Abstract
Serological analysis of recombinant cDNA expression libraries (SEREX) using tumor mRNA and autologous patient serum provides a powerful approach to identify immunogenic tumor antigens. We have applied this methodology to a case of esophageal squamous cell carcinoma and identified several candidate tumor targets. One of these, NY-ESO-1, showed restricted mRNA expression in normal tissues, with high-level mRNA expression found only in testis and ovary tissues. Reverse transcription–PCR analysis showed NY-ESO-1 mRNA expression in a variable proportion of a wide array of human cancers, including melanoma, breast cancer, bladder cancer, prostate cancer, and hepatocellular carcinoma. NY-ESO-1 encodes a putative protein of Mr 17,995 having no homology with any known protein. The pattern of NY-ESO-1 expression indicates that it belongs to an expanding family of immunogenic testicular antigens that are aberrantly expressed in human cancers in a lineage-nonspecific fashion. These antigens, initially detected by either cytotoxic T cells (MAGE, BAGE, GAGE-1) or antibodies [HOM-MEL-40(SSX2), NY-ESO-1], represent a pool of antigenic targets for cancer vaccination.
Keywords: cancer, testis antigens
Although there has been a long search for evidence regarding human immune recognition of cancer, major progress has only recently been made in the molecular definition of human tumor antigens. By defining the antigenic targets on cultured melanoma cells recognized by autologous cytotoxic T lymphocytes (CTL), an array of autoimmunogenic tumor antigens has been identified, including several melanocyte differentiation antigens (1–7) and a growing family of testicular antigens aberrantly expressed in tumors (8–10). This approach, however, cannot be easily adapted to define antigens of other tumor types, particularly the common epithelial cancers such as breast or colon cancer. This limitation stems from the fact that the approach requires the establishment of autologous target cells in culture and the isolation of stable lines of autologous CTL that recognize antigens on the cultured tumor cells. In contrast to melanoma, where a success rate of 30% or more can be expected in establishing permanent cell lines, it has been notoriously difficult to establish cell lines from a number of other tumor types (e.g., breast cancer). This obstacle is further complicated by the fact that some epithelial cell types are poorly susceptible to CTL in vitro. For these reasons, the definition of tumor antigens in other epithelial cancers recognized by the autologous host requires alternative strategies. Sahin et al. (11) have recently developed an approach that can be used to identify immunogenic protein antigens in the generality of human cancers. In this approach, prokaryotically expressed cDNA libraries are prepared from tumor specimens and immunoscreened with absorbed and diluted patients’ sera for the detection of tumor antigens that have elicited a high-titer IgG humoral response. Such a humoral response implies T cell recognition of the detected antigens by helper T cells. Thus, even though the antigens are initially identified by antibodies, the method reveals tumor products that can then be analyzed in the context of cell-mediated immunity.
Applying this methodology to esophageal squamous cell carcinoma, we describe the isolation and characterization of NY-ESO-1, a gene expressed in normal testis and ovary, with aberrant expression in malignant tumors of various types.
MATERIALS AND METHODS
RNA Extraction and Construction of cDNA Expression Library.
Total RNA was extracted from cultured cell lines and from normal and tumor tissues. A cDNA library was constructed from a case of esophageal squamous cell carcinoma of a 58-year-old female. The library was constructed in a λZAP Express vector using a cDNA library kit (Stratagene).
Immunoscreening of the cDNA Library.
The cDNA library was screened with autologous patient’s serum as described (11). Briefly, the serum was diluted 1:10, preabsorbed with transfected Escherichia coli lysate, and a 1:10 dilution of the absorbed serum (final dilution of serum, 1:100) was incubated overnight at room temperature with the nitrocellulose membranes containing the phage plaques. After washing, the filters were incubated with alkaline phosphatase-conjugated goat anti-human Fcγ secondary antibodies, and the reactive phage plaques were visualized by incubating with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium. Phagemid clones encoding human immunoglobulin sequences were subsequently eliminated during the secondary screening.
Sequence Analysis of the Reactive Clones.
The reactive clones were subcloned, purified, and, in vitro, excised to pBK-CMV plasmid forms (Stratagene). Plasmid DNA was prepared using the Wizard Miniprep DNA purification system (Promega). The inserted DNA was evaluated by EcoRI-XbaI restriction mapping, and clones representing different cDNA inserts were sequenced. The sequencing reactions were performed by DNA Services at Cornell University (Ithaca, NY) using ABI PRISM (Perkin–Elmer) automated sequencers.
Reverse Transcription–PCR (RT-PCR).
To evaluate the mRNA expression pattern of the cloned cDNA in normal and malignant tissues, gene-specific oligonucleotide primers for PCR were designed to amplify cDNA segments of 300–400 bp in length, with the estimated primer melting temperature in the range of 65–70°C. All primers were commercially synthesized (Operon Technologies, Alameda, CA). RT-PCR was performed using 35 amplification cycles in a thermal cycler (Perkin–Elmer) at an annealing temperature of 60°C.
Northern Blot Analysis.
Northern blot analysis was performed as described (12). Briefly, 20 μg of total RNA per lane was dissolved in loading buffer containing formamide and formaldehyde, heated at 65°C, separated on a 1.2% agarose gel with 3% formaldehyde and transferred to nitrocellulose paper. Subsequent hybridization to the 32P-labeled probe and washing were performed under high-stringency conditions, with the final wash of the filter carried out with 0.1× standard saline citrate (SSC; 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.1% SDS at 60°C for 15 min.
RESULTS
SEREX Analysis of an Esophageal Carcinoma cDNA Library.
An expression cDNA library of 1.6 × 106 primary clones was prepared from a specimen of well to moderately differentiated squamous cell carcinoma of the esophagus, and 8.6 × 105 phage plaques were immunoscreened (6.0 × 105 from an unamplified library; 2.6 × 105 from an amplified library). Excluding the false-positive clones encoding immunoglobulin gene fragments, 13 positive clones were identified. These clones were purified, excised in vitro, and converted to pBK-cytomegalovirus plasmid forms. The inserts were analyzed by restriction mapping and DNA sequencing. The size of the inserts ranged from 0.5 to 1.3 kb, and sequencing revealed that they were derived from eight different genes, designated NY-ESO-1 through NY-ESO-8. Of these, NY-ESO-1 was represented by four overlapping clones, NY-ESO-2 by three overlapping clones, and others by one clone each (Table 1).
Table 1.
Genes isolated from an esophageal cancer library by autologous antibody screening
| Gene | Clone | Size, bp | DNA search | Comments* |
|---|---|---|---|---|
| NY-ESO-1 | E1-5b | 679 | No strong homology | Expressed in testis and ovary |
| E1-114b | 614 | |||
| E1-153c | 670 | |||
| E1-50 | 679 | |||
| NY-ESO-2 | E1-71a | 605 | U1 sn RNP1 homolog | Previously cloned by antibody screening |
| E1-140 | 874 | |||
| NY-ESO-3 | E1-141b | 517 | Colon 3′ direct MboI cDNA: brain cDNA | gb D25606D25606 and H18368H18368, unpublished |
| NY-ESO-4 | E1A-10c | 400 | No strong homology | Ubiquitous expression in normal tissues |
| NY-ESO-5 | E1A-54 | 670 | No strong homology | Predominant expression in normal esophagus |
| NY-ESO-6 | E1B-9b | ≈1200 | Human FUS/TLS gene | Translocated in liposarcoma, t(12:16) |
| NY-ESO-7 | E1B-20f | ≈1000 | Human U1-70k sn RNP | Different from NY-ESO-2 (gb M22636M22636) |
| NY-ESO-8 | E1B-20g | ≈1300 | No strong homology | Ubiquitous expression in normal tissues |
gb, GenBank accession numbers.
See text for detailed discussion and references.
DNA sequence homology revealed that NY-ESO-2 is identical to a U1 small nuclear ribonucleoprotein (sn RNP) homolog (13), and NY-ESO-7 is identical to hU1–70k sn RNP (14). Of the other clones, NY-ESO-6 is identical to human FUS/TLS, a gene involved in the chromosome 12:16 translocation in liposarcoma (15, 16), and NY-ESO-3 is identical to sequence tags previously cloned from human colon and adult brain cDNA libraries (GenBank database accession numbers H18368H18368 and D25606D25606, unpublished). Both NY-ESO-3 and NY-ESO-6 have been previously shown to be expressed in normal human tissues, with no evidence of lineage restriction. The NY-ESO-6 cDNA sequence is derived from the 3′ untranslocated portion of FUS/TLS. Comparison of NY-ESO-6 and FUS/TLS by sequencing and Southern blot analysis showed no evidence of translocation or point mutations in the esophageal cancer used to prepare the cDNA library (data not shown).
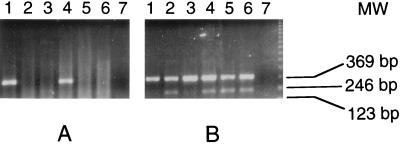
No strong homology to sequences in the DNA database was found for NY-ESO-1, 4, 5, or 8. Gene-specific primer pairs were prepared, and RNA expression patterns for these genes were examined by RT-PCR, using a limited tissue panel consisting of normal colon, kidney, testis, liver, and brain, in addition to the original tumor mRNA, which served as a positive control. NY-ESO-4 and NY-ESO-8 showed ubiquitous mRNA expression. NY-ESO-5 showed high-level expression in the original tumor, with equivocal-to-weak expression in other tissues. Subsequent tests with normal esophageal tissue also showed strong expression, suggesting that NY-ESO-5 might represent an autoimmunogenic normal differentiation marker. Of most interest, however, was NY-ESO-1, which appeared to be expressed only in testis and the tumor mRNA, but not in normal colon, kidney, liver, or brain (Fig. 1).
Figure 1.
RT-PCR analysis of NY-ESO-1 (A) and NY-ESO-4 (B) mRNA expression. Total RNA was extracted from the original esophageal cancer (lane 1) and from normal colon (lane 2), kidney (lane 3), testis (lane 4), liver (lane 5), and brain (lane 6). Negative control with no RNA was included (lane 7). NY-ESO-1 PCR primer sequences were: ESO1A, 5′-CACACAGGATCCATGGATGCTGCAGATGCGG-3′ and ESO1B, 5′-CACACAAAGCTTGGCTTAGCGCCTCTGCCCTG.
Restricted NY-ESO-1 Expression in Normal and Tumor Tissues.
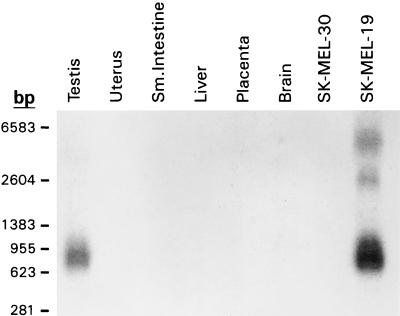
The expression of NY-ESO-1 in normal tissues was subsequently evaluated with a more complete panel by RT-PCR (Table 2). In addition to testis, NY-ESO-1 expression was found in ovary. A small amount of RT-PCR product was detected in uterine myometrium, but not in the endometrium. This reactivity, however, was seen inconsistently, suggesting a very low level mRNA expression. This interpretation was supported by the negative result on Northern blotting analysis (Fig. 2). Squamous epithelium from various sites, including normal esophagus and skin, showed no evidence of NY-ESO-1 expression. Placenta, an anatomic site previously shown to express MAGE-4 (17), a testicular antigen also aberrantly expressed in some malignancies, was negative for NY-ESO-1.
Table 2.
mRNA distribution of NY-ESO-1 in normal human tissues
| Tissue | mRNA | Tissue | mRNA |
|---|---|---|---|
| Esophagus | − | Adrenal | − |
| Brain* | − | Pancreas | − |
| Fetal brain | − | Seminal vesicle | − |
| Heart | − | Placenta | − |
| Lung | − | Thymus | − |
| Liver | − | Lymph node | − |
| Spleen | − | Tonsil | − |
| Kidney | − | Peripheral blood lymphocyte | − |
| Stomach | − | Peripheral blood lymphocyte activated† | − |
| Small intestine | − | Melanocytes | − |
| Colon | − | Thyroid | − |
| Rectum | − | Uterus | ±‡ |
| Breast | − | Testis | + |
| Skin | − | Ovary | + |
| Bladder | − | ||
Specimens from several different regions tested.
With interleukin 2 and phytohemagglutinin.
Weakly positive in some specimens, negative by Northern blot analysis (see Fig. 2).
Figure 2.
Northern blot analysis of NY-ESO-1, showing NY-ESO-1 mRNA in testis and SK-MEL-19, but not in uterus, small intestine, liver, placenta, brain, and SK-MEL-30. The major mRNA species is 0.8–0.9 kb, with minor higher molecular weight species seen in SK-MEL-19.
To evaluate NY-ESO-1 mRNA expression in tumors, a large panel of various tumor types was tested for NY-ESO-1 expression. RT-PCR results showed NY-ESO-1 mRNA expression in 23 of 67 melanoma specimens, 10 of 33 breast cancers, 4 of 16 prostate cancers, 4 of 5 bladder cancers, as well as in a proportion of other tumor types (e.g., hepatocellular carcinoma, lung cancer, ovarian cancer, etc.) (Table 3). A panel of 11 cultured melanoma cell lines was also tested; two (SK-MEL-19, SK-MEL-37) expressed NY-ESO-1, whereas the other nine were negative (SK-MEL-13, 23, 29, 30, 31, 33, 179, MZ2-MEL3.1, and MZ2-MEL2.2).
Table 3.
NY-ESO-1 mRNA expression in various human tumors by RT-PCR
| Tumor type | mRNA, positive/total | Tumor type | mRNA, positive/total |
|---|---|---|---|
| Melanoma | 23/67 | Ovarian cancer | 2/8 |
| Breast cancer | 10/33 | Thyroid cancer | 2/5 |
| Prostate cancer | 4/16 | Bladder cancer | 4/5 |
| Colon cancer | 0/16 | Burkitt lymphoma | 1/2 |
| Glioma | 0/15 | Basal cell carcinoma | 0/2 |
| Gastric cancer | 0/12 | Leiomyosarcoma | 0/2 |
| Lung cancer | 2/12 | Other sarcomas | 0/2 |
| Renal cancer | 0/10 | Pancreatic cancer | 0/2 |
| Lymphoma* | 0/10 | Seminoma | 0/1 |
| Hepatoma | 2/7 | Spinal cord tumor | 0/1 |
Non-Hodgkin, non-Burkitt types.
Northern blot analysis was performed to determine the size of the NY-ESO-1 transcript and to confirm the tissue expression pattern (Fig. 2). RNA from testis and NY-ESO-1 mRNA-positive melanoma cell line SK-MEL-19 showed a RNA transcript of ≈0.8 kb. The esophageal carcinoma specimen showed a smear in the 0.4–0.8 kb range, reflecting partial degradation of the tumor mRNA specimen (data not shown). mRNA from uterus, small intestine, liver, placenta, brain, and melanoma cell line SK-MEL-30 showed no NY-ESO-1 transcript.
Full-Length NY-ESO-1 cDNA and Putative Peptide Sequences.
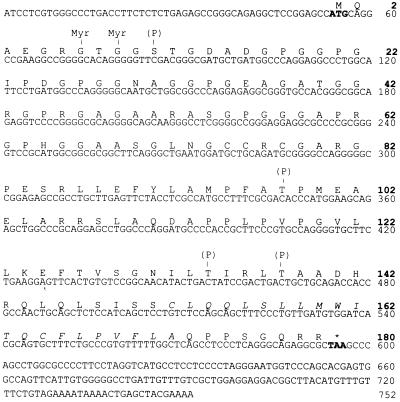
To obtain the full-size NY-ESO-1 cDNA sequence, the esophageal cDNA library was rescreened by plaque hybridization using the original cDNA clone as the nucleotide probe. Upon screening of 3 × 105 clones, six positive clones were obtained. Sequencing of the three longest clones and analysis of the ORFs revealed that all three clones contained the entire coding region, with 5′ untranslated regions of variable size. The longest clone, 747 bp in length excluding the poly(A) tail, contained the 543-bp coding region, flanked by 53 bp in the 5′ untranslated region and 151 bp in the 3′ untranslated region (Fig. 3).
Figure 3.
Nucleotide and amino acid sequences of NY-ESO-1, indicating potential N-myristoylation (Myr) and phosphorylation (P) sites. The potential transmembrane domain at the carboxyl end is italicized. (GenBank database accession number: U87459U87459.)
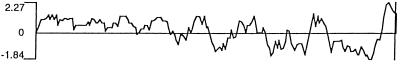
The long ORF, which is represented in the immunopositive clone, indicated that the putative NY-ESO-1 protein consists of 180 aa, 173 of which are in the immunopositive clone (Fig. 3). The putative molecular mass of NY-ESO-1 is 17,995 Da. An abundance of glycine residues was noted in the N-terminal portion of the protein, comprising 30 of the first 80 aa (4 in the remaining 100 residues). Hydrophilicity analysis (Fig. 4) indicated hydrophilic sequences in the N-terminal half of the molecule, followed by alternating hydrophobic and hydrophilic sequences. A long hydrophobic tail is present at the C terminus (amino acids 152–172) in front of a short hydrophilic tail, suggesting the possibility of a transmembrane domain. However, there was no evidence of a leader sequence in this protein. Analysis of the protein sequence for possible posttranslational modifications revealed several potential N-myristoylation sites and three phosphorylation sites, but no evidence for N-glycosylation sites (Fig. 3).
Figure 4.
Hydrophilicity plot of NY-ESO-1, indicating hydrophilic domain (positive value) in the amino terminus, and a long hydrophobic stretch (negative value) close to the carboxyl end of the putative protein sequence.
DISCUSSION
Although there have been many attempts to define the humoral response of patients to their own cancer, analyzing the specificity of the detected antibodies has been a persistent challenge. To devise a test system that would be as unambiguous as possible, an approach termed autologous typing was developed more than two decades ago [see review by Old (18)]. Patients’ sera were screened for cell-surface reactivity against autologous cultured tumor cell lines by several serological methods, and specificity was established by testing a broad range of autologous and allogeneic malignant and normal cell types in direct assays and by absorption analysis. Autologous typing was applied mainly to melanoma (19), astrocytoma (20), and renal cancer (21), due to the relative ease of establishing autologous cell lines from these tumor types. Although glycolipid (22) and glycoprotein antigens (23) were identified by this approach, the serum titers were generally too low to permit the molecular identification of the detected antigens.
The development of serological identification of antigens by recombinant expression cloning (SEREX) was designed to extend the range of autologous typing by incorporating molecular techniques of expression cloning, thus providing a general method to identify immunogenic tumor antigens in any tumor type (11). Unlike autologous typing, SEREX is not restricted to antigens expressed on the cell surface, but can dissect the immune response to intracellular antigens, a task that was generally beyond the means of conventional serology. In addition, SEREX obviates the need for establishing cultured cell lines as serological test targets, a critical advantage in the study of tumor types such as breast and colon cancers, which generally are difficult to culture. Specificity testing also enters a new realm of precision with SEREX, because once a reactive clone is identified, the cDNA can be sequenced and the pattern of mRNA expression can be readily examined.
Expression cloning and immunoscreening with human serum has been well established as an approach to identify genes encoding antigens associated with autoimmune diseases (24) or antigens related to paraneoplastic syndromes (e.g., see ref. 25). Similar approaches, however, only recently have been applied to the identification of tumor antigens. One obvious concern was that most of the genes isolated in this way would code for common cellular constituents, representing autoantigens to which humoral responses either preexisted or were elicited following tumor growth/necrosis. Indeed, two of the eight genes isolated in the present study are related to U1 sn RNPs, one of which was previously identified by the immunoscreening of a thyroid cDNA library with serum from a patient with autoimmune thyroid disease (13).
However, the application of SEREX to cancer has convincingly demonstrated the power of the technique to identify antigens of particular interest and relevance. For instance, several novel tumor products were identified in the initial SEREX analysis of renal cancer, Hodgkin disease, astrocytoma, and melanoma (11). In the case of melanoma, several antigens were isolated, including MAGE-1, tyrosinase, and a novel melanoma antigen HOM-MEL-40, which was later shown to be identical to SSX2, a gene translocated in synovial sarcoma (26, 27). Since MAGE-1 was originally detected by cytotoxic T cells (2, 4, 8), these findings confirm the notion that SEREX is capable of capturing antigens recognized by both B and T cells, serving as an alternative strategy for the identification of antigenic targets for tumor vaccination.
The present study adds another antigen, NY-ESO-1, to the growing list of SEREX-defined tumor antigens. Comparison with other immunogenic tumor antigens identified by either cell-mediated or humoral immunity indicates that NY-ESO-1 belongs to an expanding family of antigens with characteristic expression in normal testis and in cancers. The list of these human cancer/testis (CT) antigens includes the MAGE family [MAGE-1 (8), MAGE-3 (10), and others], GAGE-1 (28), BAGE (9), HOM-MEL-40 (SSX2) (11, 27), and NY-ESO-1. Mouse tumor antigen P815 (29), expressed in testis and mastocytoma, also belongs to this family. All CT antigens share the following features: (i) predominant mRNA expression in testis, but generally not in other normal tissues, (ii) gene activation and high-level mRNA expression in certain malignancies, and (iii) expression in malignancies in a lineage-nonspecific fashion. Another intriguing common feature of CT antigens is mapping to the X chromosome; this has been shown for MAGE (17), GAGE-1 (K. Arden, personal communication), SSX2 (27), and NY-ESO-1 (K. Arden, and Y.-T.C., unpublished data). BAGE has not yet been mapped.
Despite these shared characteristics, individual CT antigens also show distinct features. For example, mRNA expression in placenta was found in the case of MAGE-4, but not with other MAGE members, BAGE, SSX2, or NY-ESO-1. SSX2 is expressed in thyroid at low levels and NY-ESO-1 is expressed in ovary, whereas MAGE, BAGE, and GAGE show no expression in ovary (8, 9, 28). Analysis of mRNA expression in panels of tumor cell lines or tumor specimens also revealed no evidence for coordinated expression of various CT antigens (ref. 28; Y.-T.C., and L.J.O., unpublished data), indicating that they are regulated independently. The aberrant expression of CT antigens in tumors can best be ascribed to derepression events, allowing sporadic activation of these antigens in tumors of unrelated lineages. In this regard, residence on the X chromosome with a single active copy probably renders these genes more highly susceptible to derepression events. The finding that some CT antigens (e.g., MAGE and BAGE) are more frequently expressed in metastatic melanomas than primary lesions (9, 30) suggests that such derepression may be a later event in tumor progression.
The restricted expression of these antigens in testis, an immunologically privileged anatomical site, likely explains the potential for cancer-related immune recognition of these antigens. Of the CT antigens identified to date, four (MAGE-1, MAGE-3, BAGE, GAGE-1) have been shown to elicit cytotoxic T cell responses, and three (MAGE-1, SSX2, NY-ESO-1) have been shown to elicit humoral responses. Considering the example of MAGE-1, where both cytotoxic T lymphocyte and antibody responses have been described, it will be of interest to investigate the possible CD8 T cell response against NY-ESO-1. In addition, serological surveys can now be carried out to assess the frequency of CT-specific antibodies in patients with cancer and other diseases, and normal individuals.
Despite the detailed knowledge on the genomic and cDNA sequences of the CT antigens, nothing is known about the biologic role of these molecules. Through cell fractionation and immunohistochemical studies (31, 32), it has been shown that MAGE-1 is a cytoplasmic protein, and there is no structural evidence for a transmembrane domain in MAGE, BAGE, GAGE, or SSX2. NY-ESO-1, in contrast, contains a hydrophobic segment at its carboxyl end. This observation, in conjunction with the identification of potential N-myristoylation sites, suggests that NY-ESO-1 may be a membrane-associated protein. An antibody probe should help clarify this issue. Serological reagents are also needed to identify the nature of NY-ESO-1-positive cells in the testis and ovary, and to immunophenotype tumors for NY-ESO-1 expression (33).
ABBREVIATIONS
- SEREX
Serological analysis of recombinant cDNA expression libraries
- RT-PCR
reverse transcription–PCR
- CT
cancer/testis
- sn RNP
small nuclear ribonucleoprotein
Footnotes
References
- 1.Boon T, Cerottini J C, Van den Eynde B, van der Bruggen P, Van Pel A. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 2.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulie P G, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora J P, Renauld J C, Boon T. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins P F, El-Gamil M, Kawakami Y, Rosenberg S A. Cancer Res. 1994;54:3124–3126. [PubMed] [Google Scholar]
- 5.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Rivoltini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Sakaguchi K, Apella E, Yannelli J R, Adema G J, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R F, Robbins P F, Kawakami Y, Kang X-Q, Rosenberg S A. J Exp Med. 1995;181:799–804. doi: 10.1084/jem.181.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 9.Boel P, Wildmann C, Sensi M L, Brasseur R, Renauld J, Coulie P, Boon T, van der Bruggen P. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 10.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio J J, De Plaen E, Lethe B, Brasseur F, Boon T. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U, Türeci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Stuhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 13.Elisei R, Weightman D, Kendall-Taylor P, Vassart G, Ludgate M. J Endocrinol Invest. 1993;16:533–540. doi: 10.1007/BF03348900. [DOI] [PubMed] [Google Scholar]
- 14.Spritz R A, Strunk K, Surowy C S O, Hoch S, Barton D E, Francke U. Nucleic Acids Res. 1987;15:10373–10391. doi: 10.1093/nar/15.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabbits T H, Forster A, Larson R, Nathan P. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 16.Crozat A, Aman P, Mandahl N, Ron D. Nature (London) 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 17.De Plaen E, Arden K, Traversari C, Gaforio J J, Szikora J-P, De Smet C, Brasseur F, Van der Bruggen P, Lethe B, Lurquin C, Brasseur R, Chomez P, De Backer O, Cavenee W, Boon T. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 18.Old L J. Cancer Res. 1981;41:361–375. [PubMed] [Google Scholar]
- 19.Carey T E, Takahashi T, Resnick L A, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1976;73:3278–3282. doi: 10.1073/pnas.73.9.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfreundschuh M, Shiku H, Takahashi T, Ueda R, Ransohoff J, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1978;75:5122–5125. doi: 10.1073/pnas.75.10.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda R, Shiku H, Pfreundschuh M, Takahashi T, Li L T C, Whitmore W F, Oettgen H F, Old L J. J Exp Med. 1979;150:565–579. doi: 10.1084/jem.150.3.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe T, Pukel C S, Takeyama H, Lloyd K O, Shiku H, Li L T C, Travassos L R, Oettgen H F, Old L J. J Exp Med. 1982;156:1884–1889. doi: 10.1084/jem.156.6.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Real F X, Mattes M J, Houghton A N, Oettgen H F, Llyod K O, Old L J. J Exp Med. 1984;160:1219–1233. doi: 10.1084/jem.160.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter J B, Shoenfeld Y, editors. Autoantibodies. Amsterdam: Elsevier Science; 1996. [Google Scholar]
- 25.Dropcho E A, Chen Y-T, Posner J B, Old L J. Proc Natl Acad Sci USA. 1987;84:4552–4556. doi: 10.1073/pnas.84.13.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Türeci Ö, Sahin U, Schobert I, Koslowski M, Schmitt H, Schild H-J, Stenner F, Seitz G, Rammensee H-G, Pfreundschuh M. Cancer Res. 1996;56:4766–4772. [PubMed] [Google Scholar]
- 27.Crew A J, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson B A, Cooper C S. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van den Eynde B, Lethe B, Van Pel A, De Plaen E, Boon T. J Exp Med. 1991;173:1373–1384. doi: 10.1084/jem.173.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brasseur F, Rimoldi D, Lienard D, Lethe B, Carrel S, et al. Int J Cancer. 1995;63:375–380. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- 31.Amar-Costesec A, Godelaine D, Stockert E, Van der Bruggen P, Beaufay H, Chen Y-T. Biochem Biophys Res Commun. 1994;204:710–715. doi: 10.1006/bbrc.1994.2517. [DOI] [PubMed] [Google Scholar]
- 32.Schultz-Thater E, Juretic A, Dellabona P, Luscher U, Siegrist W, Harder F, Heberer M, Zuber M, Spagnoli G C. Int J Cancer. 1994;59:435–439. doi: 10.1002/ijc.2910590324. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y-T, Stockert E, Tsang S, Coplan K A, Old L J. Proc Natl Acad Sci USA. 1995;92:8125–8129. doi: 10.1073/pnas.92.18.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]