Abstract
N-acetyl-l-leucyl-l-leucyl-l-norleucinal (LLnL), which reversibly inhibits the proteasome in addition to other proteases, and a more specific irreversible inhibitor of the proteasome, lactacystin, were found to cause the accumulation of major histocompatibility complex (MHC) class I heavy chains in the cytosol of the β2-microglobulin-deficient cell line Daudi and the TAP-deficient cell line .174. These cell lines, which are severely impaired in their ability to fold MHC class I heavy chain, showed an accumulation of soluble class I heavy chains at different rates over a period of hours in the presence of LLnL. The accumulation of soluble class I heavy chains in the presence of either LLnL or lactacystin was easily revealed in Daudi and .174 but almost undetectable in a Daudi transfectant expressing β2-microglobulin and in 45.1, the wild-type parent of .174. The soluble class I heavy chain was also found to be devoid of its N-linked glycan and to be located in the cytosol. When the gene for ICP47, a herpes simplex virus protein that blocks the translocation of peptides into the endoplasmic reticulum, was transfected into 45.1, a similar accumulation of soluble MHC class I heavy chain was detectable. These data suggest that in cells where the MHC class I molecule is unable to assemble properly, the misfolded heavy chain is removed from the endoplasmic reticulum to the cytosol, deglycosylated, and degraded by the proteasome.
Newly synthesized major histocompatibility complex (MHC) class I heavy chains are rapidly glycosylated and associate noncovalently with β2-microglobulin (β2m) in the endoplasmic reticulum (ER). The proper folding of free and β2m-bound human class I heavy chains is believed to be aided by the chaperones calnexin (1, 2) and calreticulin (3), respectively. The resulting class I–β2m heterodimer binds peptides primarily generated from cytosolic proteins by the proteasome (reviewed in ref. 4). These peptides are transported into the ER from the cytosol in an ATP-dependent fashion by the transporters associated with antigen processing (TAP; reviewed in ref. 5), which have been shown to physically associate with peptide-free class I–β2m dimers via a novel 48-kDa glycoprotein, tapasin (3). The correct assembly of the class I heavy chain with the soluble β2m subunit and an 8- to 10-residue peptide is necessary for the transport of the complex out of the ER to the cell surface. If β2m is absent, class I heavy chain–β2m dimers fail to transport to the cell surface and are degraded (6). If peptide is absent, for example, in mutant cell lines lacking one or both of the TAP subunits, “empty” class I–β2m dimers similarly fail to be expressed on the cell surface and are degraded (7).
The precise location and mechanism of the degradation of misfolded or incompletely assembled proteins retained in the ER have been difficult to determine. Studies on the assembly and degradation of multiple subunit complexes such as the T cell receptor (8), asialoglycoprotein receptor (9), acetylcholine receptor (10), or immunoglobulins (11) have suggested that degradation occurs in the ER or in a related compartment. Recently, experiments have indicated that such degradation could occur in the cytosol. For example, the degradation in isolated microsomes of the yeast secretory protein pro-α-factor, lacking native glycosylation sites, required cytosol and ATP (12), whereas the degradation of mutant cystic fibrosis transmembrane conductance regulator molecules was shown to involve proteasomes and ubiquitination (13, 14). Cytosolic degradation of misfolded MHC class I heavy chains was also implied in mice that had a targeted disruption of TAP.1. In this study, it was shown that peptide-deficient MHC class I complexes traveled to an ER–Golgi intermediate compartment that was found to be associated with ubiquitin and ubiquitin-conjugating enzymes (15). More recently, it was found that in yeast a mutated soluble vacuolar protein, carboxypeptidase yscY, is synthesized, glycosylated, and then transported from the ER lumen to the cytosolic side of the organelle, where the glycosylated form is conjugated with ubiquitin and degraded (16). Together these observations suggest that cytosolic degradation may play an important role in ER quality control.
Wiertz et al. (17) found that the US11 gene product from cytomegalovirus caused the dislocation of MHC class I heavy chain into the cytosol and determined that its subsequent degradation was caused by proteasomes. We speculated that US11 could be targeting class I molecules to the normal site of degradation for misfolded ER proteins. To investigate this possibility, we examined the degradation of class I heavy chains in the class I processing mutants Daudi and .174. Daudi does not express β2m (18), and .174 has a deletion in its MHC that includes TAP (19). These mutations severely limit the ability of class I heavy chain to fold properly or maintain a stable conformation and travel to the cell surface. We found that a cytosolic, deglycosylated soluble form of MHC class I heavy chain accumulates in cells treated with the protease inhibitors N-acetyl-l-leucyl-l-leucyl-l-norleucinal (LLnL), which has been shown to bind the active sites of the Archebacterial proteasome (20), and lactacystin, which has been shown to covalently bind the unique threonine active sites of the proteasome (21). Accumulation is induced only in the mutant cells Daudi and .174 and not in Daudi cells expressing β2m following transfection, nor in 45.1, the wild-type parent of .174. Interestingly, accumulation in 45.1 is induced when peptide loading is abrogated by transfecting into these cells the gene for ICP47, a herpes simplex virus protein that stops peptide translocation by the TAP heterodimer (22, 23). The mechanism of degradation, the involvement of the proteasome, and the possible role of viral targeting for immune system evasion are discussed.
MATERIALS AND METHODS
Cell Lines.
The Daudi cell line (6, 18) transfected with β2m (Daudi.β2m) and Daudi cells mock-transfected with vector only were provided by R. Salter (University of Pittsburgh, Pittsburgh). The line 45.1 and its TAP-deficient derivative, .174, have previously been described (19). The gene for ICP47 was a gift from David C. Johnson (Oregon Health Sciences University, Portland, OR) and was cloned into the expression vector pMCFR-PAC (constructed by T. Novak and L. Denzin, Yale University) at the EcoRI site. Stable cell lines were created by transfecting pMCFR-PAC.ICP47 into 45.1, yielding 45.1.ICP47 by selection in medium containing 0.75 μg/ml puromycin. All other cell lines were maintained in IMDM (GIBCO/BRL) with 7% calf serum (HyClone).
Antibodies.
The mouse mAbs HC10 [anti-free human leukocyte antigen (HLA) class I heavy chain] and w6/32 (anti-conformation specific HLA class I) and the rat mAb 3B10.7 (anti-HLA class I heavy chain) were previously described (24–26). The rat anti-grp94 mAb (clone 9G10) was purchased from Affinity BioReagents (Golden, CO). The rat anti-Hsc70 mAb (1B5) was purchased from StressGen Biotechnologies (Victoria, Canada).
Flow Cytometry.
Surface expression of MHC class I complexes on 45.1 and 45.1.ICP47 was detected by flow cytometry as described (27).
Inhibitors.
The protease inhibitor Calpain Inhibitor 1 or LLnL was purchased from Calbiochem–Novabiochem (San Diego) and prepared as a 25 mM (100×) stock in dimethyl sulfoxide (DMSO). The proteasome inhibitor lactacystin was purchased from E. J. Corey (Harvard University, Cambridge, MA) and prepared as a 2 mM (100×) stock in water.
Metabolic Labeling.
Cells (60 × 106) were incubated in methionine-free medium containing 5% dialyzed fetal calf serum (FCS; HyClone) with LLnL at 250 μM or DMSO solvent as control for 1 hr at 37°C. The cells were pulse labeled with 2.0 mCi [35S]methionine (ICN; 1 Ci = 37 GBq) for 10 min in fresh methionine-free medium and chased in the continued presence of LLnL or DMSO alone with a 15-fold excess of unlabeled methionine at 37°C for the indicated times. Labeling was stopped by diluting the cells in ice-cold PBS.
Subcellular Fractionation.
Labeled or unlabeled cells (107) were pelleted and frozen–thawed in a dry ice/ethanol bath twice. The frozen pellet was resuspended in 1.0 ml of 10 mM Tris/150 mM NaCl (TBS), pH 7.4, containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM N-α-tosyl-l-lysyl-chloromethyl ketone (TLCK), 5.0 mM iodoacetamide (IAA), and 5.0 mM N-ethylmaleimide (NEM) at 4°C. The nuclei were pelleted for 10 min at 2500 × g at 4°C. The postnuclear supernatant was then centrifuged at 100,000 × g for 1 hr at 4°C. The pellet from this spin was considered the membrane fraction, and the supernatant was considered the soluble fraction. The membrane pellet was lysed for 30 min on ice in TBS containing PMSF, TLCK, IAA, and NEM, as above, with the addition of 1% Triton X-100 (Sigma). The soluble fraction was also adjusted to 1% Triton X-100 by addition of a 10% stock. Immunoprecipitations were performed from the membrane and soluble fractions, as described.
Streptolysin-O (SLO) Permeabilization of Cells.
Cells (107) were treated with LLnL or DMSO for 4 hr at 37°C, pelleted in cold PBS, and permeabilized with SLO, as described (28). Briefly, cells were resuspended in 1 ml of PBS, pH 7.4, containing 0.5 mM PMSF, 0.1 mM TLCK, 5.0 mM IAA, 5.0 mM NEM, and 250 μM LLnL. Two units of SLO (Murex Diagnostics Limited, Dartford, U.K.), which was previously activated by treatment with 4 mM DTT at 37°C for 10 min, was added to the cell suspension, which was inverted twice, and incubated on ice for 10 min. The cells were washed twice in the PBS/inhibitor solution to remove unbound SLO and incubated for 30 min at 20°C in the same solution to allow the SLO to form pores and allow the cytosol to escape. The cells were pelleted for 30 sec at 4000 rpm, and the supernatant was cleared of fragmented cells by a 100,000 × g spin, and defined as the cytosolic fraction. The pellet was frozen–thawed and resuspended in TBS, as before, and the nuclei were removed by centrifugation. This postnuclear supernatant was defined as the lumenal fraction. Triton X-100 was added to both the cytosolic and the lumenal fractions to a final concentration of 1%, and after incubation on ice for 20 min, immunoprecipitations were performed as described below.
Immunoprecipitations and Carbohydrate Cleavage.
Membrane/soluble or lumenal/cytosolic fractions prepared from labeled or unlabeled cells, as described above, were precleared for 1 hr with normal rabbit serum and protein G-Sepharose and then incubated with mAb HC10 with protein A-Sepharose, or anti-grp94 and anti-Hsc70 with protein G-Sepharose for 1 hr at 4°C. Endoglycosidase H (endo H) and N-glycosidase F (N-glyF; Boehringer Mannheim) digestions were performed as described (7) using buffers suggested by the manufacturer. For pulse–chase experiments, HC10 immunoprecipitates from the membrane and soluble fraction were heated at 100°C for 5 min in 2% SDS and 2 mM DTT in TBS, diluted 10-fold with 1% Triton X-100 in TBS with 10 mM IAA, and allowed to incubate at room temperature for 40 min. After cooling to 4°C, released class I heavy chains were reprecipitated as above with 3B10.7 and protein G-Sepharose. All immunoprecipitations were separated in an SDS/10.5% polyacrylamide gel.
Immunoblots.
Blots were performed as described (29). Briefly, samples were separated by SDS/PAGE (10.5% gel) and electroblotted onto an Immobilon membrane (Millipore). The membrane was blocked for 1 hr in PBS containing 0.05% Tween 20 and 5% dehydrated milk (Blotto), rinsed in PBS, and incubated overnight at 4°C with a 1:1 dilution of 3B10.7 hybridoma supernatant in Blotto, a 1:1000 dilution of anti-Hsc70 ascites in Blotto, or a 1:3000 dilution of anti-grp94 ascites in Blotto. Bands were visualized with horseradish peroxidase-conjugated secondary mouse anti-rat IgG, F(ab′)2 fragment-specific antibody (Jackson ImmunoResearch) and chemiluminescent substrate (Pierce). Exposure times were typically 10–15 sec.
RESULTS
LLnL and the Proteasome Inhibitor Lactacystin Cause the Accumulation of Soluble MHC Class I Heavy Chains.
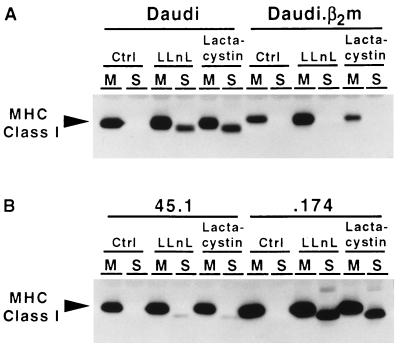
LLnL has been shown to bind the active sites of the Archebacterial proteasome (20) and to inhibit the activity of the mammalian proteasome (30, 31) in addition to other cellular proteases (31, 32). A more specific and potent inhibitor of the proteasome, lactacystin, has been found to bind irreversibly to the unique threonine active sites of the complex (21). In the experiments shown in Fig. 1, Daudi cells, which are deficient in β2m, and .174 cells, which lack the TAP heterodimer, were treated with 250 μM LLnL, 20 μM lactacystin, or DMSO as a solvent control for 2 hr at 37°C. The cells were frozen–thawed and separated into membrane and soluble fractions as described in Materials and Methods. After the addition of Triton X-100, HLA class I heavy chains were immunoprecipitated and visualized by SDS/PAGE and immunoblotting. LLnL and lactacystin induced the accumulation of a 40-kDa class I heavy chain derivative in both Daudi and .174, suggesting that the proteasome is involved in the degradation of misfolded class I heavy chains. When the repaired or normal counterparts of Daudi and .174, i.e., Daudi.β2m and 45.1, respectively, were treated with LLnL and lactacystin, the soluble 40-kDa species was undetectable in Daudi.β2m and barely detectable in 45.1. This result argues that MHC class I heavy chain can be rescued from proteasome-mediated degradation by expressing β2m in Daudi cells, allowing normal folding and assembly. It can also be diverted into this pathway by deletion of the genes for TAP, which causes the accumulation of unstable peptide-deficient class I molecules. Successful folding and assembly of MHC class I molecules in normal cells in the presence of the inhibitors also argues that the appearance of soluble molecules is not an artifact of inhibitor addition and is most likely the result of class I heavy chain misfolding in the ER.
Figure 1.
Proteasome inhibitors induce the accumulation of a MHC class I heavy chain derivative in Daudi and .174 cells but not in their wild-type counterparts. Daudi, Daudi.β2m, .174, and 45.1 cells were incubated in 250 μM LLnL, 20 μM lactacystin, or DMSO solvent control for 2 hr at 37°C and separated into membrane (M) and soluble (S) fractions by ultracentrifugation. MHC class I heavy chain was immunoprecipitated after addition of Triton X-100 using mouse antibody HC10, subjected to SDS/PAGE, transferred to Immobilon P membrane, and visualized using the rat antibody 3B10.7.
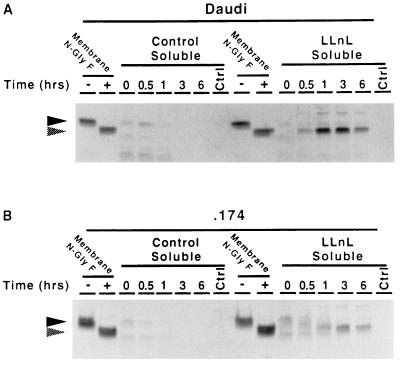
The Kinetics and Amount of Soluble MHC Class I Heavy Chain Accumulation Differ Between Daudi and .174.
To examine in more detail the degradation of misfolded MHC class I molecules, pulse–chase analysis was performed on the mutant cell lines Daudi and .174 in the presence and absence of the protease inhibitor LLnL. Both mutant cell lines are severely impaired in their ability to fold endogenous MHC class I molecules and maintain a stable conformation. However, in .174 cells, as in the related T2 cells, a substantial fraction of the encoded HLA-A2 molecules is expressed normally because it binds signal sequence-derived peptides (32–34). The cells were pulse-labeled with [35S]methionine for 10 min and chased for periods up to 6 hr, and membrane and soluble fractions were prepared. MHC class I molecules were immunoprecipitated from aliquots of the fractions using the mAb HC10, which recognizes free class I heavy chains.
In the absence of LLnL, only faint bands of apparently soluble glycosylated class I heavy chain and low molecular weight degradation products were visible in the soluble fraction of Daudi cells during early time points (Fig. 2A). In the presence of LLnL, however, an intense band, with the same mobility as membrane-bound class I heavy chain deglycosylated with N-glyF, appeared at the 0.5 hr time point. This species reached a maximum at 1 hr but remained detectable at 6 hr. It seems likely that the 40-kDa species represents class I molecules accumulating as a consequence of the proteolytic inhibitory activity of LLnL. The mutant cell line .174 behaved similarly, but the peak did not occur until 3 hr (Fig. 2B). In the absence of LLnL, a faint band of soluble and apparently glycosylated class I heavy chain could be seen at the early time points. In the presence of LLnL, a strong band with the same mobility as deglycosylated class I appeared in the soluble fraction from .174.
Figure 2.
Kinetic analysis of the accumulation of the MHC class I heavy chain derivative in Daudi (A) and .174 (B). Cells were preincubated for 1 hr with 250 μM LLnL or DMSO solvent control at 37°C, pulsed for 10 min with 2.0 mCi of [35S]methionine, and chased for 6 hr. At the indicated time points, samples were separated into membrane and soluble fractions by ultracentrifugation. MHC class I heavy chain was precipitated using the mouse antibody HC10 and then re-precipitated using the rat antibody 3B10.7 and subjected to SDS/PAGE. One-eighth of the membrane fraction from the zero time point was treated in the presence or absence of N-glyF and loaded to serve as a reference for deglycosylated class I heavy chain. Filled arrowheads indicate glycosylated class I heavy chain. Shaded arrowheads indicate deglycosylated class I heavy chain.
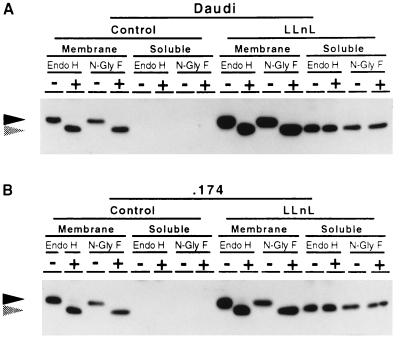
Soluble MHC Class I Heavy Chains Lack N-Linked Glycans.
Since the class I heavy chain degradation product that accumulated in the presence of LLnL had the same mobility on SDS/polyacrylamide gels as class I heavy chain devoid of N-linked glycans (Fig. 1), it seemed likely to represent deglycosylated class I heavy chains. To address this question, Daudi and .174 cells were treated with LLnL for 2 hr and separated into membrane and soluble fractions. Immunoprecipitated class I heavy chains were digested with endo H or N-glyF. Endo H cleaves N-linked glycans distal to the asparagine-linked N-acetylglucosamine residue, whereas N-glyF cleaves proximal to it. The results are shown in Fig. 3. It can clearly be seen that whereas membrane-associated MHC class I heavy chains in both cells were sensitive to digestion with either enzyme, the LLnL-induced soluble class I heavy chains were not. This result indicates that the soluble product lacks N-linked glycans. The identical mobilities in SDS/polyacrylamide gels of the soluble class I heavy chain and deglycosylated heavy chain argue that the protein is essentially intact. Similar experiments were performed using a rabbit antibody raised to the extreme C-terminal peptide of class I heavy chain with the same results (data not shown), indicating that the C terminus of the protein moiety is certainly intact.
Figure 3.
The LLnL-induced MHC class I heavy chain derivative is not glycosylated. Daudi (A) and .174 (B) cells were treated with 250 μM LLnL or DMSO solvent control for 2 hr at 37°C, separated into membrane and soluble fractions, immunoprecipitated using the mouse antibody HC10, treated with endo H or N-glyF, subjected to SDS/PAGE, transferred to Immobilin P membrane, and visualized using the rat antibody 3B10.7. Solid arrowheads indicate glycosylated class I heavy chain. Shaded arrowheads indicate deglycosylated class I heavy chain.
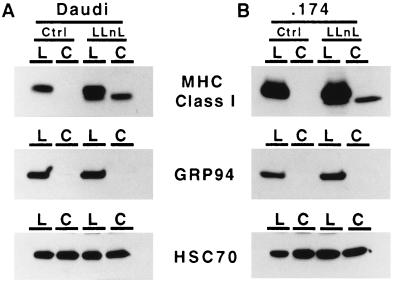
LLnL-Induced Soluble MHC Class I Heavy Chain Is in the Cytosol.
Since LLnL has been shown to inhibit peptidases in the ER (32), and the freeze–thaw technique can release soluble material from the ER, it was necessary to demonstrate that the deglycosylated and soluble class I heavy chain induced by LLnL was in the cytosol and thus accessible to the proteasome. To show this, Daudi and .174 cells were incubated with LLnL or DMSO for 4 hr at 37°C, chilled to 0°C, and incubated with SLO. After washing, the cells were incubated for 30 min at 20°C to allow pores to form. This technique results in pore formation only in the plasma membrane and not in internal vesicles, and allows cytosol to be released. Lumenal material remains within the cellular matrix and can be separated from cytosolic material by simple centrifugation. In Fig. 4, it can readily be seen that in both Daudi and .174 cells, LLnL induced the accumulation of soluble deglycosylated MHC class I heavy chains in the cytosolic fraction. The ER-retained soluble chaperone, grp94 (gp96), was also examined and found to be undetectable in the cytosolic fraction, indicating that the ER remained intact after SLO treatment. The cytosolic protein Hsc70, the constitutive form of Hsp70, was easily detected in both the cellular and the cytosolic fractions. Soluble class I heavy chains as well as cytosolic Hsc70 proteins were found in both the cellular and cytosolic fractions because only approximately 80% of the cells were permeabilized under the conditions used (data not shown). Thus both glycosylated and deglycosylated class I heavy chain bands appear in the lumenal fraction treated with LLnL.
Figure 4.
The LLnL-induced MHC class I heavy chain derivative is located in the cytosol. Daudi and .174 cells were treated with 250 μM LLnL or DMSO solvent control for 4 hr at 37°C. The cells were treated with SLO for 30 min at 20°C and separated into lumenal (L) or cytosolic (C) fractions by centrifugation. Class I heavy chains were immunoprecipitated using HC10, subjected to SDS/PAGE, and visualized using 3B10.7. The soluble, ER-retained chaperone, grp94 (gp96), was immunoprecipitated and visualized using the rat mAb clone 9G10. The soluble, cytosolic control molecule, Hsc70, was immunoprecipitated and visualized using the rat antibody 1B5.
LLnL Induces Soluble MHC Class I Heavy Chain in 45.1.ICP47.
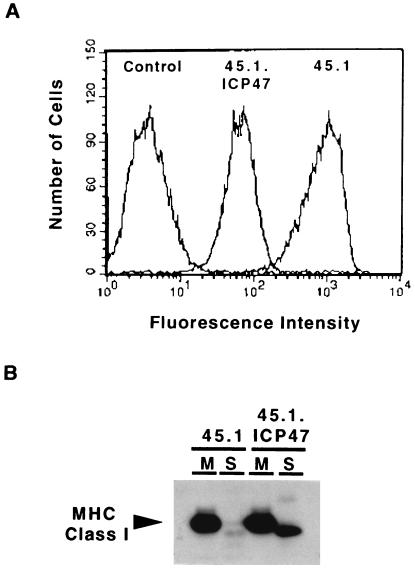
Class I heavy chains in Daudi cells do not associate with TAP (35), and in .174 cells the TAP.1/TAP.2 heterodimer is absent. Thus, one could argue that an inability to functionally interact with TAP is somehow responsible for the cytosolic proteolysis of class I molecules. To address this issue, we repeated the experiment in 45.1 cells expressing ICP47. ICP47 is an immediate early cytosolic protein encoded by herpes simplex virus that binds the TAP heterodimer and blocks the transport of peptides into the ER. In Fig. 5A, the dramatic effect of ICP47 on the surface expression of MHC class I complexes in 45.1 can be seen by flow cytometry. The 10-fold decrease in MHC class I surface expression demonstrates that ICP47 efficiently prevents peptide loading, resulting in the retention of unstable class I complexes in the ER. To determine if the accumulation of peptide-deficient class I–β2m heterodimers induced by ICP47 resulted in an increase in soluble deglycosylated class I heavy chain, 45.1 and 45.1.ICP47 were treated with LLnL for 2 hr, and membrane and soluble fractions were prepared. In the presence of LLnL, the amount of soluble MHC class I heavy chain detected in 45.1.ICP47 was clearly much greater than that found in the parent cell line 45.1 (Fig. 5B). This experiment demonstrates that simple lack of available peptides can cause MHC class I heavy chains to be diverted into the cytosol for degradation, and that the absence of TAP is not directly responsible.
Figure 5.
ICP47 induces the soluble MHC class I heavy chain derivative in 45.1. (A) The cells lines 45.1 and 45.1.ICP47 were analyzed for class I surface expression by flow cytometry using the mAb w6/32 and fluorescein-conjugated rabbit anti-mouse IgG. The control was a nonspecific isotype-matched mAb. (B) The lines 45.1 and 45.1.ICP47 were treated with 250 μM LLnL for 2 hr at 37°C and separated into membrane (M) and soluble (S) fractions by ultracentrifugation. MHC class I heavy chain was immunoprecipitated after addition of Triton X-100 using mouse antibody HC10, subjected to SDS/PAGE, transferred to Immobilon P membrane, and visualized using the rat antibody 3B10.7.
DISCUSSION
Recently, studies in yeast and mammalian cells using a variety of genetically altered and misfolded secreted or lumenal membrane-associated proteins have implicated the proteasome and the process of ubiquitination in the proteins’ degradation. One study on the degradation of misfolded or unstable MHC class I molecules suggested an ER–Golgi intermediate compartment associated with ubiquitin and ubiquitin-conjugating enzymes as the site of degradation (15). In another study, the expression of the human cytomegalovirus US11 gene product resulted in the rapid translocation of MHC class I heavy chain into the cytosol and degradation by the proteasome (17). Together these findings suggested that the class I molecules’ cytosolic degradation may be the normal result of a failure of MHC class I heavy chains to satisfy ER quality control processes. In the present study, we have examined cells expressing a viral protein that blocks MHC class I loading and mutant cell lines in which the process of class I assembly has been abrogated by a deficiency of β2m or peptide while the endogenous MHC class I genes remain unaltered. In these cells, the degradation of misfolded or unstable class I heavy chain is enhanced.
After the addition of the proteasome inhibitors LLnL and lactacystin, an intermediate in MHC class I heavy chain degradation accumulated in a soluble subcellular fraction of the mutant cell lines Daudi and .174 (Figs. 1 and 2). Importantly, this degradation intermediate was almost undetectable in the normal counterparts of these cells, Daudi.β2m and 45.1, indicating that the accumulation depended on class I heavy chain misfolding and that ER folding events influenced their processing (Fig. 1). The small amount of soluble, deglycosylated heavy chain visible in 45.1 cells upon treatment with LLnL and lactacystin may be because the supply of class I-binding peptides is reduced by proteasome inhibition, resulting in impaired assembly (31, 32). The intermediate was found to have the same mobility as deglycosylated class I heavy chain and was also found to be resistant to digestion with N-glyF, indicating the absence of N-linked glycans and suggesting that the protein portion was more or less intact (Fig. 3). Although relatively specific proteasome inhibitors were used to generate the intermediate, it was necessary to show that the accumulated soluble heavy chain was in the cytosol and accessible to the proteasome. This was clearly illustrated by release of the soluble heavy chains with the cytosolic contents of SLO-permeabilized cells (Fig. 4). We also showed that expressing ICP47 to specifically induce a peptide-deficient state caused cytosolic MHC class I heavy chains to accumulate in the presence of LLnL (Fig. 5). Taken together, our data suggest that MHC class I heavy chains, if improperly assembled with β2m and/or peptide in the ER, are dislocated into the cytosol, deglycosylated, and degraded by the proteasome.
The rate at which soluble MHC class I heavy chain accumulated in the cell line .174 (Fig. 2) was slower, and the intensity was lower than that in Daudi. Estimates from these pulse–chase experiments indicate that approximately 20% of the radiolabeled class I heavy chain was found in the soluble fraction of Daudi cells at the peak (1 hr), whereas only approximately 10% was in the soluble form in .174, peaking at 3 hr. If the accumulation depends ultimately on misfolding, the difference in kinetics and intensity between the two cells can be explained by the different defects in MHC class I assembly and the alternative pathway of signal sequence peptide loading available to HLA-A2 molecules synthesized in .174 cells. The lack of β2m in Daudi causes an early misfolding event that is absolute, making it impossible for any class I heavy chain to fold properly soon after synthesis. In .174, however, the presence of β2m enables both the HLA-A2 and the HLA-B51 heavy chains encoded by .174 to form transiently stable heterodimers. This allows a substantial fraction of the HLA-A2 molecules to bind peptides generated from signal sequences and form a stable conformation, even in absence of the TAP heterodimer. Thus, in .174, binding to β2m delays the appearance of class I heavy chain in the soluble fraction, whereas the binding of signal sequence peptides decreases the amount of class I heavy chain diverted into this pathway.
An interesting aspect of the cytosolic MHC class I heavy chain is the fact that an apparently full-length protein that includes a hydrophobic transmembrane region is soluble. This is inconsistent with the behavior of purified class I molecules and indicates to us that the material could be complexed to a chaperone. Many different chaperones are involved in protein degradation; some are involved in facilitating translocation across membranes, including the ER membrane, and others are involved in promoting degradation by the proteasome (reviewed in ref. 36). We speculate that hydrophobic regions of the class I heavy chain destined to be degraded in the cytosol may be bound by a constitutive heat shock protein, such as Hsc73. Although the proteasome is responsible for the majority of cytosolic degradation, proteins are not necessarily targeted to the proteasome by ubiquitination (37, 38). However, the class I material detected in the cytosol may be tagged with ubiquitin molecules. In the presence of LLnL or lactacystin, “laddering” of high molecular weight material reactive with the HLA class I heavy chain-specific antibody 3B10.7 can be detected when gels are overexposed. In addition, affinity-purified cytosolic class I molecules contain high molecular weight material reactive with anti-ubiquitin antibody on immunoblots (data not shown). These bands, however, are weak, and their relationship to the free soluble class I heavy chain has been difficult to determine.
The machinery responsible for the translocation of misfolded or unstable MHC class I heavy chains from the ER lumen to the cytosol is unknown. However, it has been suggested that a possible candidate is the translocon because of its configuration and position in the ER. The translocon is a large multisubunit complex responsible for the insertion and translocation of secreted or integral membrane proteins into and through the ER membrane (39). A few of the extended functions of the translocon are believed to include signal peptide cleavage, glycosylation, folding, quality control, and possibly the degradation of nascent polypeptide chains (reviewed in ref. 40). If the degradative function of the translocon is indeed mediated by the proteasome, it must have the ability to “retrotranslocate” or transport protein from the ER to the cytosol. Pseudomonas exotoxin A can enter the secretory pathway and escape into the cytoplasm from the ER apparently without ever leaving the translocon (41), providing some evidence for this possibility. It seems plausible, however, that a separate quality control mechanism, which does not involve the translocon, may cause the misfolded and unstable MHC class I heavy chains in Daudi and .174 to be transported back into the cytosol for degradation by the proteasome. Particularly in the case of .174 cells, assembly of heavy chain–β2m dimers appears to precede cytoplasmic delivery of the heavy chain for degradation. It seems unlikely that this would occur before segregation of the class I heavy chain from the translocon into the ER membrane. Resegregation of disassembled class I heavy chains into the translocon would have to occur before retrotranslocation if the translocon is indeed involved.
The enzymes mediating the deglycosylation of “retrotranslocated” proteins as well as the location of this activity are poorly characterized. Suzuki et al. (42) found N-glycanase activity apparently located in the cytosol, and Wiertz et al. (17) argued that ER localized N-glycanase activity is unlikely due to the risk of inappropriate removal of glycans from nascent or newly synthesized glycoproteins. The detection of apparently glycosylated class I heavy chain in soluble fractions at early time points during pulse–chase analysis (Fig. 2), as well as the kinetic difference between Daudi and .174, discussed earlier, also argues that deglycosylation occurs after translocation into the cytosol.
It remains unknown whether cytosolic deglycosylation and proteolysis of misfolded MHC class I heavy chains, or indeed other glycoproteins, by the proteasome is the main mechanism of their degradation after failing ER quality control. It may be an alternative pathway used in addition to an undefined ER proteolytic system. However, that cytosolic degradation of class I heavy chain can occur at all is important in that it can be used as a target by pathogens to evade the immune response. Thus, the cytomegalovirus US11 gene product may simply be taking advantage of this degradative pathway when it induces rapid MHC class I degradation. US11 may directly induce retrotranslocation or it may cause the misfolding of class I heavy chain, indirectly resulting in its diversion to the cytosol via the normal pathway. This would be analogous to the finding that ICP47 results in increased cytosolic degradation of peptide-deficient MHC class I molecules (Fig. 5). The extreme rapidity with which US11-mediated degradation occurs, however, suggests that it may play a more direct role than simply associating to cause misfolding. Whether retrotranslocation by US11 involves the translocon remains unknown. US11 or other cytomegalovirus products could target class I heavy chains for ubiquitination following translocation back into the cytosol, similar to the way that the human papilloma virus gene product E6 targets the p53 protein for ubiquitination and degradation (43). Whatever its mode of action, pathogenic targeting of this pathway may prove instrumental in deciphering the mechanism of proteasome-mediated degradation of ER proteins.
Acknowledgments
We thank Dr. R. Salter for the kind gift of β2m-transfected and mock-transfected Daudi cells and Dr. D. Johnson for the ICP47 gene. We also thank Mr. Will Stephen for his daily support and Ms. Nancy Dometios for help in preparation of this manuscript. This work was supported by National Institutes of Health Grant AI23081 and by the Howard Hughes Medical Institute. E.A.H. is also supported by the National Institutes of Health Medical Scientist Training Program.
ABBREVIATIONS
- MHC
major histocompatibility complex
- β2m
β2-microglobulin
- ER
endoplasmic reticulum
- TAP
transporters associated with antigen processing
- LLnL
N-acetyl-l-leucyl-l-leucyl-l-norleucinal
- HLA
human leukocyte antigen
- DMSO
dimethyl sulfoxide
- SLO
streptolysin-O
- endo H
endoglycosidase H
- N-glyF
N-glycosidase F
References
- 1.Williams D B, Watts T H. Curr Opin Immunol. 1995;7:77–84. doi: 10.1016/0952-7915(95)80032-8. [DOI] [PubMed] [Google Scholar]
- 2.Tector M, Salter R D. J Biol Chem. 1995;270:19638–19642. doi: 10.1074/jbc.270.33.19638. [DOI] [PubMed] [Google Scholar]
- 3.Sadasivan B, Lehner P J, Ortmann B, Spies T, Cresswell P. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.Howard J C. Curr Opin Immunol. 1995;7:69–76. doi: 10.1016/0952-7915(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 6.Sege K, Rask L, Peterson P A. Biochemistry. 1981;20:4523–4530. doi: 10.1021/bi00519a003. [DOI] [PubMed] [Google Scholar]
- 7.Salter R D, Howell D N, Cresswell P. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippincott-Schwartz J, Bonifacino J S, Yuan L C, Klausner R D. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 9.Wikstrom L, Lodish H F. J Biol Chem. 1993;268:14412–14416. [PubMed] [Google Scholar]
- 10.Blount P, Merlie J P. J Cell Biol. 1990;111:2613–2622. doi: 10.1083/jcb.111.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sitia R, Neuberger M S, Milstein C. EMBO J. 1987;6:3969–3977. doi: 10.1002/j.1460-2075.1987.tb02739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCracken A A, Brodsky J L. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 14.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 15.Raposo G, van Santen H M, Leijendekker R, Geuze H J, Ploegh H L. J Cell Biol. 1995;131:1403–1419. doi: 10.1083/jcb.131.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiller M M, Finger A, Schweiger M, Wolf D H. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 17.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 18.Rosa F, Berrissi H, Weissenbach J, Maroteux L, Fellous M, Revel M. EMBO J. 1983;2:239–243. doi: 10.1002/j.1460-2075.1983.tb01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeMars R, Rudersdorf R, Chang C, Petersen J, Strandtmann J, Korn N, Sidwell B, Orr H. Proc Natl Acad Sci USA. 1985;82:8183–8187. doi: 10.1073/pnas.82.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;26:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 21.Fenteany G, Standaert R F, Lane W S, Choi S, Correy E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 22.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 23.Fruh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampe R, Peterson P A, Yang Y. Nature (London) 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 24.Stam N J, Vroom T M, Peters P J, Pastoors E B, Ploegh H L. Int Immunol. 1990;2:113–125. doi: 10.1093/intimm/2.2.113. [DOI] [PubMed] [Google Scholar]
- 25.Barnstable C J, Bodmer W F, Braun G, Galfre G, Milstein C, Williams A F, Ziegler A. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 26.Lutz P M, Cresswell P. Immunogenetics. 1987;25:228–233. doi: 10.1007/BF00404692. [DOI] [PubMed] [Google Scholar]
- 27.Sadasivan B K, Cariappa A, Waneck G L, Cresswell P. Cold Spring Harbor Symp Quant Biol. 1995;60:267–275. doi: 10.1101/sqb.1995.060.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Androlewicz M J, Anderson K S, Cresswell P. Proc Natl Acad Sci USA. 1993;90:9130–9134. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson K S, Cresswell P. EMBO J. 1994;13:675–682. doi: 10.1002/j.1460-2075.1994.tb06306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlowski M, Cardozo C, Michaud C. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 31.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 32.Hughes E A, Ortmann B, Surman M, Cresswell P. J Exp Med. 1996;183:1569–1578. doi: 10.1084/jem.183.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson R A, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt D F, Engelhard V H. Science. 1992;268:262–264. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 34.Wei M, Cresswell P. Nature (London) 1992;356:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 35.Ortmann B, Androlewicz M J, Cresswell P. Nature (London) 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 36.Hayes S A, Dice J F. J Cell Biol. 1996;132:255–258. doi: 10.1083/jcb.132.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 38.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Nature (London) 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 39.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 40.Andrews D W, Johnson A E. Trends Biochem Sci. 1996;21:365–369. [PubMed] [Google Scholar]
- 41.Theuer C P, Buchner J, FitzGerald D, Pastan I. Cell Biol. 1993;90:7774–7778. doi: 10.1073/pnas.90.16.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. J Biol Chem. 1994;269:17611–17618. [PubMed] [Google Scholar]
- 43.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]