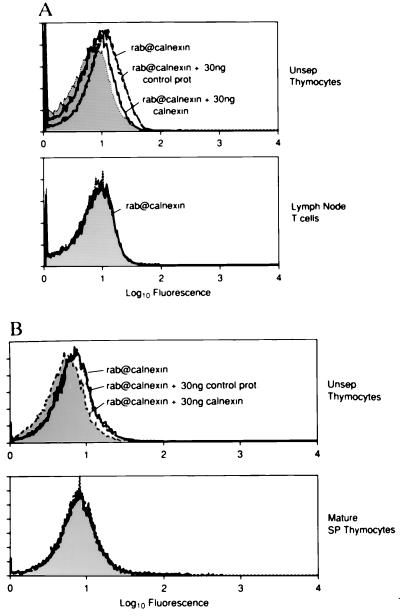
Next, we attempted to detect surface expression of the molecular chaperone calnexin on individual cells by immunofluorescence and flow cytometry (Fig. 3). We found that thymocytes stained uniformly, albeit at low levels, with an Ab that is directed against calnexin’s exodomain. This anti-calnexin staining was specific because it was blocked with 30 ng of bacterial extract containing calnexin fusion protein but was not affected by 30 ng of control bacterial extract (Fig. 3A). Importantly, expression of surface calnexin is developmentally regulated, as mature lymph node T cells did not stain detectably with the anti-calnexin Ab. To determine if calnexin were expressed on the surface of mature cells in the thymus, we enriched for the small subpopulation of mature thymocytes by depleting immature thymocytes using mAb specific for heat stable antigen (HSA). HSA was used because immature thymocytes express HSA at high levels, whereas mature thymocytes express HSA at low levels (47). In contrast to unseparated thymocytes, which are almost entirely immature and that exhibited specific anti-calnexin staining, mature HSAlo thymocytes did not exhibit any specific anti-calnexin staining (Fig. 3B). Thus, surface expression of calnexin is restricted to immature HSAhi thymocytes and is extinguished upon their further differentiation into HSAlo T cells.
Figure 3.
Calnexin’s expression on the cell surface is developmentally regulated. (A) Single-cell suspensions from fresh C57BL/6 mouse thymic and lymph node explants were either stained directly (Unsep. Thymocytes) or after depletion of non-T cells (Lymph Node T cells). (B) Single-cell suspensions of thymocytes were stained before (Unsep. Thymocytes) or after immature cells were eliminated using anti-HSA and complement (Mature SP Thymocytes). Cell suspensions were stained with a rabbit polyclonal serum directed at calnexin’s lumenal domain (rab@calnexin, solid lines) and bound Ab were visualized with fluoresceinated-goat anti-rabbit IgG. Specificity of the anti-calnexin staining was demonstrated by preincubating the anti-calnexin Ab with 30 ng of control bacterial extract (short dashes) or extract containing calnexin fusion protein (long dashes). Cells stained with preimmune serum are indicated by shaded curves.
Since surface expression of molecular chaperones is developmentally regulated, we considered that it might offer an explanation for expression on immature thymocytes of glycoproteins bearing incompletely processed N-linked carbohydrate side chains (25, 48). In particular, we thought it possible that molecular chaperones continued to associate with glycoproteins even after escape from the ER, thereby blocking accessibility to processing enzymes during transit through the Golgi. Indeed, the exodomain of calnexin can bind glycoproteins via their N-linked oligosaccharide side chains, but this binding is restricted to oligosaccharide chains containing a terminal glucose moiety, a modification that is normally removed from nascent glycoproteins before their exit from the ER (49–51). Interestingly, we found that the surface CD3 complexes expressed by immature thymocytes could be subdivided into two populations based upon the availability of their carbohydrate side chains to binding by the mannose and glucose reactive-lectin, ConA (Fig. 4). The CD3 complexes bound by ConA (ConA Bound; Fig. 4) were not associated with calnexin; however, the CD3 complexes that were not bound by ConA (ConA Unbound) were associated with calnexin, suggesting that calnexin binding to CD3 masks the CD3 oligosaccharide side chains and makes them unavailable for binding by ConA (Fig. 4). In agreement, digestion of these surface calnexin–CD3 complexes with jackbean mannosidase (which more extensively cuts oligosaccharide side chains lacking terminal glucose moieties than those containing terminal glucose moieties) revealed that surface calnexin-associated CD3γ proteins still possessed the terminal glucose residues that are normally removed within the ER (Fig. 4, glc+). Thus, calnexin association prevented processing of N-linked oligosaccharide side chains during intracellular transport. Curiously, while the surface CD3γ proteins not associated with calnexin (Fig. 4, ConA Bound) lacked glucose they were still sensitive to Endo H, possibly because they had been associated with a molecular chaperone other than calnexin that interfered with the processing of oligosaccharide side chains during transit through the Golgi. We suggest that association with molecular chaperones during intracellular transport provides one explanation for expression of proteins bearing incompletely processed N-linked carbohydrate side chains on the surface of immature thymocytes.
Figure 4.
Calnexin association with CD3 masks their carbohydrate side chains. Calnexin–CD3 complexes were immunoprecipitated from detergent extracts of surface biotinylated BW5147 thymic lymphoma cells using anti-CD3ɛ mAb (145–2C11) either before (Unfrac.) or after the extracts were fractionated using ConA-Sepharose. The population of proteins that did not bind to ConA are referred to as ConA Unbound, whereas proteins that bound to ConA and were eluted using α-methyl α-d-mannoside are referred to as ConA Bound. The resultant immune complexes were incubated in buffer alone (M) or digested with the following glycosidases: jack bean mannosidase (JB), which differentially cuts N-linked oligosaccharides depending upon the presence or absence of terminal glucose residues and endoglycosidase H (EH), which is unaffected by the presence of glucose residues. Digested proteins were blotted to immobilon membranes and the surface-labeled proteins were visualized with HRP-Av. The migration positions of mock-treated or endo H-sensitive (EHs) CD3γ proteins are indicated. Also indicated are the migration positions of CD3γ proteins that either do contain (glc+) or do not contain (glc−) terminal glucose residues.
DISCUSSION
In this report we demonstrate that calnexin, as well as many other normally “ER-resident” proteins, can escape from the ER of immature thymocytes to be expressed on the plasma membrane, indicating that ER retention is incomplete in these developmentally immature cells. Importantly, escape of resident proteins from the ER is not a property of all cells, since escaped calnexin molecules were not detected on the surface of developmentally mature T lymphocytes. Nor is escape from retention a property of all ER proteins, but rather appears to be somewhat selective. Furthermore, escape of proteins from ER retention requires ongoing protein synthesis suggesting that whereas the newly synthesized pool of ER proteins is able to escape, preformed ER proteins are retained quite efficiently.
The release of newly synthesized molecules from the ER suggests that escape is only possible temporarily before such molecules become permanently sequestered in the ER. While there are several possible explanations for the escape of resident proteins from the ER of immature thymocytes, we have schematized what we think is the most likely in Fig. 5. We think that immature thymocytes may contain an insufficient number of functional ER retention receptors either because their overall number is low, or because a subpopulation of ER retention receptors is functionally impaired. Consequently, a number of newly synthesized molecular chaperones avoid capture by ER retention receptors and so escape from the ER and reach the cell surface. It is also possible that release is a consequence of protein synthesis itself. For example, the ER-bound ribosomes engaged in protein synthesis may sterically hinder access of retention receptors to the emerging ER protein or, alternatively, a subpopulation of nascent ER proteins may adopt a conformation during folding that masks their retention signals. Whatever the mechanism underlying escape of nascent ER proteins to the surface of thymocytes, this “defect” is corrected upon development of immature thymocytes into mature functional T cells.
Figure 5.
Models for escape of ER proteins from retention. We propose that escape of ER proteins from retention requires ongoing protein synthesis and that escape is only possible for newly synthesized ER proteins. We hypothesize that the initial localization process is driven by retention (or retrieval) signals, e.g., KDEL and lys-lys-X-X and their cognate receptors, which may be diminished in number or alternatively may be functionally compromised in immature thymocytes as a result of posttranslational modifications, association with other proteins, etc. However, once nascent molecular chaperones have been bound by retention/retrieval receptors they are permanently held within the ER, possibly by protein:protein interactions independent of the retention signals.
Our observations that immature thymocytes are unable to completely retain resident proteins within their ER provides an explanation for surface expression of partial receptor complexes on immature thymocytes, which are otherwise sequestered within the ER of developmentally mature T cells (25). The presence of such partial receptor complexes has important developmental consequences because they are able to transduce differentiative signals in immature thymocytes that are vital for early thymocyte development (17, 18, 21–23). In particular, dimers of CD3γɛ and CD3δɛ appear on the surface of immature thymocytes in association with calnexin and are able to transduce intracellular signals that drive immature CD4−CD8− thymocytes to differentiate into more mature CD4+CD8+ thymocytes (21–25). In fact, it is possible that surface expression of pre-T cell receptor complexes by immature thymocytes is also a consequence of inefficient ER retention (19, 20). It is also tempting to speculate that the ER resident molecular chaperones that have been released to the surface of immature thymocytes may themselves play an important role in thymocyte development. In particular, it is possible that molecular chaperones like calnexin and CRT, through their ability to bind carbohydrate ligands (49–52), may promote interactions between developing thymocytes and thymic stromal cells, an important aspect of their codependent differentiation.
In conclusion, this study demonstrates that a variety of molecules previously thought to reside exclusively in the ER are in fact expressed at low levels on the surface of immature thymocytes. Indeed, incomplete ER retention may not be unique to immature thymocytes, but may be a feature of other developmentally immature cells as well. Experiments are currently in progress that may provide a more thorough understanding of how proteins can avoid the ER retention machinery and reach the surface of immature cells.
Acknowledgments
We wish to thank Dr. Cynthia J. Guidos for providing the VL3-3M2 thymic lymphoma cell line and Drs. Kelly P. Kearse and Cynthia J. Guidos for critical reading of this manuscript.
ABBREVIATIONS
- ER
endoplasmic reticulum
- Ab
antibody(ies)
- GRP
glucose regulated protein
- KDEL
lys-asp-glu-leu
- BFA
Brefeldin A
- CHX
cycloheximide
- ConA
concanavalin A
- HBSS
Hanks’ balanced salt solution
- CRT
calreticulin
References
- 1.Gething M J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 2.Melnick J, Argon Y. Immunol Today. 1995;16:243–250. doi: 10.1016/0167-5699(95)80167-7. [DOI] [PubMed] [Google Scholar]
- 3.Hammond C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsson T, Warren G. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelham H R B. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza J C, Hardwick K G, Dean N, Pelham H R B. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 7.Lewis M J, Sweet D J, Pelham H R B. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- 8.Hardwick K G, Lewis M J, Semenza J, Dean N, Pelham H R B. EMBO J. 1990;9:623–630. doi: 10.1002/j.1460-2075.1990.tb08154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosson P, Letourneur F. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 10.Letourneur F, Gaynor E C, Hennecke S, Demolliere C, Duden R, Emr S D, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 11.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro S, Pelham H R B. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 13.Murakami K, Mihara K, Omura T. J Biochem. 1994;116:164–175. doi: 10.1093/oxfordjournals.jbchem.a124489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson T, Hoe M H, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger E G, Warren G. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro S. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisz O A, Swift A M, Machamer C E. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levelt C N, Eichmann K. Immunity. 1995;3:667–672. doi: 10.1016/1074-7613(95)90056-x. [DOI] [PubMed] [Google Scholar]
- 18.Fehling H J, Krotkova A, Saint-Ruf C, von Boehmer H. Nature (London) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 19.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling H J, von Boehmer H. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 20.Groettrup M, Ungewiss K, Azogui O, Palacios R, Owen M J, Hayday A C, von Boehmer H. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- 21.Levelt C N, Mombaerts P, Iglesias A, Tonegawa S, Eichmann K. Proc Natl Acad Sci USA. 1993;90:11401–11405. doi: 10.1073/pnas.90.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinkai Y, Alt F. Int Immunol. 1994;6:995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs H, Vandeputte D, Tolkamp L, deVries E, Borst J, Berns A. Eur J Immunol. 1994;24:934–939. doi: 10.1002/eji.1830240423. [DOI] [PubMed] [Google Scholar]
- 24.Wiest D L, Burgess W H, McKean D, Kearse K P, Singer A. EMBO J. 1995;14:3425–3433. doi: 10.1002/j.1460-2075.1995.tb07348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiest D L, Kearse K P, Shores E W, Singer A. J Exp Med. 1994;180:1375–1382. doi: 10.1084/jem.180.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 27.David V, Hochstenback F, Rajagopalan S, Brenner M B. J Biol Chem. 1993;268:9585–9592. [PubMed] [Google Scholar]
- 28.Hochstenback F, David V, Watkins S, Brenner M B. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan S, Xu Y, Brenner M B. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- 30.Letourneur F, Klausner R D. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 31.Letourneur F, Hennecke S, Demolliere C, Cosson P. J Cell Biol. 1995;129:971–978. doi: 10.1083/jcb.129.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos C J. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- 33.Hyman R, Stallings V. J Natl Cancer Inst. 1974;52:429–437. doi: 10.1093/jnci/52.2.429. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber K L, Bell M P, Huntoon C J, Rajagopalan S, Brenner M B, McKean D J. Int Immunol. 1994;6:101–111. doi: 10.1093/intimm/6.1.101. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y H, Sabitini D D, Kreibich G. J Cell Biol. 1990;111:1335–1342. doi: 10.1083/jcb.111.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prehn S, Herz J, Hartmann E, Kurzchalia T V, Frank R, Roemisch K, Dobberstein B, Rapoport T A. Eur J Biochem. 1990;188:439–445. doi: 10.1111/j.1432-1033.1990.tb15421.x. [DOI] [PubMed] [Google Scholar]
- 37.Leo O, Foo M, Sachs D H, Samelson L E, Bluestone J A. Proc Natl Acad Sci USA. 1987;84:1374–1378. doi: 10.1073/pnas.84.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson J R, Ora A, Van P N, Helenius A. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nash P D, Opas M, Michalak M. Mol Cell Biochem. 1994;135:71–78. doi: 10.1007/BF00925962. [DOI] [PubMed] [Google Scholar]
- 41.Lee A. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- 42.Melnick J, Dul J L, Argon Y. Nature (London) 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 43.Haas I G, Wabl M. Nature (London) 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 44.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann E, Gorlich D, Kostka A, Otto A, Kraft R, Knespel S, Burger E, Rapoport T A, Prehn S. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 46.Kreibich G, Ulrich B L, Sabitini D D. J Cell Biol. 1978;77:464–487. doi: 10.1083/jcb.77.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsdell F, Jenkins M, Dinh Q, Fowlkes B J. J Immunol. 1991;147:1779–1785. [PubMed] [Google Scholar]
- 48.Kishi H, Borgulya P, Scott B, Karjalainen K, Traunecker A, Kaufman J, von Boehmer H. EMBO J. 1991;10:93–100. doi: 10.1002/j.1460-2075.1991.tb07924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ware F E, Vassilakos A, Peterson P A, Jackson M R, Lehrman M A, Williams D B. J Biol Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- 50.Kearse K P, Williams D B, Singer A. EMBO J. 1994;13:3678–3686. doi: 10.1002/j.1460-2075.1994.tb06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammond C, Braakman I, Helenius A. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]