Abstract
Treatment of human cells with 2′,5′ oligoadenylate covalently linked to antisense (2–5A-antisense) results in the selective cleavage of targeted RNA species by 2–5A-dependent RNase L. Here we show that 2–5A-antisense containing stabilizing modifications at both termini are effective in suppressing the replication of respiratory syncytial virus (RSV) in human tracheal epithelial cells. The affinity of 2–5A-antisense for different regions in the RSV M2 and L mRNAs was predicted from a computer-generated model of the RNA secondary structure. The most potent 2–5A-antisense molecule caused a highly effective, dose-dependent suppression of RSV yields when added to previously infected cells. In contrast, control oligonucleotides, including an inactive dimeric form of 2–5A linked to antisense, 2–5A linked to a randomized sequence of nucleotides, and antisense molecules lacking 2–5A, had minimal effects on virus replication. The specificity of this approach was shown by reverse transcriptase-coupled PCR analysis of RSV M2, P, and N mRNA and of cellular glyceraldehyde-3-phosphate dehydrogenase mRNA. The RSV M2 mRNA amounts were depleted after treating RSV-infected cells with 2–5A-antisense targeted to this mRNA, whereas the amounts of the other RNA species were unchanged. These studies demonstrate that 2′,5′ oligoadenylate covalently linked to antisense (2–5A-antisense) can effectively suppress RSV replication by directing the cellular RNase L to selectively degrade an essential viral mRNA.
Respiratory syncytial virus (RSV), a nonsegmented, negative-strand RNA virus in the pneumovirus genus of Paramyxoviridae, is a major cause of lower respiratory disease, resulting in 90,000 hospitalizations and 4500 deaths per year in infants and young children in the United States (1). Of growing concern are outbreaks of RSV within institutionalized elderly (2) and immunodeficient (1) adults. There is only one approved anti-RSV compound, ribavirin, which reduces virus shedding in infected children, but has no effect on mortality or duration of hospitalization (3). In the search for alternative antiviral strategies, we have investigated the potential of novel chimeric antisense molecules, 2–5A-antisense, in the control of RSV infections.
Zamecnik and Stephenson were the first to report an antiviral effect of antisense oligonucleotides by showing that replication of the retrovirus Rous sarcoma virus could be inhibited by the addition of oligonucleotides complementary in sequence to reiterated terminal sequences of the viral 35S RNA (4). Subsequently, antisense oligonucleotides have been used against a wide range of different types of viruses, including the negative-strand RNA viruses such as rabies virus (5) and influenza virus (6), positive-strand RNA viruses such as dengue virus (7), DNA viruses including hepatitis B (8) and herpes simplex virus type 1 (9), and the retrovirus HIV-1 (10, 11). Despite encouraging results, significant challenges remain in the development of antisense oligonucleotides as antiviral agents. For instance, oligonucleotides applied to infected cells must penetrate the cell membrane, enter the intracellular compartment that contains the viral nucleic acid, then bind and inactivate or inhibit the function of the viral nucleic acid before the oligonucleotides themselves are degraded. Therefore, oligonucleotides that are stable and can function at very low concentrations to bring about the rapid and catalytic destruction of the viral RNA targets will have inherent advantages compared with other types of antisense.
2–5A-antisense represents a class of chimeric oligonucleotides designed to bind to and activate the ubiquitous intracellular endoribonuclease RNase L (12). A 5′-phosphorylated 2′,5′ linked oligoadenylate (2–5A) is required to activate the catalytic activity of RNase L (reviewed in ref. 13). 2–5A-antisense chimeras contain 2–5A covalently attached through butanediol phosphate linkers to antisense oligonucleotide. In nature, RNase L functions as a component of the 2–5A system (14), implicated in the antiviral activity of interferon against picornaviruses (15–19), vaccinia virus (20, 21), and reovirus (22). In the 2–5A-antisense approach, RNase L is converted to a highly specific endoribonuclease capable of selectively cleaving individual RNA targets. This previously was demonstrated in cell-free systems against a modified HIV-1 vif mRNA (12) and against an mRNA target encoding the double-stranded RNA-activatable protein kinase, PKR (23). When applied to HeLa cells, the 2–5A-antisense resulted in the ablation of PKR mRNA and enzymatic activity (24). The previous demonstration of elevated levels of RNase L after RSV infections of human alveolar macrophages (25) suggested that RSV might be susceptible to antiviral approaches involving RNase L. Here we show that the recruitment and activation of RNase L to an RSV mRNA with 2–5A-antisense oligonucleotides results in a potent inhibition of RSV replication in human tracheal epithelial cells.
MATERIALS AND METHODS
Chemical Synthesis of 2–5A-Antisense Chimeras and Control Oligonucleotides.
Chimeric oligonucleotides for this study were synthesized and purified using modifications of previously published procedures (12, 24, 26–28). Solid supports, 3′-O-dimethoxytrityl-N6-benzoyl-2′-deoxyadenosine-5′-lcaa-CPG (500 Å; lcaa-CPG indicates long chain alkylamino-controlled pore glass), 3′-O-dimethoxytrityl-N4-benzoyl-2′-deoxycytidine-5′-lcaa-CPG (500 Å), 3′-O-dimethoxytritylthymidine-5′-lcaa-CPG, and 3′-O-dimethoxytrityl-N2-isobutyryl-2′-deoxyguanosine-5′-lcaa-CPG (500 Å) were obtained from Glen Research (Sterling, VA) and were used to synthesize oligonucleotides with an inverted 3′-3′ terminal phosphodiester bond. Antisense phosphorothioate oligodeoxynucleotide was prepared on Applied Biosystems 380B synthesizer using tetraethylthiuram disulfide and β-cyanoethyl phosphoramidite chemistry (29). The generic structure of 2–5A-antisense is sp5′A2′p(5′A2′p)n -[O(CH2)4Op]2–5′dN3′p(5′dN3′p)m5′dN3′p-3′pdN5′ (n = 1, 3) (for specific structures see Table 1). All of the 2–5A-antisense chimeras described herein were characterized by HPLC, capillary gel electrophoresis, and digestion with snake venom phosphodiesterase (26–28).
Table 1.
Nomenclature and sequences of oligonucleotides
| RSV mRNA targeted | Oligonucleotide | Sequence |
|---|---|---|
| M2 | spA4-antiRSV3′-3′A/(8251–8270) | sp(5′A2′p)3A-Bu2-5′-ATAGTGTGTTCTTTTGATT-3′-3′A5′ |
| M2 | spA4-antiRSV3′-3′T/(8261–8279) | sp(5′A2′p)3A-Bu2-5′-GATTGAAATATAGTGTGT-3′-3′T5′ |
| M2 | spA4-antiRSV3′-3′T/(8281–8299) | sp(5′A2′p)3A-Bu2-5′-ATGGTTATTTGGGTTGTT-3′-3′T5′ |
| M2 and L | spA4-antiRSV3′-3′A/(8530–8547) | sp(5′A2′p)3A-Bu2-5′-CTATCGGTTAGATAAAC-3′-3′A5′ |
| L | spA4-antiRSV3′-3′C/(8561–8578) | sp(5′A2′p)3A-Bu2-5′-CTCTGAGAAAGAGATAA-3′-3′C5′ |
| L | spA4-antiRSV3′-3′G/(8599–8618) | sp(5′A2′p)3A-Bu2-5′-GATAAGGACCATTGAATAT-3′-3′G5′ |
| M2 | spA2-antiRSV3′-3′T/(8281–8299) | sp5′A2′p5′A-Bu2-5′-ATGGTTATTTGGGTTGTT-3′-3′T5′ |
| None | spA4-scramble3′-3′T/(8281–8299) | sp(5′A2′p)3A-Bu2-5′-TTGTTATGTTGGGATTTG-3′-3′T5′ |
| M2 | antiRSV(P-S)/(8281–8299) | (PS)5′-ATGGTTATTTGGGTTGTTT-3′ |
| M2 | antiRSV3′-3′T/(8281–8299) | 5′-ATGGTTATTTGGGTTGTT-3′-3′T5′ |
The numbers in parentheses indicate nucleotide numbers in the RSV strain A2 genome (GenBank accession no. M74568M74568 and refs. 30 and 31). The nucleotide sequences, except for that of spA4-scramble3′-3′T/(8281–8299), are complementary (or antisense) to the RSV mRNAs. Bu, butanediol linkers; P-S indicates all of the internucleotide linkages were phosphorothioates.
Computer Analysis of RNA Secondary Structure.
The RNA structure analyses were done with the mfold program, which determines the secondary structures of minimum free energy for RNA molecules based on published values of stacking and loop destabilizing energies (32). mfold is a public domain program run on a package by Genetics Computer Group (Madison, WI) using the National Institutes of Health Division of Computer Research and Technology’s Convex Mainframe. The energies used by M. Zuker’s program first were described by Salser (33) and now are defined by Turner and colleagues (34).
Cell Culture and RSV Propagation.
A human tracheal epithelial cell line, 9HTE (35), and an African green monkey kidney cell line, CV-1 (American Type Culture Collection), which is highly permissive to RSV infection, were cultured in minimal essential medium (MEM) supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, MEM amino acids solution, MEM nonessential amino acids solution, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (GIBCO/BRL).
RSV strain A2 (American Type Culture Collection) was propagated in CV-1 cells. CV-1 monolayers were infected at a multiplicity of infection (moi) of 0.2 plaque-forming units per cell and cultured 50 h in OptiMEM (GIBCO/BRL) in a 5% CO2/95% air atmosphere at 37°C. Cells then were washed twice in MEM and subsequently covered in OptiMEM, 100 units/ml penicillin, 100 μg/ml streptomycin, 50 mM Hepes (pH 7.5), 100 mM MgSO4, and 1.5% glycerol. After 5 h at 37°C, cells were scraped and sonicated as previously described (25). Aliquots of cell sonicates were flash-frozen in ethanol/dry ice within 20 min of scraping. Several aliquots then were thawed and titered by plaque assay on CV-1 cells in 96-well microtiter plates as previously described (36). Briefly, virus sonicates were serially diluted in chilled phosphate-buffered saline and transferred to 70% confluent monolayers of CV-1. Monolayers were exposed to virus for 2 h, then washed once in MEM and overlaid with prewarmed Eagle’s minimal essential medium (EMEM, BioWhittaker) containing 2% fetal bovine serum, 200 units/ml penicillin, 200 μg/ml streptomycin, 0.5 μg/ml amphotericin B, and 0.5% agarose. After 5 days with 5% CO2 at 37°C, cells were fixed in 10% formalin for 2 h, the agarose plugs were removed, and 0.2% crystal violet in 10% formalin was added for 1 min. Stained monolayers then were washed in water to remove excess dye, and the number of syncytia (plaques) was quantified under a low-power microscope. The range of virus yield from this procedure was 106 to 107 plaque-forming units per ml.
Oligonucleotide Treatment of 9HTE Cells After RSV Infection.
Infection of 9HTE cells was performed as previously described (37). Briefly, subconfluent monolayers were exposed to CV-1-propagated RSV (moi = 2) diluted in MEM and 2% fetal bovine serum at 37°C in 5% CO2. After 2 h, the virus was removed, and cells were washed once with MEM and 2% fetal bovine serum followed by the addition of culture medium. Oligonucleotides immediately were added 2 h postinfection, and they were added again at 12 h postinfection. Progeny virus was harvested at 24 h postinfection as previously described (36). Cells were washed twice and scraped in the culture medium, and the suspension was sonicated for 10 sec on ice. The titer of infectious virus then was determined by plaque assays on the CV-1 indicator cells. Viral yield reduction data is presented as the average of the percent reduction of infectious virus particles ± standard error calculated using the computer program StatView 4.1 (Abacus Concepts, Berkeley, CA).
Reverse Transcriptase (RT)-Coupled PCR.
9HTE cells, 2 × 105, were infected and then washed as described above (moi = 2) and treated at 2 h postinfection with 3.3 μM 2–5A-antisense. RNA was collected at 8 h postinfection, before release of infectious virions, by RNAzol treatment as described by the manufacturer (Tel-Test, Friendswood, TX). Isolated RNA (≈1 μg) was incubated with 100 pmols oligo(dT) or specific primers for antigenomic RNA. RNA and primers were heated to 70°C for 10 min, then cooled rapidly on ice for 10 min. A final RT-reaction volume of 30 μl contained 300 μM each dNTP, 200 units of SuperScript RT (GIBCO/BRL), 50 mM Tris·HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM DTT. Reverse transcription was allowed to proceed for 1 h at 37°C.
PCR were performed using 50-μl Hot-Start tubes (Molecular Bio-Products, San Diego) with 25 μl of lower buffer containing 40 mM Tris·HCl (pH 8.4), 100 mM KCl, 4 mM MgCl2, 1 mM each dNTP, and 200 pmol of the appropriate (+) primers and 50 pmol of the appropriate (−) primers (See Table 2). Twenty-five microliters of Hot-Start upper buffer contained 5 units of Taq DNA polymerase (GIBCO/BRL) and 1/10th of the cDNA from the RT reaction. Thirty cycles of PCR were performed for 20 sec at 92°C, 20 sec at the annealing temperatures indicated in Table 2, and 20 sec at 72°C. Aliquots of the RT-PCR mixtures were analyzed on 1% agarose/Tris borate/EDTA gels using ethidium bromide fluorescence. PCR product sizes were confirmed by comparison of their mobilities on gels with those of DNA markers. In addition, cDNA from the RT reactions was serially diluted 1:3 into Hot-Start tubes (same constituents as above), and the PCRs were stopped after 25 cycles to determine a limiting dilution of input cDNA for comparing mRNA levels. Densitometry was performed with a Sierra Scientific high-resolution charge-coupled device camera (Sunnyvale, CA) and the program NIH Image 1.6. Equivalent-sized boxes were used to quantitate the numbers of pixels in the fields after background subtraction.
Table 2.
Oligodeoxyribonucleotide primers used for PCR and RT reactions
| Target | Sequence | Annealing temp., °C | Product size, bp |
|---|---|---|---|
| G3PDH(+) | 5′-AAATCCCATCACCATCTTC-3′ | 56 | 762 |
| G3PDH(−) | 5′-CACCACCCTGTTGCTGTAG-3′ | ||
| RSV (M2+) | 5′-AAACAATCAGCATGTGTTG-3′ | 46 | 587 |
| RSV (M2−) | 5′-AATGTAACGATGTGGTGAG-3′ | ||
| RSV (N+) | 5′-ACAACACATCGTCAAG-3′ | 46 | 577 |
| RSV (N−) | 5′-ATTCATAAACCTCAAC-3′ | ||
| RSV (P+) | 5′-AAACAACAGGGCTACTA-3′ | 46 | 575 |
| RSV (P−) | 5′-TTTCACTTTCCTCATTC-3′ | ||
| RSV (L−) | 5′-GAGCTTTATTAGCAGCATC-3′ | RT primer | |
The target genes are indicated on the left, followed by the target sequence and the optimal annealing temperature as determined by the program oligo (version 4.1, National Biosciences, Plymouth, MN). G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
RESULTS
Selection of Oligonucleotide Binding Sites and Design of 2–5A-Antisense.
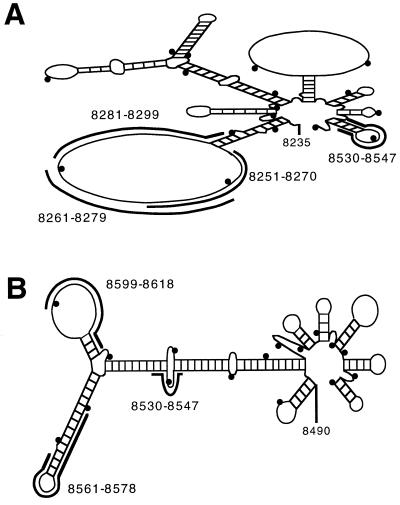
To select antisense-binding sites in RSV RNAs, a computer-assisted analysis of the secondary structure of the RSV RNA sequence was performed. The diagram of a portion (nucleotides 8235–8560) of the predicted structure of the RSV M2 mRNA shows a large loop and potential binding sites for 2–5A-antisense species complementary to nucleotides 8251–8270, 8261–8279, and 8281–8299 (Fig. 1A). The M2 mRNA was targeted, because it contains this large predicted loop and encodes a transcription elongation factor (38). In addition, a 2–5A-antisense species was targeted to a smaller predicted loop in the M2 mRNA 3′ untranslated region (nucleotides 8530–8547); due to a gene overlap, this oligonucleotide also targets a predicted bulge in the RSV L mRNA (Fig. 1 A and B). The L mRNA was separately targeted, because it encodes the viral polymerase essential for viral replication (Fig. 1B). Two 2–5A-antisense species were made against the 5′-region of the RSV L mRNA, one to a predicted 17-nucleotide loop (nucleotides 8599–8618), and the other to a predicted hairpin structure (nucleotides 8561–8578).
Figure 1.
Computer prediction of RNA secondary structure of RSV mRNAs. (A) Predicted secondary structure of a 3′region of the RSV M2 mRNA (nucleotides 8235–8560), (B) Structure of a 5′-region of the RSV L mRNA (nucleotides 8490–8800). 2–5A-antisense binding sites are indicated by thick, black bands. The entire M2 mRNA contains nucleotides 7599–8561, and the entire L mRNA contains nucleotides 8490–15070. Small black dots are shown in the diagram after every 20 nucleotides beginning with nucleotide 8240 (A) and beginning with nucleotide 8500 (B). ΔG values were −33.6 and −39.3 kcal per mol for the structures shown in (A) and (B), respectively.
The 2–5A-antisense species were modified at both termini to reduce their rates of enzymatic decay. Previously, we showed that replacing the 5′-phosphate group with a 5′-thiophosphate group on 2–5A protected against 5′-phosphatase activity without affecting activation of RNase L (27). Furthermore, 2–5A-chimeras with a 5′-thiophosphate were effective in targeting the PKR mRNA for degradation (24). In addition, the terminal internucleotide linkage in the DNA moieties of the chimeras was inverted from a 3′,5′ linkage to a 3′,3′ linkage. These modifications substantially improved the stabilities of the oligonucleotides toward snake venom phosphodiesterase and human serum exonuclease activities (G.L., W.X., and P.F.T., unpublished observations).
Treatment of Human Tracheal Epithelial Cells with 2–5A-Antisense Oligonucleotides Suppressed RSV Yields.
The effects of 2–5A-antisense compounds on the replication of the A2 strain of RSV were determined in the established human trachael epithelial cell line, 9HTE. These cells were observed to contain basal levels of RNase L by a specific, radiolabeled 2–5A crosslinking assay (ref. 39 and data not shown). Because we observed similar antiviral effects when the oligonucleotides were added beginning either 4 h before infection or 2 h after infection, a treatment protocol was developed in which cells were treated only after infection (data not shown). In the most effective treatment regimen, 9HTE cells were infected with RSV at an moi of 2 followed by treatment with 2–5A-antisense at 2 h and 12 h postinfection. One treatment (at 2 h postinfection) was relatively ineffective, whereas three oligonucleotide treatments (at 2 h, 8 h, and 16 h postinfection) did not significantly enhance antiviral activity compared with just two treatments at 2 h and 12 h postinfection (data not shown). Virus was harvested at 24 h postinfection, and titers were measured by plaque assays on CV-1 indicator cells.
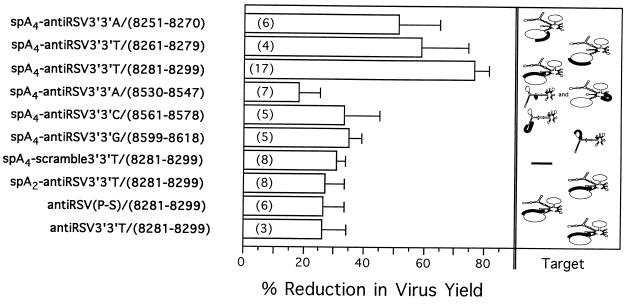
The 2–5A-antisense species that had the greatest antiviral activities (averages of 51% to 77% reduction in viral yields with treatments of 3.3 μM oligonucleotide) were those that targeted the predicted 50-nucleotide loop in the M2 mRNA (the upper three compounds shown in Fig. 2). Furthermore, it was apparent that the anti-M2 chimera, spA4-antiRSV3′-3′T/(8281–8299), produced a greater level of antiviral activity than the other two oligonucleotides targeted to the same loop, spA4-antiRSV3′-3′A/(8251–8270) and spA4-antiRSV3′-3′T/(8261–8279) (Fig. 2). Substantially lower antiviral effects were observed with the 2–5A-antisense chimeras directed against other regions in RSV M2 and L mRNAs. The oligonucleotide directed to the M2/L overlap region, spA4-antiRSV3′-3′A/(8530–8547), showed only 18 ± 7% inhibition, whereas the 2–5A-antisense directed to the L mRNA hairpin and small loop, spA4-antiRSV3′-3′C/(8561–8578) and spA4-antiRSV3′-3′G/(8599–8618), showed only 33 ± 11% and 35 ± 4% inhibition, respectively (Fig. 2).
Figure 2.
The antiviral potency of the oligonucleotides is dependent on both the presence of a functional 2–5A group and the sequence of the antisense cassette. Oligonucleotides were added twice to RSV-infected cells at 3.3 μM per dose as described. The horizontal bars show the average reduction in infectious RSV yields as determined by viral plaque assays. The standard errors are indicated by the vertical lines connected to the bars. The numbers shown in parenthesis within the bars are the numbers of experiments used to obtain the data. The percent reduction in virus yield is defined as [1−(the number of viral plaques obtained from the oligonucleotide-treated cells/the number of viral plaques obtained from the untreated cells) × 100]. (Left) The oligonucleotides used. (Right) The predicted structures of the RNA targets and the oligonucleotide binding sites (bold lines) correspond to those shown in Fig. 1.
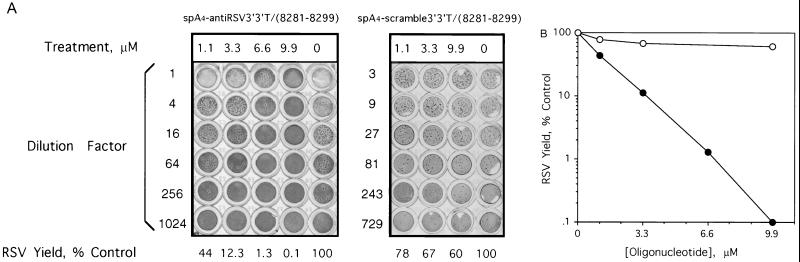
Undiluted progeny virus from the RSV-infected, untreated 9HTE cells destroyed the CV-1 cell monolayer, whereas spA4-antiRSV3′-3′T/(8281–8299) treatments of the RSV-infected 9HTE cells reduced viral yields to the extent that most of the indicator cells remained viable (Fig. 3A, top row). The syncytia formed discrete plaques that were counted under a microscope to measure the antiviral effects (Fig. 3A). The dose–response curve for this experiment and two others showed ≥97% reduction in viral yields, whereas the average reduction from a total of six experiments performed using 9.9 μM treatments of spA4-antiRSV3′-3′T/(8281–8299) was 88 ± 6%. A 2–5A-antisense chimera with the same base composition as spA4-antiRSV3′-3′T/(8281–8299), but with a randomized (scrambled) DNA sequence, spA4-scramble3′-3′T/(8281–8299), produced 33% and 40% inhibition of virus yield with 3.3 μM and 9.9 μM oligonucleotide treatments (Fig. 3). Because spA4-scramble3′-3′T/(8281–8299) contains an intact, functional 2–5A domain, this level of inhibition may have been due to nonspecific RNase L activity.
Figure 3.
Dose-dependent suppression of infectious RSV by the 2–5A-antisense species, spA4-antiRSV3′-3′T/(8281–8299) in comparison to the effect of spA4-scramble3′-3′T/(8281–8299). (A) Viral plaque assays of CV-1 indicator cells was performed using RSV harvested from infected 9HTE cells that were untreated or treated with different amounts of spA4-antiRSV3′-3′T/(8281–8299) or spA4-scramble3′-3′T/(8281–8299) as described. The doses of oligonucleotides and the dilutions of the crude preparations of RSV that were added to the indicator cells are shown. (B) Graph of the data from A. •, spA4-antiRSV3′3′T(8281-8299); ○, spA4-scramble3′3′T(8281-8299).
Authentic 2–5A Was Required to Enhance the Antiviral Activity of Antisense.
To establish the contribution of the 2–5A moiety of the chimeras to the anti-RSV effects, spA2-antiRSV3′-3′T/(8281–8299) was synthesized, which contained only two instead of the usual four 2′,5′-linked adenylyl residues (Fig. 2). Chains of three or more 2′,5′-linked adenylyl residues are required for RNase L activation, therefore dimeric forms of 2–5A are inactive (reviewed in ref. 40). Also, phosphorothioate antiRSV(P-S)/(8281–8299) and phosphodiester antiRSV3′-3′T/(8281–8299) that lack 2–5A moieties were produced. Treatments of infected cells with 3.3 μM doses of spA2-antiRSV3′-3′T/(8281–8299), antiRSV(P-S)/(8281–8299), and antiRSV3′-3′T/(8281–8299) gave only 31 ± 3%, 26±8%, and 26 ± 8% reduction in viral yields (Fig. 2). These results demonstrated that the antiviral potency of spA4-antiRSV3′-3′T/(8281–8299) was dependent on the presence of functional 2–5A and on the antisense sequence.
2–5A-Antisense Caused a Selective Decrease in the Levels of the Targeted mRNA in Infected Cells.
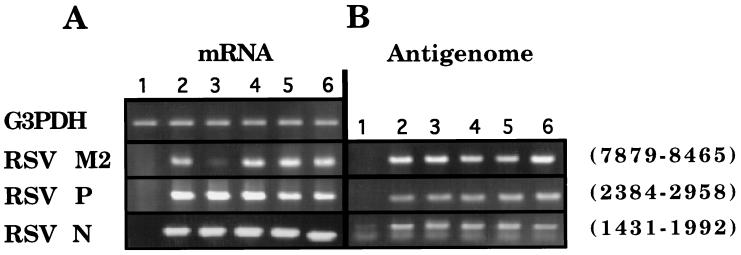
It was apparent from these data that the recruitment and activation of RNase L was principally responsible for the potency of spA4-antiRSV3′3′T/(8281–8299) against RSV. To further explore the role of RNase L in the antiviral mechanism, cellular amounts of the M2 RNA target were compared with other viral and cellular RNAs. RT-PCR was performed on RNA isolated from RSV-infected (moi = 2) and uninfected 9HTE cells with and without treatment (3.3 μM at 2 h postinfection) with spA4-antiRSV3′-3′T/(8281–8299) or several other antisense compounds (Fig. 4). The RSV M2 mRNA (harvested at 8 h postinfection) was converted to cDNA using oligo(dT) primers, and then was amplified by PCR (from nucleotides 7879–8465). M2 mRNA from the RSV-infected cells produced a clearly visible DNA product (Fig. 4A, lane 2). In contrast, there was a substantially reduced amount of M2 mRNA detected from the RSV-infected cells treated with spA4-antiRSV3′-3′T/(8281–8299) (Fig. 4A, lane 3). The other oligonucleotides, spA2-antiRSV3′-3′T/(8281–8299), spA4-scramble3′-3′T/(8281–8299), and antiRSV(P-S)/(8281–8299), had no effect on levels of the M2 mRNA (Fig. 4A. lanes 4–6). None of the oligonucleotides altered levels of the RSV N or P mRNAs or cellular glyceraldehyde-3-phosphate dehydrogenase mRNA (Fig. 4A). Therefore, spA4-antiRSV3′-3′T/(8281–8299) was remarkably selective for the RSV M2 mRNA. Furthermore, the specific degradation of M2 mRNA by spA4-antiRSV3′-3′T/(8281–8299) was dependent on the presence of the tetrameric 2–5A moiety, thereby confirming the involvement of RNase L.
Figure 4.
RT-PCR analysis reveals a selective decrease in the levels of the targeted viral M2 mRNA. RT-PCR analysis of mRNAs (A) and the antigenomic (+) strand of RSV RNA (B) is shown. RNA was analyzed from either uninfected cells (lane 1) or RSV-infected 9HTE cells (lanes 2–6). Single doses of oligonucleotides (3.3 μM) added at 2 h postinfection were as follows: lane 3, spA4-antiRSV3′-3′T/(8281–8299); lane 4, spA2-antiRSV3′-3′T/(8281–8299); lane 5, spA4-scramble3′-3′T/(8281–8299); and lane 6, antiRSV(P-S)/(8281–8299). RNA samples were collected at 8 h postinfection for RT reactions, which were primed with either oligo(dT) (for analysis of mRNAs) or with an oligonucleotide complementary to a sequence in the RSV L gene for the (+) antigenomic strand (see Table 2). The RSV bases that were amplified are indicated to the right. An image of the ethidium bromide-stained gel is shown. G3PDH, cellular glyceraldehyde-3-phosphate dehydrogenase mRNA.
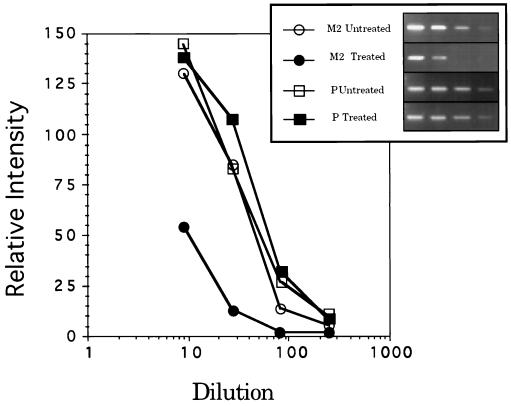
To determine whether the antigenomic (+) RSV RNA strand was affected, cDNA synthesis was primed with an oligodeoxynucleotide in the L gene (residues 9594–9611), thereby converting the (+) replicative intermediate RNA or polycistronic mRNA to cDNA. This cDNA was used as a template for PCR reactions that were identical to those primed with oligo(dT) (Fig. 4B). The results show that the (+) antigenomic RNA strand was not affected at 8 h postinfection by treatment with these compounds, perhaps because the RSV N protein binds tightly to the (+) strand, making it inaccessible to antisense and RNase L (3). To obtain further evidence for the specific decrease in the M2 mRNA, the cDNAs from RT reactions were serially diluted for PCR. In this way, the quantity of the PCR products would be proportional to the input cDNA concentrations over a range of dilutions. The levels of M2 mRNA in spA4-antiRSV3′-3′T/(8281–8299)-treated, RSV-infected 9HTE were determined to be reduced by about 80% compared with levels found in control infected cells (Fig. 5). The levels of RSV P mRNA were unchanged after treatment of the infected cells with spA4-antiRSV3′-3′T/(8281–8299), thus demonstrating the specificity of 2–5A-antisense for its target RNA (Fig. 5).
Figure 5.
PCR after serial dilutions of cDNA shows a relative decrease in amounts of the targeted RSV M2 mRNA. PCR products from the RSV M2 (○, •) and P (□, ▪) mRNAs were separated on agarose gels and stained with ethidium bromide. Quantitation of bands plotted in the semilogarithmic plot shown was by densitometry. RNA was from either untreated infected cells (○, □) or infected cells treated at 2 h postinfection with 3.3 μM spA4-antiRSV3′-3′T/(8281–8299) (•, ▪). RNA was harvested at 8 h postinfection. (Inset) An image of the PCR products from untreated and spA4-antiRSV3′-3′T/(8281–8299)-treated cultures in ethidium bromide-stained gels.
DISCUSSION
Here we have shown that 2–5A-antisense chimeric oligonucleotides can function as effective anti-RSV agents when added to previously infected cells in culture. These results were dependent upon improvements in both the protection of the oligonucleotides against enzymatic decay and in the computer-assisted selection of RNA target sites. The 5′-thiophosphorylation of 2–5A blocks phosphatase digestion without impairing RNase L activation, whereas inversion of the last 3′,5′ phosphodiester bond in the DNA portion to a 3′,3′ linkage stabilizes against 3′,5′-exonuclease activity in serum (ref. 27 and G.L., W.X. and P.F.T., unpublished observations). The most potent antiviral activities were attained by targeting the 50-nucleotide predicted loop in the M2 mRNA with spA4-antiRSV3′-3′A/(8251–8270), spA4-antiRSV3′-3′T/(8261–8279), and spA4-antiRSV3′-3′T/(8281–8299); the last of these was most potent. The relative inefficiency of 2–5A-antisense species directed against the RSV L mRNA may have been due to an absence of large loop structures (Figs. 1 and 2). The RSV M2 and L mRNAs have a region of overlap that is complementary to the antisense arm of spA4-antiRSV3′-3′A/(8530–8547), the least active compound (Figs. 1 and 2). The inactivity of this compound may have been due to both a lack of an accessible target sequence and to its being sequestered by aborted RSV L transcripts terminating at the M2 gene-end signal (40). The M2 mRNA contains an open reading frame (ORF) for a 22-kDa internal virion protein (ORF1, nucleotides 7606–8188) implicated in transcriptional elongation and a downstream 90-codon ORF2 (nucleotides 8159–8431), the protein product of which has not been detected in pneumovirus-infected cells (3, 38). Recent evidence suggests that M2 ORF2 has an inhibitory effect on transcription (38). It is likely that the antisense chimeras used here will result in the down-regulation of both M2(ORF1) and M2(ORF2) through the endonucleolytic cleavage of this mRNA by RNase L followed by general exoribonuclease action. An approximately 80% decrease in M2 RNA levels was observed after application of a single 3.3 μM dose of spA4-antiRSV3′-3′T/(8281–8299) to RSV-infected cells for 6 h (Figs. 4 and 5). The treatment was remarkably selective in that RSV N and P mRNAs and cellular glyceraldehyde-3-phosphate dehydrogenase mRNA levels were not reduced. It would appear from these results that the M2 protein(s) is an important factor for RSV replication. The M2 gene is unique to the pneumovirus subfamily of the Paramyxoviridae (3).
Our findings support previous reports that covalent linkage of 2–5A to an antisense oligonucleotide enhances rates of decay for the target RNAs in cells through the action of RNase L (12, 23, 24). In particular, the data show that spA4-antiRSV3′-3′T/(8281–8299) attracts RNase L to complementary sequences in RSV M2 mRNA, causing its degradation. The evidence includes the inactivity of antisense compounds in which the 2–5A portion was either absent or rendered nonfunctional and by the linking of 2–5A to an irrelevant scrambled nucleotide sequence. For instance, a chimeric oligonucleotide containing a disabled 2–5A moiety, spA2-antiRSV3′-3′T/(8281–8299), did not cause M2 RNA levels to decline and was a poor anti-RSV agent. These same results indicate that RNase H, which does not rely on 2–5A for its activity against the RNA strands in RNA·DNA hybrids, was unlikely to have been involved in the specific decline in M2 RNA levels after 3.3 μM treatment with spA4-antiRSV3′-3′T/(8281–8299). A phosphorothioate-linked antisense of the identical sequence also failed to induce M2 RNA decay and was weakly active against RSV (Figs. 2 and 4). Similarly, antiRSV3′-3′T(8281–8299), lacking a 2–5A/linker, showed only weak activity against RSV (Fig. 2). The 2–5A-chimeras—e.g. as spA4-antiRSV3′-3′T/(8281–8299)—did not cause the general decay of RNA, probably because they are relatively inefficient activators of RNase L (23). Nevertheless, when target RNAs were brought into close proximity to RNase L, the enzymatic activity was sufficient to cause the rapid cleavage of the RNA target. The antigenomic or replicative intermediate (+) strand was not degraded in 2–5A-antisense-treated infected cells, probably because the positive-sense strand is tightly encapsidated (Fig. 5).
These results show that 2–5A-antisense can be a highly effective agent for suppressing ongoing RSV infections of cells in culture. The therapeutic potential of 2–5A-antisense is dependent on the development of an effective method for delivery of the drug to the infected cells. In this regard, controlling RSV infections provides an attractive goal for the future development of this technology, because antisense complexes could be administered by any of several different routes, including inhalation of an aerosol (the same method currently used to deliver ribavirin), nasal drops, and intravenous or intraperitoneal injections.
Acknowledgments
We thank James R. Panuska (Case Western Reserve University, Cleveland) for the generous gifts of RSV and 9HTE cells and for advice, and A. Banerjee and B. R. G. Williams (Cleveland Clinic) and Robert Sidwell (Utah State University) for comments made during preparation of the manuscript. This investigation was supported by U.S. Public Health Service Grant 1 PO1 CA 62220 awarded by the Department of Health and Human Services, National Cancer Institute, and by funds from Atlantic Pharmaceuticals, Inc. (to R.H.S.) and by a Cooperative Research and Development Agreement between the National Institutes of Health and Atlantic Pharmaceuticals, Inc.
ABBREVIATIONS
- RSV
respiratory syncytial virus
- MEM
minimal essential medium
- moi
multiplicity of infection
- RT
reverse transcriptase
- ORF
open reading frame
References
- 1.Anonymous Morbid Mortal Wkly Rep. 1995;44:900–902. [PubMed] [Google Scholar]
- 2.Falsey A R, Cunningham C K, Barker W H, Kouides R W, Yuen J B, Menegus M, Weiner L B, Bonville C A, Betts R F. J Infect Dis. 1995;172:389–394. doi: 10.1093/infdis/172.2.389. [DOI] [PubMed] [Google Scholar]
- 3.Collins P L, McIntosh K, Chanock R M. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, et al., editors. Philadelphia: Lippincott; 1996. pp. 1313–1351. [Google Scholar]
- 4.Zamecnik P C, Stephenson M L. Proc Natl Acad Sci USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Z F, Wickstrom E, Jiang M, Corisdeo S, Yang J, Dietzschold B, Koprowski H. Antisense Nucleic Acid Drug Dev. 1996;6:87–93. doi: 10.1089/oli.1.1996.6.87. [DOI] [PubMed] [Google Scholar]
- 6.Leiter J M E, Agrawal S, Palese P, Zamecnik P C. Proc Natl Acad Sci USA. 1990;87:3430–3434. doi: 10.1073/pnas.87.9.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raviprakash K, Liu K, Matteucci M, Wagner R, Riffenbach R, Carl M. J Virol. 1995;69:69–74. doi: 10.1128/jvi.69.1.69-74.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu G Y, Wu C H. J Biol Chem. 1992;267:12436–12439. [PubMed] [Google Scholar]
- 9.Smith C C, Aurelian L, Reddy M P, Miller P S, Ts’o P O P. Proc Natl Acad Sci USA. 1986;83:2787–2791. doi: 10.1073/pnas.83.9.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamecnik P C, Goodchild J, Taguchi Y, Sarin P S. Proc Natl Acad Sci USA. 1986;83:4143–4146. doi: 10.1073/pnas.83.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisziewicz J, Sun D, Metelev V, Zamecnik P, Gallo R C, Agrawal S. Proc Natl Acad Sci USA. 1993;90:3860–3864. doi: 10.1073/pnas.90.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torrence P F, Maitra R K, Lesiak K, Khamnei S, Zhou A, Silverman R H. Proc Natl Acad Sci USA. 1993;90:1300–1304. doi: 10.1073/pnas.90.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman R H. In: Ribonucleases: Structure and Function. D’Alessio G, Riordan J F, editors. New York: Academic; 1997. in press. [Google Scholar]
- 14.Kerr I M, Brown R E. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams B R G, Golgher R R, Brown R E, Gilbert C S, Kerr I M. Nature (London) 1979;282:582–586. doi: 10.1038/282582a0. [DOI] [PubMed] [Google Scholar]
- 17.Chebath J, Benech P, Revel M, Vigneron M. Nature (London) 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 18.Rysiecki G, Gewert D R, Williams B R G. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 19.Hassel B A, Zhou A, Sotomayor C, Maran A, Silverman R H. EMBO J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteban M, Benavente J, Paez E. Virology. 1984;134:40–51. doi: 10.1016/0042-6822(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 21.Goswami B B, Sharma O K. J Biol Chem. 1984;259:1371–1374. [PubMed] [Google Scholar]
- 22.Nilsen T W, Maroney P A, Baglioni C. Mol Cell Biol. 1983;3:64–69. doi: 10.1128/mcb.3.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maitra R K, Li G, Xiao W, Dong B, Torrence P F, Silverman R H. J Biol Chem. 1995;270:15071–15075. doi: 10.1074/jbc.270.25.15071. [DOI] [PubMed] [Google Scholar]
- 24.Maran A, Maitra R K, Kumar A, Dong B, Xiao W, Li G, Williams B R G, Torrence P F, Silverman R H. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 25.Panuska J R, Merolla R, Rebert N A, Hoffmann S P, Tsivitse P, Cirino N M, Silverman R H, Rankin J A. J Clin Invest. 1995;96:2445–2453. doi: 10.1172/JCI118302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesiak K, Khamnei S, Torrence P F. Bioconjugate Chem. 1993;4:467–472. doi: 10.1021/bc00024a008. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, Li G, Lesiak K, Dong B, Silverman R H, Torrence P F. Bioor Med Chem Lett. 1994;4:2609–2614. [Google Scholar]
- 28.Xiao W, Player M R, Li G, Zhang W, Lesiak K, Torrence P F. Antisense Nucleic Acid Drug Dev. 1996;6:247–258. doi: 10.1089/oli.1.1996.6.247. [DOI] [PubMed] [Google Scholar]
- 29.Zon G, Stec W J. In: Oligonucleotides and Analogues: A Practical Approach. Eckstein F, editor. London: Oxford Univ. Press; 1991. pp. 87–108. [Google Scholar]
- 30.Collins P L, Olmsted R A, Spriggs M K, Johnson P R, Buckler-White A J. Proc Natl Acad Sci USA. 1987;84:5134–5138. doi: 10.1073/pnas.84.15.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mink M A, Stec D S, Collins P L. Virology. 1991;185:615–624. doi: 10.1016/0042-6822(91)90532-g. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 33.Salser W. Cold Spring Harbor Symp Quant Biol. 1977;42:985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- 34.Freier S M, Kienzek R, Jaegar J A, Sugimoto N, Caruthers M H, Neilson T, Turner D H. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruenert D C, Basbaum C B, Welsh M J, Li M, Finkbeiner W E, Nadel J A. Proc Natl Acad Sci USA. 1988;85:5951–5955. doi: 10.1073/pnas.85.16.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirino N M, Panuska J R, Villani A, Taraf H, Rebert N A, Merolla R, Tsivitse P, Gilbert I A. J Gen Virol. 1993;74:1527–1537. doi: 10.1099/0022-1317-74-8-1527. [DOI] [PubMed] [Google Scholar]
- 37.Merolla R, Rebert N A, Tsivitse P T, Hoffmann S P, Panuska J R. Am J Respir Crit Care Med. 1995;152:1358–1366. doi: 10.1164/ajrccm.152.4.7551395. [DOI] [PubMed] [Google Scholar]
- 38.Collins P L, Hill M G, Cristina J, Grosfeld H. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan-Sorden N L, Lesiak K, Bayard B, Torrence P F, Silverman R H. Anal Biochem. 1990;184:298–304. doi: 10.1016/0003-2697(90)90684-2. [DOI] [PubMed] [Google Scholar]
- 40.Torrence P F, Xiao W, Li G, Khamnei S. Curr Med Chem. 1994;1:176–191. [Google Scholar]