Abstract
We have examined mechanisms involved in gene transfer, protein expression, and antigen presentation after direct administration of retroviral vectors using a variety of antigen systems. We have identified transduced infiltrating cells at the injection site, and the majority of the infiltrating cells were of the monocyte/macrophage lineage. We found that the splenic dendritic cell fraction contained proviral DNA, expressed antigenic proteins, and was able to present antigens efficiently to the immune system. Furthermore, the dendritic cell fractions from retroviral vector-immunized mice were able to prime naive T cells in vitro, and adoptive transfer of in vitro-transduced dendritic cell fractions elicited antigen-specific cytotoxic T lymphocytes. These data suggest a role for dendritic cells in induction of immune responses elicited by retroviral vector-mediated gene transfer.
We have developed Moloney murine leukemia virus-based retroviral vectors encoding therapeutic genes and have been studying the induction of immune responses mediated by these retroviral vectors. We have demonstrated that successful induction of immune responses could be achieved in mice (1), nonhuman primates (2), and humans (3), by injection of ex vivo vector-transduced autologous fibroblasts. These studies led us to investigate whether immune responses could be elicited by direct administration of retroviral vector by intramuscular injection. We have recently demonstrated induction of immune responses after direct administration of retroviral vectors intramuscularly in mice, rhesus monkeys, and baboons (4).
Direct injection of retroviral vector offers several advantages over the ex vivo approach, primarily the elimination of the extensive efforts involved in generating autologous ex vivo fibroblast cell lines for each patient. With the ex vivo approach, the target antigen is exclusively expressed by transduced fibroblasts; however, direct administration of retroviral vectors can lead to the transduction of cells near the injection site as well as those cells residing in tissues to which vector can be transported. Consequently, it was not clear which cells were transduced by the retroviral vector, and which cells were responsible for presentation of retroviral vector-encoded antigens. In this paper, we have attempted to identify the vector-transduced cells and cells capable of presenting antigen to delineate mechanisms for induction of immune responses after direct injection of retroviral vectors.
We have used retroviral vectors encoding HIV env/rev, E. coli β-galactosidase (β-gal), chicken ovalbumin, and firefly luciferase to identify the subsets of cells involved in antigen presentation and induction of immune responses. These four different antigen systems were used to verify that the mechanisms of in vivo transduction and induction of immune responses are comparable among retroviral vectors derived from the same packaging cell line and backbone construct regardless of the specific antigen they encode. Furthermore, each of these retroviral vectors offers unique advantages, such as the availability of extremely sensitive detection systems or antibodies suitable for immunohistochemistry. Using these assays, we not only detected cells belonging to each of these subsets, but have also estimated their relative numbers. In this paper, we discuss the implications of these data in conjunction with the potential mechanisms of antigen presentation following direct administration of retroviral vector.
MATERIALS AND METHODS
Retroviral Vectors.
The nonreplicating, amphotropic murine retroviral vectors encoding HIV IIIB env/rev (N2-IIIBenv), bacterial β-gal (N2-β-gal), firefly luciferase (N2-luci), and chicken ovalbumin (N2-ova) were used. Generation of retroviral vector backbone and production of high titer vectors [>1 × 107 colony-forming units (cfu)/ml] were described previously (4, 5). Purified, formulated high titer retroviral vector preparations were used in all experiments.
Immunizations, in Vitro Cytotoxic T Lymphocyte (CTL) Induction, and Cytotoxicity Assays.
Six- to eight-week-old female BALB/c (H-2d) or C57BL/6 (H-2b) mice from Harlan–Sprague–Dawley were used in all experiments. On days 1, 4, and 7, mice were injected intramuscularly in both gastrocnemius muscles with 100 μl each of retroviral vector. For adoptive transfer experiments, BC-env fibroblasts stably transduced with N2-IIIBenv, or ex vivo-transduced dendritic cell fractions, were injected once intraperitoneally to naive recipient mice. One week later, spleens were harvested, and splenocyte fractions were mixed with irradiated stimulators at a 50:1 ratio and assayed 7 days later as described (4).
Immunohistochemistry.
Antibodies to the following leukocyte markers were used: CD4 (6), CD8 (7), B220 (8), macrophages (9), activated macrophages (10), and dendritic cells (11–13). Muscles from injection sites were excised and rapidly frozen. Cryostat sections at 5–6 μm were cut, air dried, and fixed in acetone for 5 min. The sections were incubated with previously titrated optimal concentrations of antibodies diluted in TBS (pH 7.4) for 60 min, washed in TBS, and incubated with biotinylated anti-rat or rabbit IgG (Jackson ImmunoResearch) for 30 min. The sections were then incubated with streptavidin-conjugated alkaline phosphatase (Jackson ImmunoResearch) for 30 min, and the bound alkaline phosphatase was detected with the substrate Fast-violet. The sections were lightly counterstained with Meyer’s hematoxylin and were photographed (×200) as described (14).
Splenocyte Fractionation by Percoll Density Gradients.
Splenocytes from naive or immunized mice were fractionated into B cells, T cells, macrophages, and dendritic cells by the method described previously (15) and/or sorting by a fluorescence-activated cell sorter (FACS). Resulting B cells, T cells, and macrophage fractions were shown to be homogenous as determined by their representative marker profiles of B220, Thy-1.2, and F4/80, respectively. The cell fractions that adhere to plastic immediately after the cell isolation, but not after overnight culturing, are mostly dendritic cells, as determined by staining with several dendritic cell antibodies, NLDC-145, 33D1, and N418, although these cells can include B220+ B cells (up to 20%) and F4/80+ monocyte/macrophages (up to 30%). No Thy-1.2-positive cells were detected from dendritic cell fractions by FACS analysis.
PCRs, 5-Bromo-4-chloro-3-indosyl β-galactopyranoside (X-Gal) Staining, and B3.Z Assays.
Cells from lymph nodes and spleen fractions were processed according to the method previously described (16, 17). Briefly, 500 ng of DNA in a 10-μl aliquot was run in each PCR using the following primers: NEO-47 (5′-CAA GAT GGA TTG CAC GCA GG) and NEO-48 (5′-CCC GCT CAG AAG AAC TCG TC). The reaction products were Southern blotted and analyzed with a digoxygenin-labeled probe for the presence of neomycin-resistance gene sequence. Copy numbers were estimated by comparison to the positive controls, with numbers between 1 and 1 × 105 copies per reaction.
For X-Gal staining, cultures were fixed with 2% formaldehyde for 5 min at room temperature, followed by washing with PBS. The cells were overlayed with a solution of 1 mg/ml X-Gal/5 mM potassium ferrocyanide/5mM potassium ferricyanide/2 mM MgCl2. The plates were examined microscopically for the presence of LacZ-expressing (blue) cells after an overnight incubation at 37°C.
B3.Z cells are T cell hybridomas that recognize chicken ovalbumin in the context of H-2Kb molecules (18, 19). They contain a plasmid containing the bacterial LacZ gene fused to the minimal promoter of the human interleukin 2 gene and, therefore, are LacZ-inducible upon antigenic stimulation. Antigen-presenting cells are either cells transfected with ovalbumin genes or fractionated splenocytes from ovalbumin retroviral vector-immunized mice. Individual cultures containing 5 × 106 B3.Z cells and 1 × 107 antigen-presenting cells were incubated overnight in 12-well cultures followed by X-Gal staining to assess the level of B3.Z activation. The numbers of antigen-presenting cells in each fraction were estimated from the standard curve generated from cultures containing known numbers of EL4 cells transfected with ovalbumin genes, E.G7-OVA cells.
Ex Vivo Transduction of the Dendritic Cell Fraction and Adoptive Transfer.
Naive mice were killed, and the dendritic cell fraction was prepared as described above and put into media containing granulocyte/macrophage colony-stimulating factor (500 units/ml) and interleukin 4 (1000 units/ml). DA/KT-1, the producer cell line for N2-IIIBenv, was seeded in transwells (Corning Costar) with a pore size of 0.45 μm, which does not allow any leakage of producer cells across the membrane. The dendritic cell fraction was added to low adherence tissue culture wells (Corning Costar) and transwells containing DA/KT-1 were placed above the dendritic cell fraction. After 3 days of cocultivation, the recovered cells were washed three times with PBS before injection into naive recipient mice. Spleens from recipient mice were harvested 7 days later and assayed for cytotoxicity as described above.
RESULTS
Infiltrating Leukocytes at the Injection Sites Are Transduced with Retroviral Vector.
We were interested in defining the types of cells that are transduced after direct administration of retroviral vectors in muscle and examined the injection sites by immunohistochemistry. Mice were immunized in the gastrocnemius muscles with a retroviral vector encoding chicken ovalbumin (N2-ova, a total of 2.4 × 107 cfu per mouse). We selected ovalbumin as the primary system to analyze the injection sites because of the availability of antibodies suitable for immunohistochemistry. We also utilized HIV env/rev retroviral vector (N2-IIIBenv) and Escherichia coli β-gal retroviral vector (N2-β-gal) systems to verify the phenotypes of the infiltrating cells obtained using N2-ova (data not shown).
After three injections, mice were killed and injection site muscles were cryosectioned and analyzed by immunohistochemistry using antibodies against ovalbumin and a variety of leukocyte markers (Fig. 1). We found a large number of infiltrating cells in the endomesium near the injection site, and some of the infiltrating mononuclear cells expressed ovalbumin proteins (Fig. 1 a–c). Expression of retroviral vector-encoded proteins in infiltrating cells was further confirmed from cells recovered from muscles injected with retroviral vectors encoding luciferase (N2-luci) and ovalbumin (N2-ova) by luciferase assays and B3.Z assays, respectively, as described in Materials and Methods (data not shown).
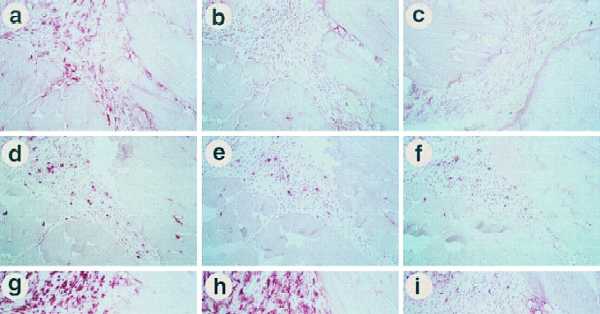
Figure 1.
Immunohistochemistry examination of the injection site. Mice were injected with either N2-ova or N2-IIIBenv intramuscularly in both legs on days 1, 4, and 7, and were killed on day 10. The following antibodies were used to stain serial sections: anti-ovalbumin (a), normal rabbit serum (b), anti-ovalbumin (c), anti-CD4 (d), B220 (e), anti-CD8 (f), F4/80 (g), N418 (h), and NLDC-145 (i). (a, b, and d–i) Sections from N2-ova-injected animals. (c) Section from an N2-IIIBenv-injected animal. The sections shown in a and c were mounted adjacent to each other on a single slide and processed simultaneously to assess background levels of the anti-ovalbumin antibodies against retroviral vector-injected muscles. We also examined the intradermal route of injection that induces comparable immune responses, and again the injection site showed comparable infiltration with the same phenotypes of cells present (data not shown).
Next, we examined the phenotypes of the infiltrating cells using antibodies against several lymphocyte markers to address which cells were potential effectors of immune responses (Fig. 1 d–f). We found that a significant number of CD4+ cells and few B cells infiltrated the injection sites. In agreement with this result was the presence of prominent CD45 staining in adjacent sections (data not shown). In this particular experiment, we also found some CD8+ infiltrating cells. However, in many other experiments using N2-ova, N2-IIIBenv, and N2-β-gal, we generally observed minimal, if any, infiltration of the CD8+ cells at the injection sites. This relative absence of CD8+ cells at the injection site agrees with reported absence of CTL in nonlymphoid organs during immune response (20). Nonetheless, the absence of CD8+ cells is particularly interesting since the spleens from these mice demonstrated consistently positive CTL responses against the antigenic protein encoded by the corresponding retroviral vector.
Finally, we examined the phenotypes of the infiltrating cells using antibodies against various monocyte/macrophage and dendritic cell markers to address infiltration of potential antigen-presenting cells (Fig. 1 g–i). F4/80+ monocyte/macrophages were identified as the most prominent infiltrating cells at the injection sites. Consistent with the presence of monocyte/macrophages and B cells was our observance of prominent major histocompatibility complex class II staining in adjacent sections (data not shown). Many of the F4/80+ cells were activated, as determined by their strong staining using the N418 antibody directed against CD11c, commonly expressed on activated monocyte/macrophages and dendritic cells (10). However, the majority of N418+ cells appear to be of the monocyte/macrophage lineage since the dendritic cell-specific markers, NLDC-145 and MIDC-8, detected only a few cells (Fig. 1i and data not shown).
Splenic Dendritic Cell Fractions Contain Cells That Express Retroviral Vector-Encoded Proteins.
Previously, biolocalization studies using DNA isolated from whole organs indicated that PCR signals were detected sporadically in spleen and lymph nodes (16, 17). In addition, several studies suggest that tumor-specific T cells are primed by bone marrow-derived cells in the lymphoid organs rather than by tumor itself (21, 22). Therefore, we decided to investigate proviral DNA integration, protein expression, and antigen presentation in the leukocyte populations from lymph nodes and spleens.
The limited sensitivity of PCR using DNA isolated from whole organs led us to investigate PCR from fractionated spleen cells, based on the assumption that each splenocyte population might be transduced disproportionately (see Discussion). Splenocytes from mice immunized with retroviral vectors were pooled and fractionated into B cells, T cells, macrophages, and dendritic cell fractions as described in Materials and Methods. Spleens from mice immunized with N2-luci (6 × 107 total cfu per mouse) were fractionated and analyzed by PCR using primers specific for the neomycin-resistance marker, a sequence common to all our retroviral vectors. A positive PCR signal was detected in the dendritic cell fraction and not in any other splenocyte fraction. Quantitative PCR was performed on the dendritic cell fraction, and the frequency of positive cells was estimated as 1 copy per 500 ng of genomic DNA, which corresponds to 1 copy per 7.5 × 104 cells. From this result, the minimal number of proviral DNA-containing cells per spleen, consisting of 1 × 108 cells, was estimated to be 1300 (Table 1). Furthermore, we were able to detect luciferase activity in the same dendritic cell fractions.
Table 1.
Estimated frequencies of cells per spleen
| Cell type | Models*
|
||
|---|---|---|---|
| Luciferase | β-gal | Ovalbumin | |
| Provirus-containing | 1300 (0.0013) | N/A | N/A |
| Protein-expressing | |||
| Experiment 1 | N/A | 2000† (0.0020) | N/A |
| Experiment 2 | 2400† (0.0024) | ||
| 1600† (0.0016) | |||
| 800‡ (0.0008) | |||
| Antigen-presenting | N/A | N/A | 300 (0.0003) |
Three model systems used to estimate the frequencies of each subset of cells were compared. A total of 2–6 × 107 cfu was injected in each mouse. All mice were injected with an identical regimen. N/A, not available.
Values are the number of transduced splenocytes per mouse; numbers in parentheses indicate the percentage of the spleen.
“Dendritic” cells.
Monocytes/macrophages.
Because we detected proviral DNA and luciferase activity in splenocytes, we wished to analyze vector-derived protein expression in detail. BALB/c mice were injected three times with N2-β-gal (4 × 107 total cfu per mouse), and transduced cells were directly visualized by X-Gal staining. The spleens were pooled and fractionated as described above, followed by X-Gal staining. In agreement with the PCR data, the dendritic cell fraction showed β-gal-positive cells at a frequency of 2000–2400 cells per mouse spleen (Table 1). In another experiment, dendritic cell fractions were sorted into dendritic cells, B cells, and monocytes by double staining with B220 and F4/80 antibodies, followed by X-Gal staining to estimate number of β-gal-expressing cells in each cell type. Among these cells expressing β-gal, we found approximately 800 monocytes (B220−/F4/80+) and 1600 dendritic cells (B220−/F4/80−). In addition, the macrophage fractions and cells from inguinal and popliteal lymph nodes of these mice also showed some X-Gal staining, although the numbers were greatly reduced compared with the dendritic cell fraction. The B and T cell fractions from N2-β-gal-immunized mice and all the cell fractions from N2-IIIBenv-immunized mice were negative by X-Gal staining.
Dendritic Cell Fraction from Retroviral Vector-Immunized Mice Can Present Antigens.
Because we demonstrated the presence of protein-expressing cells in the dendritic cell fractions, we then wished to estimate the frequency of antigen-presenting cells among these protein-expressing cells. We used N2-ova in these experiments to take advantage of the unique B3.Z cell line, which produces β-gal upon recognition of the ovalbumin peptide in the context of H-2Kb major histocompatibility complex molecules (18, 19). C57BL/6 mice were injected with N2-ova, and their pooled spleens were fractionated into B, T, macrophage, and dendritic cell fractions. The potential antigen-presenting cells were incubated overnight with B3.Z cells, and these cultures were then stained with X-Gal to estimate the level of B3.Z activation. Unfractionated splenocytes, the dendritic cell fraction, and occasionally, macrophages were able to induce β-gal production in B3.Z cells and, therefore, appeared to present antigen after direct vector administration. When B3.Z cells were incubated with splenocyte fractions from N2-IIIBenv-immunized mice, no β-gal activity was observed.
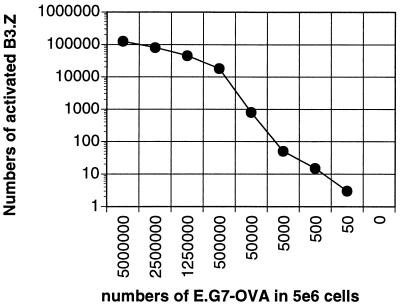
To estimate the relative abundance of ovalbumin peptide-presenting cells, a standard curve was generated from the frequency of activated blue B3.Z cells incubated with increasing numbers of E.G7-OVA cells, an EL4 thymoma cell line expressing the ovalbumin gene (Fig. 2). In the experiment shown, 1.25 × 105 B3.Z cells were activated after incubation with 5 × 106 E.G7-OVA cells, and this ratio was maintained throughout a wide range of numbers of antigen-presenting cells. The number of antigen-presenting cells in the dendritic cell fraction was estimated by comparison to this standard curve, and an average of about 300 antigen-presenting cells per spleen were detected from animals injected with a total of 2.4 × 107 cfu of N2-ova per mouse (Table 1). Pooled splenocytes from these mice contained ovalbumin-specific CTLs and gave 40% lysis in CTL assays at a 100:1 effector-to-target cell ratio (data not shown).
Figure 2.
Estimation of antigen-presenting cells. The numbers of activated B.3Z cells observed after incubation with the indicated numbers of E.G7-OVA cells mixed with EL4 cells are shown. Total number of cells from EL4 and E.G7-OVA mixtures were 5 × 106 throughout the titration range.
Dendritic Cell Fraction Contains Antigen-Presenting Cells That Can Prime Naive T Cells in Vitro.
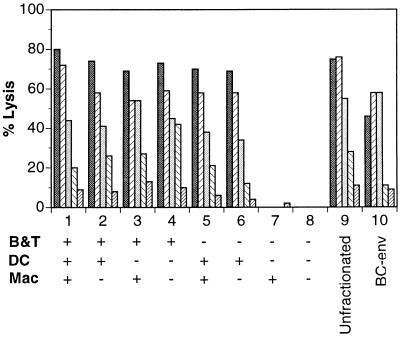
The above data indicated that the dendritic cell fraction from retroviral vector-immunized animals was capable of presenting antigenic peptides. Furthermore, it is well documented that dendritic cells pulsed with antigenic proteins or peptides can efficiently prime immune responses against viral and tumor antigens (23–28). Therefore, we wished to further examine the dendritic cell fractions from retroviral vector-immunized animals by evaluating their ability to prime naive spleens in vitro. Mice were immunized with N2-IIIBenv, which appears to induce the most vigorous T cell responses among our different retroviral vectors. The splenocytes were fractionated into three groups; B + T cells, macrophages, and dendritic cell fractions. Naive littermates were similarly processed into three groups. Fractions from immunized and naive animals were mixed to reconstitute typical splenocyte populations consisting of B + T cells, dendritic cell fractions, and macrophages (Fig. 3). Pooled unfractionated splenocytes from immunized animals were included as a positive control. In addition, splenocytes from mice immunized intraperitoneally with BC-env cells, syngeneic cells expressing HIV env/rev proteins, were included as another positive control.
Figure 3.
In vitro induction of immune responses by dendritic cell fractions from retroviral vector-immunized mice. Five mice were immunized with retroviral vectors encoding N2-IIIBenv by three injections, and their B + T cells, dendritic cells, and macrophage fractions were prepared as described in Materials and Methods. Five littermates were processed in an analogous manner. Each of the reconstituted splenocyte cultures contains B + T cells (4 × 107), dendritic cells (1 × 107), and macrophage (6 × 106) fractions and irradiated stimulators (1/50 of total responders). + and −, Fractions from immunized or naive mice, respectively. Each set of bars represents net CTL lysis at effector-to-target cell ratios from 100, 30, 10, 3, and 1.
The first four combinations included B + T cell fractions from immunized mice and were able to stimulate T cells, presumably because they have already been primed in vivo. We did not separate out B cells from the B + T cell fraction since the B cells from N2-ova-immunized mice did not contain any antigenic peptide-presenting cells, as judged from the B3.Z assays (see above). Only the dendritic cell fraction was able to induce in vitro priming of naive B + T cell populations (Fig. 3, lane 6). In contrast, the mature macrophage fractions, i.e., those cells that were adherent to plastic, were unable to prime naive splenocytes (Fig. 3, lane 7), although in the previous experiments, we found a small percentage of macrophages that were able to present ovalbumin peptides to B3.Z cells.
These data demonstrate that the dendritic cell fraction contains antigen-presenting cells that can initiate cytotoxic T cell priming in vitro. Dendritic cells were previously identified as the only population of cells that can initiate immune responses in vitro (29–32). Therefore, we reasoned that although the dendritic cell fraction contains other cell types such as B cells and monocytes, retroviral vector administration probably results in the transduction of dendritic cells.
Ex Vivo-Transduced Dendritic Cell Fraction Can Elicit T Cell Responses in Naive Recipients.
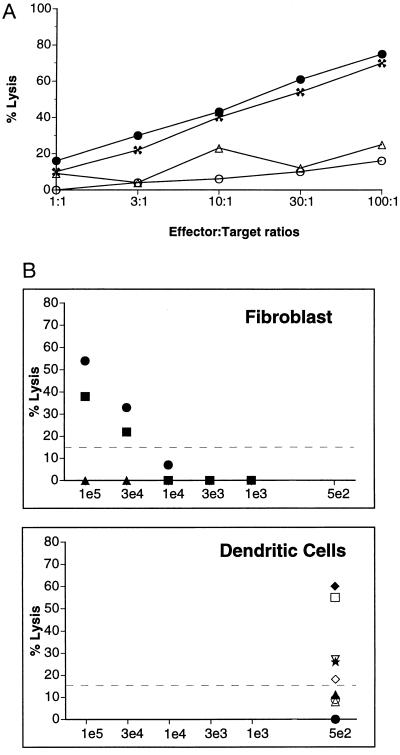
The above data suggest that the in vivo-transduced dendritic cell fraction is responsible for inducing cytotoxic T cell priming in vivo and in vitro. To extend this finding, we further examined the role of these cells by using ex vivo transduction (Fig. 4). We attempted to simulate the in vivo transduction of dendritic cells and macrophages after direct vector administration by injecting the ex vivo-transduced dendritic cell fraction. From naive mice, we separated low density cell fractions and then removed most of the contaminating B cells by an initial adherence step. The remaining cells containing the dendritic cell fraction were incubated with a cocktail of cytokines to promote cell division as described (33, 34). The proliferating cells were transduced with N2-IIIBenv using transwell cultures as described in Materials and Methods. Cells remaining after 3 days of cocultivation were examined by FACS analysis and were found to be mainly dendritic cells and monocytes (>85%). Aliquots from the transduced dendritic cell fraction were injected intraperitoneally into naive hosts, and 1 week later, CTL assays were initiated. The recipient mice generated CTL responses, confirming the role of these cells in in vivo induction of immune responses (Fig. 4A).
Figure 4.
Adoptive transfer of immune responses by ex vivo-transduced dendritic cell fractions. (A) CTL responses from naive recipients of ex vivo-transduced dendritic and monocyte cells examined by adoptive transfer. A total of 1 × 106 cells were injected per naive recipient. The splenocytes from two individual recipient mice were tested in a standard CTL assay using BC-env (• and ✖) and BC/β-gal (▵ and ○) targets. Macrophage cultures processed in a similar fashion did not elicit CTL activity. (B) Comparison between fibroblast and dendritic cells in inducing immune responses. Naive recipient mice were injected once intraperitoneally with either BC-env (three mice each per dose) or in vitro transduced dendritic cell fractions (nine mice total) as described above. Each symbol represents an individual animal. Net CTL lysis at effector-to-target cell ratios of 100 was plotted against the estimated number of cells expressing antigen. The number of dendritic cells expressing HIV env/rev was estimated from luciferase expression assays. Mice that registered 15% net lysis were considered to be positive for induction of immune responses. At fibroblast doses below 1 × 104, there were three mice per group whose symbols overlap.
Because any cell can presumably induce immune responses by the ex vivo method if given enough quantity or if injected in the region of an appropriate lymphoid organ (35), it is important to assess the relative efficacy of the transduced dendritic cell fraction as compared with other cell types. Therefore, the ability of the transduced dendritic cell fraction to induce CTL responses was compared with that of syngeneic fibroblasts expressing the same antigen and administered using identical injection routes and schedules. The transduction efficiency of the dendritic cell fraction is estimated to be at least 0.1%, based on N2-luci transduction. Specifically, a parallel preparation of the dendritic cell fraction was transduced with N2-luci using an identical protocol. The resulting cells were assayed for luciferase activity and were compared with signals from luciferase-expressing fibroblast cells normalized based on protein content. Based on this transduction efficiency, the recipient mice in the adoptive transfer experiments received an average of 5 × 102 transduced cells from a total of 5 × 105 injected cells. As shown in Fig. 4B, the levels of CTL response induced by these ex vivo-transduced dendritic cell fractions are comparable to those by injection of 3 × 104 to 1 × 105 BC-env fibroblasts expressing the same antigens. From this calculation, a single transduced cell from the dendritic cell fraction is equivalent to 60–200 fibroblasts in the induction of CTL responses, although we cannot exclude the possibility that less efficient antigen presentation by fibroblasts reflects differences in trafficking to lymphoid organs.
DISCUSSION
In this paper, we have shown that antigen presentation after direct administration of retroviral vectors can be mediated by cells found in splenic dendritic cell fractions. We envision a mechanism wherein the cells that ultimately get transduced at the injection site might have been initially recruited by the inflammatory reaction caused by needle injury. In support of this idea, we found that mice injected with needles without any injectate or injected with the formulation buffer used in retroviral vector preparations showed reduced but comparable patterns of leukocyte infiltration. After transduction, the cells are likely to drain through the lymph nodes to the spleen to present antigen. However, we cannot exclude trafficking of the retroviral vectors to lymphoid organs followed by transduction in situ. A similar mechanism for induction of immune responses by naked DNA-mediated gene transfer has also been proposed (36). In agreement with our observation, it was recently reported that cells of hematopoietic origin are essential for induction of CTL and antibody after naked DNA immunization (37, 38).
To traverse different antigen systems and to estimate the number of splenocytes in each group of impacted cells, we used vectors with similar titers and identical injection regimens. In the analyses, we made the following assumptions. (i) All aspects of transduction, protein expression, and antigen presentation are comparable among the different model antigen systems used. (ii) Transduction frequency is proportional to the total colony-forming units injected regardless of the protein encoded by the retroviral vector. (iii) All proviral DNA-containing cells can express proteins.
The cells containing proviral DNA can be detected by PCR of DNA from various sources such as whole organs, purified cell populations, or in situ from tissue sections. In our previous biolocalization studies using DNA from whole organs, we injected a total of 2 × 106 cfu of N2-IIIBenv retroviral vector per mouse and found sporadic PCR signals in spleen (16, 17). The numbers of N2-IIIBenv-transduced cells from these studies can be estimated by extrapolating from the numbers of β-gal-expressing cells in N2-β-gal-injected mice. Because 2000 β-gal-positive cells per spleen were found from mice injected with a total of 4 × 107 cfu of N2-β-gal, we can estimate that there were approximately 100 HIV env/rev-expressing cells in our previous biolocalization experiments. We estimate the sensitivity of PCR detection to be approximately 1 positive cell per 1 × 105 cells, i.e. at least 1000 transduced cells from an average spleen of 1 × 108 cells are required to register a positive PCR signal. Therefore, an amount of 100 HIV env/rev-transduced cells, corresponding to a transduction frequency of 0.1 positive cell per 1 × 105, is believed to be below the threshold of PCR detection.
In this paper, we have attempted to increase the probability of detecting PCR signals by using 6 × 107 cfu per mouse, which should increase the frequency of protein-expressing cells in the spleen to 3 cells per 1 × 105. Furthermore, we also fractionated the splenocytes, expecting different subpopulations to be transduced disproportionately. Indeed, we were able to detect cells containing proviral DNA in the dendritic cell fraction, at a frequency of 1.3 cells per 1 × 105. The fact that PCR signals were detected only in the dendritic cell fraction suggests that this fraction must be preferentially transduced in retroviral vector-injected animals and, indeed, that splenocyte fractionation was necessary to detect PCR signals.
Among the cells containing proviral DNA, some will express proteins, which can be detected by histochemistry, immunohistochemistry, or luciferase assays. Using N2-β-gal, we have also found other protein-expressing cells, primarily myofibers (data not shown). Although we cannot exclude the contribution of transduced myofibers to induction of immune responses, the long-term persistence of protein expression in transduced myofibers (>200 days) as well as their low cell surface expression of major histocompatibility complex and accessory molecules render them an unlikely candidate for induction of immune responses. Furthermore, this notion is supported by the lack of CD8+ cell infiltration in the injection site.
Nonetheless, we had to consider the fact that these four different model systems are not always comparable in expressing proteins or inducing immune responses. For example, the intracellular locations of these antigens are all distinct, such that it is not clear whether ovalbumin, a secreted protein, is processed comparably to β-gal, a cytoplasmic protein. Indeed, we have found several discrepancies in protein expression between these two antigens. Although muscle fibers are the most prominently transduced tissue in N2-β-gal-injected animals, we have yet to find strongly stained fibers in N2-ova-injected animals. Conversely, we detected very few transduced infiltrating cells from N2-β-gal-injected animals, despite similar degrees of infiltration, yet we found considerable numbers of ovalbumin-stained cells in N2-ova-injected animals by immunohistochemistry. This discrepancy between ovalbumin and β-gal systems might be due to the fact that secreted ovalbumin could be phagocytosed by infiltrating cells. If so, this group of cells may represent a distinct class of antigen-presenting cells that present antigen acquired through phagocytosis without expressing the protein.
The estimation of the numbers of cells in each group has limitations other than extrapolation among different retroviral vector systems. First, the estimated numbers are derived from spleens of retroviral vector-injected animals harvested at one specific time, i.e., 3 days after the last injection. Therefore, the estimated numbers will probably fluctuate relative to the migration of the transduced cells to and from the spleen, and the kinetics and strength of the incipient immune responses. In addition, since the number of antigen-presenting cells in the spleen of retroviral vector-injected mice was estimated from a standard curve generated using ovalbumin-expressing thymoma cells (E.G7-OVA) in B3.Z assays (Fig. 2), it is impossible to assess whether the E.G7-OVA cells are equivalent in their capacity for antigen presentation compared with splenocytes.
It is of great interest to define the precise phenotypes of cells transduced by retroviral vectors, of those expressing antigens, and of those that contribute to antigen presentation. In the current study, the low yield of the dendritic cell fraction and subsequent transduction efficiency, together with difficulties in maintaining in vitro cultures, prevented us from investigating this issue beyond the data shown in Table 1. In addition, various cells belonging to dendritic and monocyte/macrophage lineages were shown by others to express relatively ill-defined and temporally changing cell surface markers (15, 39, 40). This made it difficult to precisely define the phenotypes of cells involved in antigen presentation. Therefore, the cells in the dendritic cell fraction are likely, but may not be, the only cells involved in antigen presentation. Nonetheless, we feel that the methods used in this paper are the first attempts at this type of in vivo quantitation of tranduced cells, and the results illustrate the rarity of in vivo antigen-presenting cells.
Acknowledgments
E.G7-OVA and B3.Z cells were kindly provided by Drs. Michael Bevan and Nilabh Shastri, respectively. We thank the following individuals for their valuable assistance to this project: Drs. Stephen Chang, Linda Karavodin, and Jim McCormack for critical review of the manuscripts; Drs. Michael Irwin and Michael Bevan for discussions; Dr. Maria Donoghue for muscle anatomy; and Tammi Howard and Darcina Knapp for expert vivarium assistance. We also thank Ann Beever for excellent help with graphics. This study was funded in conjunction with The Green Cross Corp., Osaka, Japan.
ABBREVIATIONS
- CTL
cytotoxic T lymphocyte
- cfu
colony-forming units
- X-Gal
5-bromo-4-chloro-3-indosyl β-galactopyranoside
- β-gal
β-galactosidase
References
- 1.Warner J F, Anderson C-G, Laube L, Jolly D J, Townsend K, Chada S, St. Louis D. AIDS Res Hum Retroviruses. 1991;7:645–655. doi: 10.1089/aid.1991.7.645. [DOI] [PubMed] [Google Scholar]
- 2.Laube L S, Burrascano M, DeJesus C E, Howard B D, Johnson M A, Lee W L T, Lynn A E, Peters G, Ronlov G S, Townsend K S, Eason R L, Jolly D J, Merchant B, Warner J F. Hum Gene Ther. 1994;5:853–862. doi: 10.1089/hum.1994.5.7-853. [DOI] [PubMed] [Google Scholar]
- 3.Ziegner U H M, Jolly D J, Mento S J, Galpin J, Prussak C E, Hartnett D E, Bohart C, Barber J R, Peters G, Klump W, Sajjadi N, Merchant B, Warner J F. AIDS. 1995;9:43–50. doi: 10.1097/00002030-199501000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Irwin M J, Laube L S, Lee V, Austin M, Chada S, Anderson C-G, Townsend K, Jolly D J, Warner J F. J Virol. 1994;68:5036–5044. doi: 10.1128/jvi.68.8.5036-5044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chada S, DeJesus C E, Townsend K, Lee W T L, Laube L, Jolly D J, Chang S M W, Warner J F. J Virol. 1993;67:3409–3417. doi: 10.1128/jvi.67.6.3409-3417.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dialynas D P, Wilde D B, Marrack P, Pierres A, Wall K A, Havran W, Otten G, Loken M R, Pierres M, Kappler J, Fitch F W. Immunol Rev. 1983;74:29–56. doi: 10.1111/j.1600-065x.1983.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 7.Sarmiento M, Glasebrook A L, Fitch F W. J Immunol. 1980;125:2665–2672. [PubMed] [Google Scholar]
- 8.Coffman R L, Weissman I L. Nature (London) 1981;289:681–683. doi: 10.1038/289681a0. [DOI] [PubMed] [Google Scholar]
- 9.Austyn J M, Gordon S. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 10.Metlay J P, Witmer-Pack M D, Agger R, Crowley M T, Lawless D, Steinman R M. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nussenzweig M C, Steinman R M, Witmer M D, Gutchinow M A. Proc Natl Acad Sci USA. 1982;79:161–165. doi: 10.1073/pnas.79.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraal G, Breel M, Janse M, Bruin G. J Exp Med. 1986;163:981–997. doi: 10.1084/jem.163.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breel M, Mebius R, Kraal G. Eur J Immunol. 1987;17:1555–1559. doi: 10.1002/eji.1830171105. [DOI] [PubMed] [Google Scholar]
- 14.Surh C D, Gao E-K, Kosaka H, Lo D, Ahn C, Murphy D B, Karlsson L, Peterson P, Sprent J. J Exp Med. 1992;176:495–505. doi: 10.1084/jem.176.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girolomoni G, Simon J C, Bergstresser P B, Cruz P D C. J Immunol. 1990;145:2820–2826. [PubMed] [Google Scholar]
- 16.Sajjadi N, Kamantigue E, Edwards W, Howard T, Jolly D, Mento S, Chada S. Hum Gene Ther. 1994;5:693–699. doi: 10.1089/hum.1994.5.6-693. [DOI] [PubMed] [Google Scholar]
- 17.Kamantigue E, Wilson E, Chada S, Brumm D, Austin M, Irwin M, Mento S, Kowal K, Sajjadi N. Gene Ther. 1996;3:128–136. [PubMed] [Google Scholar]
- 18.Karttunen J, Shastri N. Proc Natl Acad Sci USA. 1991;88:3972–3976. doi: 10.1073/pnas.88.9.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karttunen J, Sanderson S, Shastri N. Proc Natl Acad Sci USA. 1992;89:6020–6024. doi: 10.1073/pnas.89.13.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ando K, Guidotti L G, Cerny A, Ishikawa T, Chisari F V. J Immunol. 1994;153:482–488. [PubMed] [Google Scholar]
- 21.Huang A Y C, Golumbek P, Ahmadzadeh M, Jeffee E, Pardoll D, Levetsky H. Science. 1992;264:961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 22.Maass G, Schmidt W, Berger M, Schilcher F, Koszik F, Schneeberger A, Stingl G, Birnstiel M L, Schweighoffer T. Proc Natl Acad Sci USA. 1995;92:5540–5544. doi: 10.1073/pnas.92.12.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Bruijn M L H, Nieland J D, Schumacher T N M, Ploegh H L, Kast W M, Melief C J M. Eur J Immunol. 1992;22:3013–3020. doi: 10.1002/eji.1830221137. [DOI] [PubMed] [Google Scholar]
- 24.Bender A, Bui L K, Feldman M A V, Larsson M, Bhardwaj N. J Exp Med. 1995;182:1663–1671. doi: 10.1084/jem.182.6.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flamand V M, Sornasse T, Thielemans K, Demanet C, Bakkus M, Bazin H, Tielemans F, Leo O, Urbain J, Moser M. Eur J Immunol. 1994;24:605–610. doi: 10.1002/eji.1830240317. [DOI] [PubMed] [Google Scholar]
- 26.Hsu F J, Benike C, Fagnoli F, Liles T M, Czerwinski D, Taide B, Engelman E G, Levy R. Nat Med. 1996;2:52–58. doi: 10.1038/nm0196-52. [DOI] [PubMed] [Google Scholar]
- 27.Zitvogel L, Mayordomo J I, Tjandrawan T, DeLeo A B, Clarke M R, Lotze M T, Storkus W J. J Exp Med. 1996;183:87–97. doi: 10.1084/jem.183.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paglia P, Chiodono C, Rodulfo M, Columbo M P. J Exp Med. 1996;183:317–322. doi: 10.1084/jem.183.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inaba K, Metlay J P, Crowley M T, Steinman R M. J Exp Med. 1990;172:631–640. doi: 10.1084/jem.172.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair S, Zhou F, Reddy R, Huang L, Rouse B T. J Exp Med. 1992;175:609–612. doi: 10.1084/jem.175.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macatonia S E, Patterson S, Knight S C. Immunology. 1991;74:399–406. [PMC free article] [PubMed] [Google Scholar]
- 32.Elbe A, Schleischitz S, Strunk D, Stingl G. J Immunol. 1994;153:2878–2889. [PubMed] [Google Scholar]
- 33.Inaba K, Steinman R M, Witmer-Pack M, Aya H, Inaba M, Sudo T, Wolpe S, Schuler G. J Exp Med. 1992;175:1157–1167. doi: 10.1084/jem.175.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sallusto F, Lanzavecchia A. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kundig T M, Bachman M F, DiPaolo C, Simard J J L, Battegay M, Lother H, Gessner A, Kuhlcke K, Ohashi P S, Hengartner H, Zinkernagel R M. Science. 1995;268:1343–1347. doi: 10.1126/science.7761853. [DOI] [PubMed] [Google Scholar]
- 36.Pardoll D W, Beckerleg A M. Immunity. 1995;3:165–169. doi: 10.1016/1074-7613(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 37.Doe B, Selby M, Barnett S, Baenziger J, Walker C M. Proc Natl Acad Sci USA. 1996;93:8578–8583. doi: 10.1073/pnas.93.16.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Condon C, Watkins S C, Celluzzi C M, Thompson K, Falo L D. Nat Med. 1996;2:1122–1128. doi: 10.1038/nm1096-1122. [DOI] [PubMed] [Google Scholar]
- 39.Hoefsmit E C M, Duijvestijn A M, Kamperdijk E W A. J Immunobiol. 1982;161:255–265. doi: 10.1016/S0171-2985(82)80081-8. [DOI] [PubMed] [Google Scholar]
- 40.Crowley M, Inaba K, Whitmer-Pack M D, Gezelter S, Steinman R M. J Immunol Methods. 1990;133:55–66. doi: 10.1016/0022-1759(90)90318-p. [DOI] [PubMed] [Google Scholar]