Abstract
Cytotoxic T lymphocytes (CTLs) are thought to play a crucial role in the termination of the acute primary HIV-1 syndrome, but clear evidence for this presumption has been lacking. Here we demonstrate positive selection of HIV-1 proviral sequences encoding variants within a CTL epitope in Nef, a gene product critical for viral pathogenicity, during and after seroconversion. These positively selected HIV-1 variants carried epitope sequence changes that either diminished or escaped CTL recognition. Other proviruses had mutations that abolished the Nef epitope altogether. These results provide clear evidence that CTLs exert selection pressure on the viral population in acute HIV-1 infection.
Infection with HIV-1 is often heralded by a clinical illness characterized by rash, fever, and lymphadenopathy. Simultaneously, HIV-1 RNA is detectable in the plasma at concentrations as high as 22 × 106 copies per ml (1). In most patients studied, the viremia diminishes by 10- to 200-fold within a few weeks (1–3). This suppression, which is as effective as that achieved by single anti-retroviral drugs (4, 5), has been attributed to the immune response and is thought to be mediated by cytotoxic T lymphocytes (CTLs). Although CTLs directed against HIV first become detectable as the viremia declines (6, 7), there has been no direct evidence that this cellular immune response can modulate the natural history of HIV-1 infection. Some authors have doubted whether CTLs play any role in controlling persistent viral infections (8, 9). Here, by detailed genetic analysis, we show that CTLs exert a significant selection force on the virus population during primary HIV-1 infection and cause the rapid emergence of escape variants.
MATERIALS AND METHODS
Blood Donor.
Donor SC2 is a 36-year-old homosexual male with a history of regular unprotected sexual intercourse with men known to be HIV-seropositive. A previous HIV antibody test (Jan. 10, 1995) had been negative. In October 1995, he was admitted to hospital with a sore throat, fever, headache, and photophobia; symptoms persisted until mid-November. Blood sampled on Oct. 13, 1995 tested HIV p24 antigen-positive (>100 pg/ml), antibody-negative (determined by standard ELISA methods; Abbott); a second sample 6 days later tested HIV p24 antigen-positive (>500 pg/ml), HIV gp41 antibody-positive, and HIV p31 antibody-, p24 antibody-, p17 antibody-, and gp36 antibody-negative. Together these results imply that donor SC2 acquired HIV-1 in September or early October 1995.
HLA Class 1 Determination.
HLA class I typing was performed using sequence-specific PCRs as described previously (10, 11).
Viral Load Measurement.
Plasma HIV-1 RNA was quantified using a commercially available reverse transcriptase-PCR technique (Amplicor; Roche Diagnostics).
Generation of Bulk Cultured Lymphocytes.
Heparinized blood samples (40–60 ml) were separated by Ficoll–Hypaque density gradient centrifugation, and the peripheral blood mononuclear cells (PBMCs) were washed and resuspended in RPMI-1640 medium (GIBCO) with 10% fetal calf serum (R10). Bulk cultures were prepared by the addition of autologous phytohemagglutinin-activated lymphocytes to the PBMCs as previously described (12).
Generation of CTL Lines.
CTL lines were derived from donor PBMCs or bulk cultures as previously described (12).
Cytotoxicity Assays.
Target cells in cytotoxicity assays were autologous or HLA-matched Epstein–Barr virus-transformed B cells prepulsed with 10 μM peptide or infected with recombinant vaccinia virus (rVV) and labeled with 51Cr (Amersham). Peptides were synthesized using standard fluorenylmethoxycarbonyl (Fmoc) techniques and were >90% pure as determined by HPLC (Research Genetics, Huntsville, AL and Multiple Sclerosis Society Peptide Laboratory, Oxford, U.K.). Vaccinia infections were effected at 3–5 plaque-forming units per cell (1 h) followed by 16 h of incubation before 51Cr labeling. The construction of rVV expressing p17 Gag has been described previously (13); rVV expressing influenza nucleoprotein was used to infect control target cells. Targets were plated out at 5 × 103 cells per well in round-bottomed microtiter plates (Nunclon, Naperville, IL) and incubated at 37°C with either R10, effector cells at the effector:target (E:T) ratios shown (see Fig. 1) or 5% Triton X-100 in a total volume of 150 μl. All assays were performed in duplicate. Supernatant was assayed at 4–6 h, and specific lysis was calculated as 100 × (experimental lysis − spontaneous lysis)/(maximum lysis − spontaneous lysis). Spontaneous release was <30% of detergent release for all assays shown.
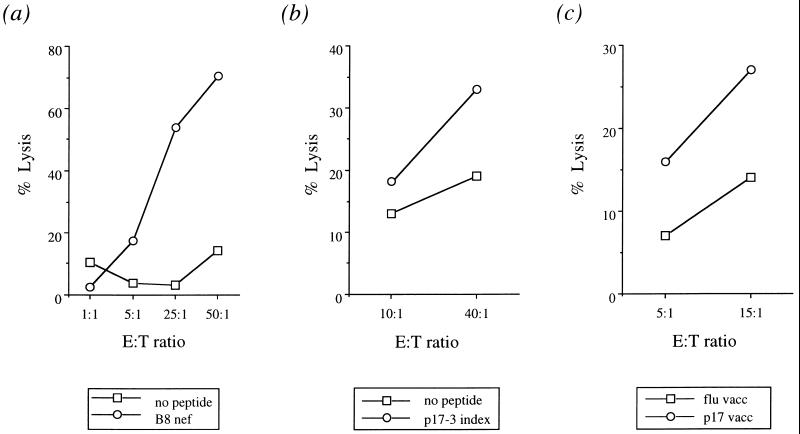
Figure 1.
Recognition of CTL epitopes by donor SC2 bulk cultured lymphocytes. (a) HLA B8 Nef (FLKEKGGL) response (Nov. 3, 1995). (b and c) HLA B8 p17-3 Gag (GGKKKYKL) response (Oct. 19, 1995). Screening peptides used were: GGKKKYKL (p17-3 Gag; ref. 13), GEIYKRWII (p24-13 Gag; ref. 13), DCKTILKAL (p24-20 Gag; ref. 13), GPKVKQWPL (Pol; ref. 14), and FLKEKGGL (Nef; P.J.R.G., unpublished work).
Functional HLA Binding Assays.
Target cells were pulsed with 10 μM peptide for 1 h at 1.5, 5, 10, 16, and 24 h before the assay, then washed twice and incubated in R10 until labeling with 51Cr 1.5 h before cytotoxicity assay.
PCRs, Product Cloning, and Sequencing.
Genomic DNA was extracted directly from fresh uncultured PBMCs used to set up the bulk cultures (Puregene, Gentra Systems). Proviral nef and p17 gag were amplified using nested PCRs. Primers for the nef PCR were: 5′-GTAGCTGAGGGGACAGATAG-3′/5′-TGCTAGAGATTTTCCACAC-3′ for the primary reaction, and 5′-GAAGAATAAGACAGGGCT-3′/5′-AGGCTCAGATCTGGTCTAAC-3′ for the secondary reaction. The primary reaction mix contained 1 μg of DNA, 2.5 units of Taq polymerase, 10 μl of 10× KCl buffer containing 0.015 M MgCl2 (Bioline, London), 0.2 mM each dNTP, and 0.5 μM each primer in a total volume of 100 μl. Cycling parameters were 5 min at 94°C, 1 min at 52°C, and 1 min at 72°C, followed by 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C for 30 cycles. The secondary reaction used 1 μl of primary product; conditions were as for the primary except that the annealing temperature was 50°C. Primers for the p17 gag PCR were: 5′-ACTAGCGGAGGCTAG-3′/5′-GTTCTAGGTGATATGG-3′ for the primary reaction, and 5′-CAGTTCTAGATCTAGTAATTTTGGCTGACC-3′/5′-GCTAGAATTCCATGGGTGCGAGAGCGT-3′ for the secondary reaction. The primary reaction mix contained 1 μg of DNA in buffer containing 2.5 units of Taq polymerase, 0.05 M KCl, 0.01 M Tris (pH 8), 0.003 M MgCl2, 0.2 mM each dNTP, and 1 μM each primer. Cycling parameters were 5 min at 94°C, 1 min at 48°C, and 1 min at 72°C, followed by 1 min at 94°C, 1 min at 48°C, and 1 min at 72°C for 35 cycles. The secondary reaction used 1 μl of primary product; conditions were as for the primary except that the annealing temperature was 67°C. PCR products were cloned into the T-vector system (Amersham), and epitopes were sequenced with insert-specific primers (5′-GCCTGGCTAGAAGCA-3′ for HLA B8 nef and 5′-TGATGTACACAATAGAG-3′ for HLA B8 p17-3 gag) using the United States Biochemical Sequenase version 2.0 protocol.
RESULTS
The tissue type of donor SC2 was determined as HLA A1, B7/B8, Cw0701/Cw0702, DR4/DR16. Serial CD4+ lymphocyte counts and viral load measurements are shown in Table 1. Synthetic peptides representing known HIV-1 CTL epitopes restricted by these HLA molecules were pulsed on to B lymphoblastoid cell lines, and the pulsed cells were then used to screen bulk cultured lymphocytes for HIV-1-specific CTL activity at each of four time points. A strong response to a HLA B8-restricted epitope in Nef (FLKEKGGL; Nov. 3, 1995) and a weaker response to a HLA B8-restricted epitope in p17 Gag (GGKKKYKL, p17-3; Oct. 19, 1995) were detected (Fig. 1). A response to the Nef epitope was subsequently detectable in CTL lines grown from later time points (data not shown). Uncultured peripheral blood leukocyte proviral DNA from four time points was amplified, cloned, and analyzed for sequence variation within and near the regions encoding the two targeted epitopes (Fig. 2).
Table 1.
Serial CD4+ lymphocyte counts and plasma HIV-1 RNA loads from donor SC2
| Sample date | CD4+, × 106 per liter | Viral load, copies per ml |
|---|---|---|
| Oct. 19, 1995 | 384 | 7,600,000 |
| Nov. 3, 1995 | 518 | ND |
| Jan. 30, 1996 | ND | 715,000 |
| April 2, 1996 | 330 | 700,000 |
ND, not done.
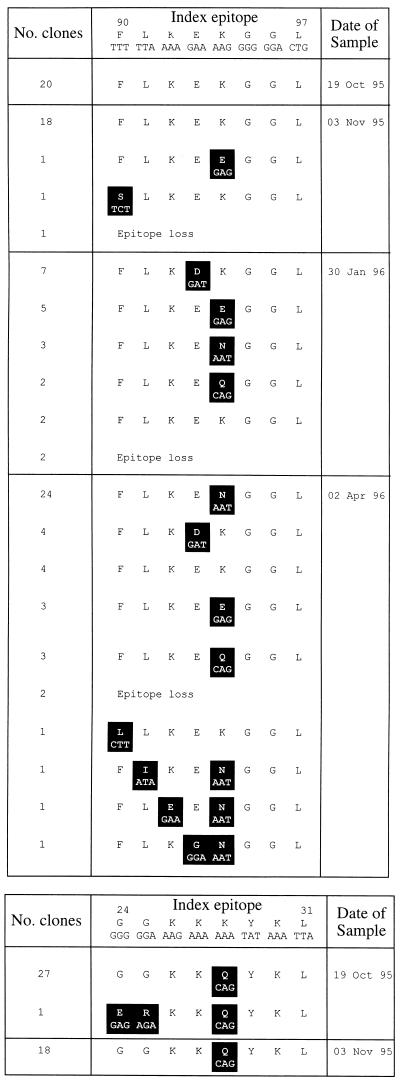
Figure 2.
Proviral sequence variation in HIV-1 epitopes from SC2. (a) HLA B8 Nef. (b) HLA B8 p17-3 Gag. Variant residues are highlighted.
Serial analysis of nef sequences demonstrated amino acid sequence variation confined chiefly to the epitope FLKEKGGL, suggesting positive selection for viruses bearing variants within this region of the gene (Fig. 3a). In contrast, the sequence of this nef region was invariant at amino acid level in a total of 27 clones from two time points from a HLA B8-negative donor with primary infection and in a total of 51 clones from two time points from a HLA B8-negative donor soon after seroconversion (data not shown).
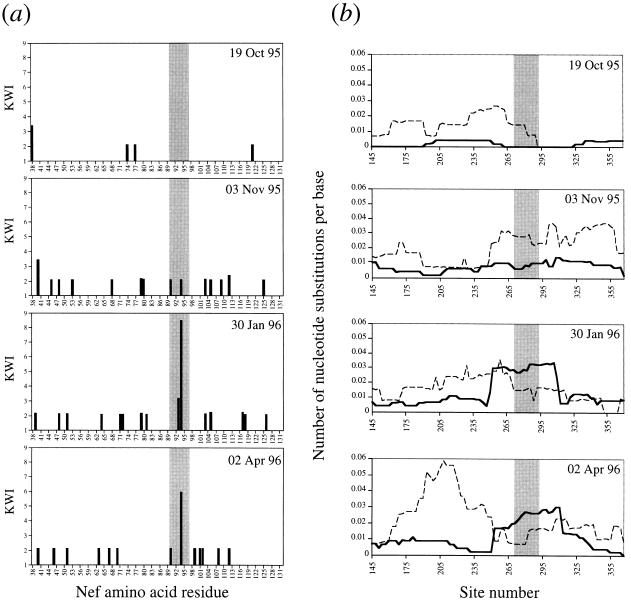
Figure 3.
Nucleotide and amino acid sequence variation in the region of Nef containing the HLA B8 epitope. For donor SC2, a number of clones from each time point (19 from each of the first three time points, 18 from the fourth time point) were sequenced further to incorporate regions flanking the epitope (nucleotides 114–395, amino acids 38–132, numbered according to the nef reference sequence HIV-LAI). (a) The Kabat–Wu index (KWI), calculated at each amino acid position as the number of different residues/frequency of the commonest residue, provides a measure of variability; an index of 1 indicates that no variation was detected at the relevant position (15). The HLA B8 epitope (amino acids 90–97) is shaded. (b) The rates of nonsynonymous (dN) and synonymous (dS) nucleotide substitutions were calculated in a sliding window of 60 nt over all pairwise comparisons between clones at a given time point, according to the method of Nei and Gojobori (16). The window is advanced in steps of three nucleotides, and the values of dN and dS are plotted against the central nucleotide in the window. dN rises sharply at the border of the region encoding the HLA B8 epitope, and exceeds dS over this region. The apparent extension of positive selection beyond the limits of the epitope reflects window width, not flanking mutations. Dotted line, synonymous substitutions; solid line, nonsynonymous substitutions.
To assess selection intensity, we examined the ratio of the rate of nonsynonymous to synonymous nucleotide mutations (dN/dS), where a ratio >1 indicates positive selection (16). With this approach, we found that dN exceeded dS in the region encoding the B8 Nef epitope on Jan. 30, 1996 and April 2, 1996, while an anti-Nef CTL response was still detectable on June 25, 1996 (Fig. 3b). The value of dN/dS for the whole amplified fragments did not exceed 0.84. Elsewhere in the amplified sequence, dN exceeded dS only at the 3′ end on Jan. 30, 1996; this region corresponds to a HLA B7-restricted epitope (amino acids 102–112; P.J.R.G., unpublished observation).
Two sequences were found in the p17-3 gag epitope (Fig. 2b). Both were variant compared with the consensus index sequence (GGKKKYKL; ref. 17).
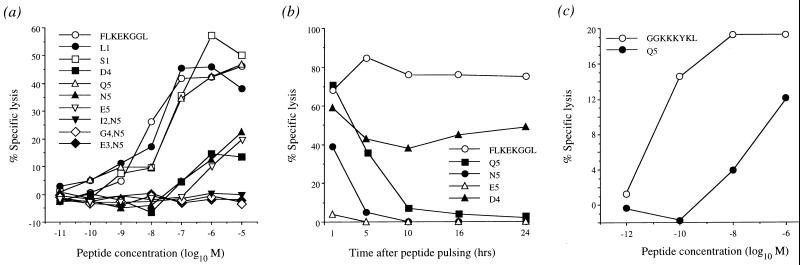
CTL lines specific for the index HLA B8 Nef epitope (FLKEKGGL) were derived from donor SC2 (Nov. 3, 1995 blood sample) and used to test the effects of the detected epitope variation on CTL recognition. Some variants (N5, E5, Q5, and D4) were poorly recognized, while others (I2/N5, E3/N5, and G4/N5) escaped recognition completely (Fig. 4a). The majority of variants encoded mutations at the HLA B8 anchor position 5 (Fig. 2a), and these changes resulted in reduced binding efficiency (Fig. 4b). Similarly, a HLA-B8 p17-3 Gag index-specific CTL line from donor SC2 showed diminished recognition of the p17-3 Q5 variant (Fig. 4c).
Figure 4.
(a) Altered recognition of variant Nef epitopes by donor SC2 CTLs. 51Cr-labeled HLA B8-matched targets were added directly to 96-well plates containing peptide, and therefore peptide was present at the concentrations drawn for the duration of the assay. Targets and peptide were incubated at room temperature for 15 min before the addition of effector cells. (b) Functional HLA binding assay. Target cells pulsed with variant peptides containing mutations at position 5 (a HLA B8 anchor position) were not recognized by donor SC2 CTLs after 6 h (N5 and E5) and 10 h (Q5), suggesting that these peptides bind poorly and have high off-rates. (c) The p17-3 Gag Q5 variant is poorly recognized by donor SC2-derived GGKKKYKL-specific CTLs.
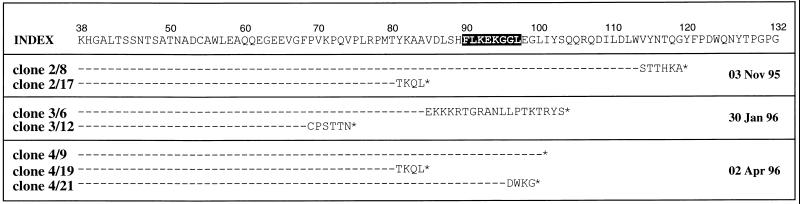
In addition to variants within the HLA B8 Nef epitope, a number of proviral clones contained mutations that abolished this epitope altogether (Fig. 5).
Figure 5.
Truncations and frameshifts within donor SC2 nef clones (deduced amino acid sequence). Identical single nucleotide deletions (clone 2/17, clone 4/19) resulting in premature termination of the open reading frame 6 aa before the B8 epitope are present at two separate time points, suggesting proviral replicative capacity. Index refers to proviral sequence from 19/10/95. ∗, Stop codon. Shading denotes the epitope. These sequences were not included in either the Kabat–Wu or dN/dS analyses.
DISCUSSION
Naturally occurring mutations in HIV-1 CTL epitopes have been described that reduce or abolish CTL recognition (13, 18). However, the role of CTLs in controlling the growth of HIV-1 in the host remains controversial, in part because there is no definitive evidence that CTL activity has a direct effect on the viral genotype. During primary infection, there is a temporal correlation between the development of anti-HIV-1 CTL responses and the decline of initial viremia (6, 7). We reasoned that if CTLs are important in limiting viral replication, then evidence of this effect should be apparent during such a period of intense CTL activity on a large viral population. Here we demonstrate positive selection for CTL escape mutations in a HIV-1 Nef epitope in a recent seroconverter; this selection was confined to the epitope and occurred in the presence of a strong CTL response directed against it. These results support the contention that CTLs play an important role in controlling viral growth in the host.
When the anti-Nef CTL response was first detected (Nov. 3, 1995), a deletion and two novel epitope sequences were present in nef in the proviral sequences. Subsequently, as the viral load fell, the array of escape variants expanded. Several months after the seroconversion illness, anti-Nef CTLs were detectable, while few HIV-1 clones in peripheral blood contained the Nef epitope sequence originally detected in this patient. Analysis of the genetic variation responsible for these epitope changes revealed that nucleotide substitutions productive of amino acid change exceeded silent substitutions within the epitope region, providing strong evidence of positive selection at this site in Nef (Fig. 3b). We cannot formally exclude the possibility that the observed nucleotide substitutions were selected by forces other than the immune response, such as structural constraints at the RNA or protein level (19). However, the confinement of the nonsynonymous substitutions and the high dN/dS ratio to the epitope encoding region, the absence of such substitutions in the control subjects, and the demonstration that the variant peptides failed to be recognized by the patient’s own CTLs indicate that CTL-mediated selection is the more likely selection force. In addition, variants that failed to escape CTL recognition (S1, L1) were poorly represented in the provirus population (Figs. 2a and 4a).
Further evidence of CTL-mediated positive selection in this individual came from the observation of deletions in the region of nef encoding the HLA B8-restricted epitope (Fig. 5). This observation is reminiscent of the epitope deletions detected following the infusion of an expanded CTL clone that recognized a single HLA A3 Nef epitope into a HIV-1-infected individual (20).
At the time of the first blood sample (Oct. 19, 1995), only variant virus sequences were detectable in the p17-3 Gag epitope (Fig. 2b). This suggests that the anti-p17-3 Gag CTL response, elicited earlier by the prototype sequence, had already driven out escape variants to fixation; however, since we did not detect index sequence in DNA proviruses, infection with virus bearing the p17-3 Q5 variant and stimulation of a crossreactive response cannot be excluded.
Using dN/dS in a comparison between HIV isolates, Seibert et al. (21) found evidence of positive selection in Env but not in Gag and concluded that anti-Gag CTL do not exert significant selection on the virus. It is presumed that positive selection on Env is antibody-mediated, as in the case of influenza A virus surface glycoproteins (gp; ref. 22). However, calculation of dN and dS in a sliding window can be used to delimit precise regions within a gene that are subject to locally focused selection (23). This technique can reveal significant positive selection in gene segments, even if the value of dN/dS averaged over the whole gene does not suggest that selection is occurring—for example, in the regions of major histocompatibility complex class I genes that encode the antigen recognition site (24).
In a systematic study of database sequences, Endo et al. (23) estimated that dN exceeded dS over the whole gene in 17/3595 (0.5%) groups of related genes. Of these 17 gene groups, 9 were the surface proteins of parasites or viruses, which are exposed to antibody-mediated selection. These results emphasize that positive selection operating over the full length of a gene is an uncommon event in evolution, particularly where the gene encodes an intracellular protein. However, positive selection has been demonstrated over the full length of the Tax (transactivator) protein of the human T cell leukemia virus type 1 (HTLV-1); this protein is the dominant target antigen of the intense anti-HTLV-1 CTL response (25). The Tax protein, like the Nef protein of HIV-1, is not expressed in an intact form on the surface of infected cells and is therefore removed from the pressure of antibody-mediated selection. It is likely that the positive selection in this case is exerted by the anti-Tax CTL response; however, CTLs in HTLV-1 infected individuals frequently recognize several epitopes in Tax (26), and therefore the high dN/dS ratio is less clearly localized to a given CTL epitope.
Selection of CTL escape variants in vivo was first demonstrated by Pircher et al. (27) in lymphocytic choriomeningitis virus (LCMV) infection. In this study, the diversity of the CTL response was experimentally limited by using mice transgenic for a T cell receptor specific for the LCMV gp antigen, thereby producing conditions in which the CTLs exerted intense selection pressure on the gp epitope in isolation. The restricted T cell diversity that has been observed during the primary CTL response to HIV-1 may facilitate escape in a similar way to the LCMV model (28).
Sequencing of HIV-1 in donors with primary infection has demonstrated substantial homogeneity, particularly in the gp120 region of env; however, some rapid sequence diversification has also been observed, especially within p17 gag and nef (29–31). Previous studies correlating the decline of initial viremia with dominant CTL responses have failed to identify escape variants within a targeted epitope in gp41 (32). However, a recent study found escape sequences in a gp160 epitope 7 weeks after infection in a patient with a monospecific immunodominant response (33).
In this study, we have demonstrated positive selection of CTL escape variants within Nef during primary HIV-1 infection. Although the phenotypic effects of these immune-selected Nef variants remain untested, viruses carrying nef gene defects have been associated with attenuated pathogenicity (34).
We conclude that HIV-1 variants that escape CTL recognition have a selective advantage during the early phase of infection. These mutant viruses may represent forms which help propagate the viremia that persists after seroconversion.
Acknowledgments
We thank Sarah Garrard, Ru Tan, and Tim Rostron for technical assistance, Toshinori Endo for supplying the computer program for sliding window analysis of dN/dS, and Andrew McMichael for helpful discussion and advice. This work was supported by the Wellcome Trust (D.A.P., P.K., A.K.S.. and R.E.P.) and the Medical Research Council (P.J.R.G.).
ABBREVIATIONS
- CTL
cytotoxic T lymphocyte
- gp
glycoprotein(s)
References
- 1.Piatak M, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K-C, Hahn B H, Shaw G M, Lifson J D. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 2.Clark S J, Saag M S, Decker W D, Campbell-Hill S, Roberson J L, Veldkamp P J, Kappes J C, Hahn B H, Shaw G M. N Engl J Med. 1991;324:954–960. doi: 10.1056/NEJM199104043241404. [DOI] [PubMed] [Google Scholar]
- 3.Daar E S, Moudgil T, Meyer R D, Ho D D. N Engl J Med. 1991;324:961–964. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M, Hahn B H, Saag M S, Shaw G M. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 5.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 6.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevan M J, Braciale T J. Proc Natl Acad Sci USA. 1995;92:5765–5767. doi: 10.1073/pnas.92.13.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari C, Bertoletti A, Fiaccadori F, Chisari F V. Semin Virol. 1996;7:23–30. [Google Scholar]
- 10.Browning M J, Krausa P, Rowan A, Bicknell D C, Bodmer J G, Bodmer W F. Proc Natl Acad Sci USA. 1993;90:2842–2845. doi: 10.1073/pnas.90.7.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunce M, Fanning G C, Welsh K I. Tissue Antigens. 1995;45:81–90. doi: 10.1111/j.1399-0039.1995.tb02422.x. [DOI] [PubMed] [Google Scholar]
- 12.Nixon D F, Townsend A R M, Elvin J G, Rizza C R, Gallwey J, McMichael A J. Nature (London) 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 13.Phillips R E, Rowland-Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R M, Rizza C R, McMichael A J. Nature (London) 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 14.Meier U-C, Klenerman P, Griffin P, James W, Koeppe B, Larder B, McMichael A, Phillips R. Science. 1995;270:1360–1362. doi: 10.1126/science.270.5240.1360. [DOI] [PubMed] [Google Scholar]
- 15.Wu T T, Kabat E A. J Exp Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 17.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N, editors. Human Retroviruses and AIDS. Group T-10, Los Alamos, NM: Theor. Biol. and Biophys.; 1995. [Google Scholar]
- 18.Couillin I, Culmann-Penciolelli B, Gomard E, Choppin J, Levy J P, Guillet J-G, Saragosti S. J Exp Med. 1994;180:1129–1134. doi: 10.1084/jem.180.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domingo E, Diez J, Martinez M A, Hernandez J, Holguin A, Borrego B, Mateu M G. J Gen Virol. 1993;74:2039–2045. doi: 10.1099/0022-1317-74-10-2039. [DOI] [PubMed] [Google Scholar]
- 20.Koenig S, Conley A J, Brewah Y A, Jones G M, Leath S, Boots L J, Davey V, Pantaleo G, Demarest J F, Carter C, Wannebo C, Yannelli J R, Rosenberg S A, Lane H C. Nat Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- 21.Seibert S A, Howell C Y, Hughes M K, Hughes A L. Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 22.Shu L L, Bean W J, Webster R G. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo T, Ikeo K, Gojobori T. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- 24.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 25.Niewiesk S, Daenke S, Parker C E, Taylor G, Weber J, Nightingale S, Bangham C R M. J Virol. 1994;68:6778–6781. doi: 10.1128/jvi.68.10.6778-6781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker C E, Nightingale S, Taylor G P, Weber J, Bangham C R M. J Virol. 1994;68:2860–2868. doi: 10.1128/jvi.68.5.2860-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pircher H, Moskophidis D, Rohrer U, Buerki K, Hengartner H, Zinkernagel R M. Nature (London) 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 28.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, Fauci A S. Nature (London) 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh-Brown A J, Simmonds P. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang S, Shlesinger Y, Daar E S, Moudgil T, Ho D D, Chen I S Y. AIDS. 1992;6:453–460. doi: 10.1097/00002030-199205000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Safrit J T, Andrews C A, Zhu T, Ho D D, Koup R A. J Exp Med. 1994;179:463–472. doi: 10.1084/jem.179.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borrow, P., Lewicki, H., Wei, X., Horwitz, M. S., Peffer, N., Myers, H., Nelson, J. A., Gairin, J. E., Hahn, B. H., Oldstone, M. B. A. & Shaw, G. M. (1997) Nat. Med., in press. [DOI] [PubMed]
- 34.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, et al. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]