Abstract
The ETV6/CBFA2 (TEL/AML1) fusion gene occurs as a result of the chromosome translocation t(12;21)(p13;q22) in up to 30% of children diagnosed with B cell precursor (cd10+, cd19+) acute lymphoblastic leukemia. Leukemic cells that have acquired the t(12;21) usually demonstrate loss of the remaining normal ETV6 (TEL) allele. Using reporter gene assays we have functionally characterized both the normal ETV6 and ETV6/CBFA2 fusion proteins in the regulation of the MCSFR proximal promoter. Neither ETV6 or ETV6/CBFA2 has any significant, detectable effect on the promoter by itself. However, both ETV6 and ETV6/CBFA2 inhibit the activation of the promoter by CBFA2B(AML1B) and C/EBPa. We have shown that a 29-bp region of the MCSFR promoter containing the binding sites for CBFA2B and C/EBPa is sufficient for the inhibition by ETV6 and ETV6/CBFA2. Mutational analysis of the MCSFR promoter revealed that binding of both CBFA2B and C/EBPa to their respective sites is necessary for the inhibition by ETV6 and ETV6/CBFA2. Deletion of the helix–loop–helix (HLH) region from the cDNAs of ETV6 and ETV6/CBFA2 decreased but did not completely abrogate the ability of either construct to inhibit promoter activation. We also found that the ETS DNA binding region of ETV6 is necessary for inhibition of the promoter. Addition of ETS1 and FLI1, two ETS family members that have homology in the 5′ HLH region, but not Spi1, an ETS family member without the 5′ HLH region, also inhibited reporter gene expression. Our data show that the inhibition mediated by ETV6 and ETV6/CBFA2, in the context of the MCSFR promoter, depend on interactions with other proteins, not just CBFA2B. Our results also indicate that the transactivation characteristics of ETV6/CBFA2 are a combination of positive and negative regulatory properties.
Translocations and deletions of chromosome 12 are among the most commonly occurring cytogenetic abnormalities in acute childhood leukemia (1–4). The chromosome translocation t(12;21)(p13;q22) has been shown to occur in up to 30% of children diagnosed with B cell lineage acute leukemia (5–7). The t(12;21) juxtaposes the ETV6 (also known as TEL) gene on chromosome 12, a member of the ETS E26-transforming-specific family, to the CBFA2 (also known as AML1) gene on chromosome 21 to create the ETV6/CBFA2 fusion gene (8, 9). In addition to the translocation, most patients who have the t(12;21) also lose the remaining ETV6 allele (7, 10–13). Thus, the progression of the disease in these patients appears first to require the acquisition of the t(12;21) followed by the loss of the remaining ETV6 allele. As both ETV6 and ETV6/CBFA2 are putative transcription factors, one might speculate that the acquisition of the t(12;21) and the deletion of ETV6 alter the expression patterns in leukemic cells that result in unregulated growth. To develop a molecular model of disease progression in this specific leukemia, the transactivating characteristics of ETV6 and ETV6/CBFA2 must be determined.
Expression of the ETV6/CBFA2 fusion gene results in a transcript containing the 5′ 1-kb coding region of ETV6 in-frame with virtually the entire CBFA2 coding region (8, 9). The fusion transcript contains several defined domains that may provide important functional properties to the fusion protein. The normal ETV6 gene contains two identifiable domains based on its homology to other ETS proteins. The ETS domain, which has been shown to be necessary for DNA binding in other ETS proteins, is in the 3′ end of the gene and is not included in the ETV6/CBFA2 fusion. DNA binding by the ETS domain of ETV6 has not yet been demonstrated and, thus, no ETV6 consensus binding site has been determined. Another domain identified by its homology to other ETS proteins is the helix–loop–helix (HLH) domain in the 5′ coding region. A recent study has demonstrated that the ETV6/CBFA2 fusion product acts as a repressor on the T cell receptor beta enhancer and that its repressor activity is dependent on the presence of the HLH domain (14).
CBFA2 contributes two characterized domains to the fusion gene. The first is the runt domain, so called because of its homology to the Drosophila runt gene (15, 16). The runt domain has also been identified in the Drosophila lozenge gene (17). Both runt and lozenge have been shown to be important in the developmental pathways of Drosophila. In CBFA2, the runt domain is important in both DNA binding and protein–protein interactions. The second characterized domain is a transactivating domain. This unique domain does not share homology with other transactivating proteins, but is necessary for the transactivating properties of CBFA2 (18). The CBFA2 gene is expressed in several splice variants (19, 20). One of the splice variants, CBFA2A contains only the runt homology domain and lacks the transactivation domain, whereas another splice variant, CBFA2B, contains both characterized domains (19, 21). The ETV6/CBFA2 fusion transcript isolated from patients contains both functional domains of the CBFA2 gene (8, 9). This fact does not preclude the possibility that the fusion gene is alternatively spliced like CBFA2, so that two possible splice variants of the fusion gene are possible. Regardless of possible splice varients, CBFA2 provides a putative DNA binding domain for the ETV6/CBFA2 fusion. Therefore, if the ETV6/CBFA2 protein maintains DNA binding abilities, it is possible that the targets of the fusion protein will be the same as CBFA2.
The expressed product of the normal CBFA2 gene is one of the two components of the core binding factor (CBF). The second component of CBF is CBFB. CBFA2 has been shown to contribute both the DNA binding and transactivating properties of CBF. CBFB has been hypothesized to stabilize the CBF complex (22, 23). CBF has been shown to be important in the regulation of several hematopoietic genes (18, 24, 25, 26, 27, 28). One of these genes, the monocyte colony stimulating factor receptor (MCSFR), has a well-characterized proximal promoter that contains a CBF binding site as well as an adjacent site to which the CAATT enhancer binding protein alpha (C/EBPa) binds (29). Both sites are necessary for full activation of the promoter. Reporter genes downstream of the MCSFR proximal promoter are synergistically activated when cotransfected with both CBFA2 and C/EBPa (29).
The MCSFR proximal promoter upstream of a reporter gene has been used to characterize other CBFA2 fusion proteins. The AML1/ETO fusion has been shown to activate the promoter, whereas the AML1/EVI1 fusion inhibits its activation (30). In this paper we also use the MCSFR proximal promoter to characterize the transactivating properties of the ETV6 and ETV6/CBFA2 gene products.
MATERIALS AND METHODS
Plasmids.
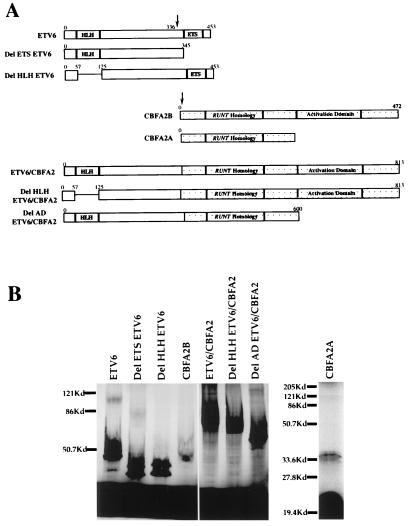
The hexamer synthetic promoter was made by placing six copies of a 29-bp region containing the CBF and C/EBPa binding sites (CAAGATTTCCAAACTCTGTGGTTGCCTTGCCTA) upstream of a thymidine kinase minimal promoter. Site-specific mutations in the CBF m(−190) (−88 5′-CAAGATTTCCAAACTCTctaaggtaccTGCCTA-3′) or C/EBPa m(−210) (−88 5′-CAggtaccatAAACTCTGTGGTTGCCTTGCCTA-3′) binding sites were made in the MCSFR proximal promoter (26). The ETV6 plasmid was provided by S. K. Bohlander (Institut fuer Humangenetik, Goettingen, Germany). The ETV6/CBFA2 (Fig. 1A) construct was made by ligating the 5′ coding region of ETV6 and the 3′ CBFA2B coding region to a linker made by PCR amplification of the breakpoint region of ETV6/CBFA2 cDNA. Thus, the ETV6/CBFA2 cDNA includes the first 333 amino acids of ETV6 fused to all but the first 5 codons of the CBFA2B coding region as reported previously (8). The del ETS ETV6 (Fig. 1A) was made by digesting the ETV6/CBFA2 construct with XbaI, which cuts ETV6/CBFA2 at the breakpoint fusion between the coding regions of the two genes. The ETV6 coding region (without an ETS domain) was then subcloned into a eukaryotic expression vector (SR alpha murine virus vector). The del AD ETV6/CBFA2 (Fig. 1A) was constructed by digesting the ETV6/CBFA2 construct with BamHI. This allowed us to separate the 5′ 1840-bp coding region containing tETV6 and the runt domain of CBFA2 from the 3′ coding region containing the activation domain of CBFA2. We subcloned the 5′ fragment in a eukaryotic expression vector (SR alpha murine virus vector). del HLH ETV6 (Fig. 1A) was made by amplifying the ETV6 coding region 5′ to the HLH from nucleotides 1 to 194 and ligating it in-frame to a PCR amplified region of the coding region of ETV6 3′ to the HLH domain (nt 397 to the end of ETV6). del HLH ETV6/CBFA2 (Fig. 1A) was then constructed by digesting del HLH ETV6 and ETV6/CBFA2 with a unique restriction enzyme and ligating the 5′ end of ETV6 without the HLH region to ETV6/CBFA2. The finished constructs were in vitro translated (Promega TnT coupled rabbit reticulocyte lysate) to confirm appropriately migrating products (Fig. 1B).
Figure 1.
(A) Diagrammatic representation of the various cDNA constructs used. Arrows indicate location of the breakpoint in the coding regions. Numbers above the constructs refer to the amino acid residues. HLH, helix–loop–helix domain; ETS, ETS DNA binding domain. (B) Autoradiographic representation of an SDS/PAGE separation of the in vitro translated constructs.
Cell Lines and Transfection Assays.
We used NIH 3T3 cells grown in DMEM/10% fetal bovine serum/1% Penn/Strep. Transfection experiments were performed using a calcium phosphate transfection kit (Invitrogen). We cotransfected 8 μg of reporter and 5 μg of effector plasmid(s) per experiment. Cells were harvested after 48 hr and assayed with a Promega luciferase assay kit. The luciferase assays were corrected for transfection efficiency using a β-galactosidase assay.
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSAs were performed as described (26).
RESULTS
ETV6 and ETV6/CBFA2 Inhibit Reporter Gene Activation by CBFA2B and C/EBPa.
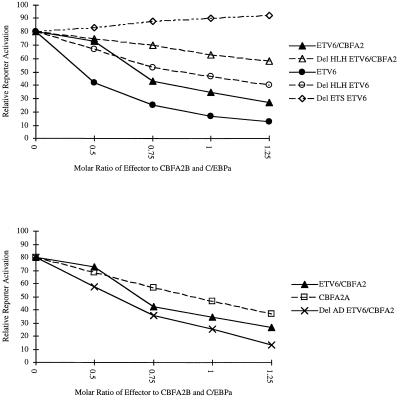
In our initial experiments we cotransfected either ETV6 or ETV6/CBFA2 into NIH 3T3 cells with the MCSFR proximal promoter upstream from a luciferase gene. Neither ETV6 (Fig. 2 A and B) or ETV6/CBFA2 (data not shown) changed the expression of luciferase from background. We then cotransfected ETV6 or ETV6/CBFA2 with the reporter gene and CBFA2B and C/EBPa. Together, CBFA2B and C/EBPa activated the promoter approximately 80-fold (Fig. 2A). Our results demonstrated that addition of equimolar concentrations of ETV6/CBFA2 to CBFA2B and C/EBPa decreased promoter activation from 80-fold to less than 30-fold (Fig. 2A). As expected in titration experiments, we found that increasing amounts of ETV6/CBFA2 resulted in increasing levels of promoter inhibition (Fig. 3 Upper).
Figure 2.
Activation of the MCSFR proximal promoter by CBFA2B and ETV6 or ETV6/CBFA2 with or without C/EBPa. (A) ETV6 has no detectable effect on reporter gene by itself. Together CBFA2B and C/EBPa synergistically activate the reporter gene approximately 80-fold. This synergistic activation is inhibited by the addition of either ETV6 or ETV6/CBFA2. (B) In the absence of C/EBPa, CBFA2B only activates the reporter gene 5-fold above background. The cotransfection of ETV6 or ETV6/CBFA2 in the absence of C/EBPa slightly increases reporter gene activation. (C) C/EBPa activates the MCSFR promoter about 15-fold in the absence of cotransfected CBFA2. Both ETV6 and ETV6/CBFA2 inhibit the activation of the promoter by C/EBPa, even when CBFA2 is not included in the cotransfection assay. Note also that the cotransfection of ETV6, together with ETV6/CBFA2 in the presence of C/EBPa, inhibits the promoter to a relative activation of 0.035-fold.
Figure 3.
Inhibition of activation of the MCSFR promoter by increasing amounts of ETV6 or ETV6/CBFA2 plasmid. (Upper) Shows that increasing amounts of either ETV6 or ETV6/CBFA2 increases the level of reporter gene inhibition. It also shows that the removal of the HLH domain of either ETV6 or ETV6/CBFA2 results in a decrease in the ability of the respective constructs to inhibit promoter activation. (Lower) Results of cotransfecting the CBFA2 splice varient CBFA2A or the mutant ETV6/CBFA2 construct lacking the activation domain into the cotransfection system. The del AD ETV6/CBFA2 is able to inhibit reporter gene expression more than the full-length ETV6/CBFA2. CBFA2A, lacking the activation domain of CBFA2, is also able to decrease the synergistic activation of the promoter by CBFA2B and C/EBPa.
Moreover, we found that cotransfection with ETV6 also inhibited the activation of the promoter by CBFA2B and CEBPα from 80-fold to less than 15-fold (Fig. 2A). Titration experiments showed that increased concentrations of ETV6 caused increasing inhibition of promoter activation (Fig. 3 Upper). To identify the important interacting components necessary for the inhibition of reporter gene expression by ETV6 and ETV6/CBFA2, we repeated the cotransfection assays, but did not include C/EBPa in the transfections. Instead of inhibition, we noted that both ETV6 and ETV6/CBFA2 slightly activated the reporter gene expression (Fig. 2B). We found that both ETV6 and ETV6/CBFA2 also slightly activate the expression of a reporter gene on a thymidine kinase minimal promoter and therefore attribute the slight increase in reporter gene expression to nonspecific activation (data not shown). We also performed the cotransfection experiments including C/EBPa, but not CBFA2B. C/EBPa activated the promoter 15-fold above background (Fig. 2C). Both ETV6 and ETV6/CBFA2 inhibited promoter activation by C/EBPa (Fig. 2C). These results indicate that the ability of either ETV6 or ETV6/CBFA2 to inhibit the activation of the MCSFR proximal promoter depends on the presence of C/EBPa. Addition of ETV6 together with ETV6/CBFA2 decreased further the activation of the promoter (Fig. 2C).
A 29-bp Region Containing Binding Sites for CBFA2B and C/EBPa Is Sufficient to Demonstrate Inhibition by ETV6 and ETV6/CBFA2.
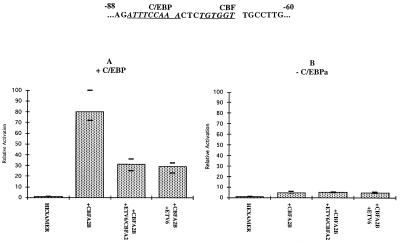
The MCSFR proximal promoter is a 400-bp region that contains several characterized cis elements (29, 31). To define the cis elements required to show the inhibitory properties of ETV6 and ETV6/CBFA2, we repeated the cotransfection assays using only a small region of MCSFR proximal promoter. This 29-bp region has both the CBF and C/EBPa binding sites but excludes other previously characterized elements (Fig. 4). Again, we noted that CBFA2B and C/EBPa together activated reporter gene expression about 80-fold above background. Addition of ETV6 or ETV6/CBFA2 to this system showed the inhibition seen with the entire MCSFR proximal promoter, namely from an 80-fold activation by CBFA2B and C/EBPa to less than 30-fold (Fig. 4A). We therefore concluded that this 29-bp region contained the elements necessary for ETV6 and ETV6/CBFA2 to mediate their inhibition of reporter gene activation by CBFA2B and C/EBPa.
Figure 4.
Activation of the hexamer reporter, a 29-bp region of the MCSFR proximal promoter containing the CBF and C/EBPa binding sites. (A) Addition of both CBFA2B and C/EBPa results in the synergistic activation of the hexamer reporter. Cotransfection of either ETV6 or ETV6/CBFA2 results in the inhibition of reporter gene activation. (B) Inhibition by ETV6 and ETV6/CBFA2 is not detectable in the absence of C/EBPa.
Functional Binding Sites for both CBFA2B and C/EBPa Are Necessary to Demonstrate Inhibition by ETV6 or ETV6/CBFA2.
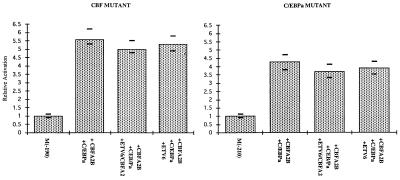
To further dissect the necessary promoter elements for inhibition by ETV6 and ETV6/CBFA2, we repeated the experiments with promoters containing mutations in either the CBF binding site or the C/EBPa binding site. We found that neither mutant promoter activated above 10-fold upon the addition of CBFA2B and C/EBPa (Fig. 5). We also noted that the addition of either ETV6 or ETV6/CBFA2 had no effect on the level of activation on either mutated promoter even in the presence of both CBFA2B and C/EBPa (Fig. 5). We conclude that ETV6 and ETV6/CBFA2 inhibit the synergy between CBFA2B and C/EBPa.
Figure 5.
Activation of site-specific mutated promoters. If either the CBF site [m(−190) Left] or the C/EBPa site [m(−210) Right] is mutated synergistic activation of the reporter gene does not occur. Furthermore, the addition of ETV6 or ETV6/CBFA2 to either system does not produce a significant effect.
Deletion of the HLH Domain of ETV6 or ETV6/CBFA2 Decreases but Does Not Completely Abrogate Inhibition.
When we cotransfected HLH deletion mutants of both ETV6 and ETV6/CBFA2 (Fig. 1) with the MCSFR promoter and CBFA2B and C/EBPa we found that the inhibiting ability of both ETV6 and ETV6/CBFA2 was diminished, but not completely abrogated. Equimolar concentrations of the del HLH ETV6 plasmid reduced promoter activation from 80-fold to 40-fold (Fig. 3 Upper). This is compared with the ability of normal ETV6 to inhibit promoter activation from 80-fold to less than 15-fold. Equimolar concentrations of del HLH ETV6/CBFA2 reduced reporter gene activation from 80-fold to less than 60-fold (Fig. 3 Upper), whereas nonmutated ETV6/CBFA2 inhibited reporter gene activation from 80-fold to less than 30-fold. The results of titration experiments with either del HLH ETV6 or del HLH ETV6/CBFA2 demonstrated increasing levels of inhibition (Fig. 3 Upper). We also found that both plasmids had slight, nonspecific activating properties on a thymidine kinase minimal promoter (data not shown). Therefore, we conclude that in this system the HLH domain of ETV6 appears to be important, but not sufficient to mediate the full inhibitory function of ETV6.
An ETS Deleted Mutant of ETV6 Does Not Inhibit Activation of the Promoter.
A mutant ETV6 cDNA lacking the ETS domain cotransfected with the reporter gene and CBFA2B and C/EBPa did not inhibit reporter gene activation (Fig. 3 Upper). These results suggest that the DNA binding property of ETV6 may be important to the ability of ETV6 to inhibit reporter gene expression in this system. Although the del ETS ETV6 was expressed from the same expression vector as the ETV6 and ETV6/CBFA2, and in vitro translation assays confirmed expression of the protein, we cannot rule out the possibility that shortening of the protein may affect stability, lifetime, and nuclear accumulation. Experiments with tagged proteins and monoclonal antibodies are currently underway to address this point.
Deletion of the Activation Domain of ETV6/CBFA2 Results in Increased Inhibition.
The activation domain of CBFA2 was present in our expression plasmid and this domain has been shown to be necessary for activation of target genes by CBFA2B. CBFA2A, a splice variant of the CBFA2 gene without the transactivation domain, inhibits the activation of the MCSFR proximal promoter by CBFA2B (Fig. 3 Lower). To characterize the role of the activation domain in the ETV6/CBFA2 fusion, we constructed an ETV6/CBFA2 mutant with the activation domain of CBFA2B removed [del AD ETV6/CBFA2 (Fig. 1)]. We found that this mutant significantly inhibited the activation of the MCSFR reporter gene by CBFA2B and C/EBPa. In fact, the del AD ETV6/CBFA2 inhibited reporter gene expression more dramatically than nonmutated ETV6/CBFA2 (Fig. 3 Lower).
Effects of Other ETS Family Members on the MCSFR Promoter.
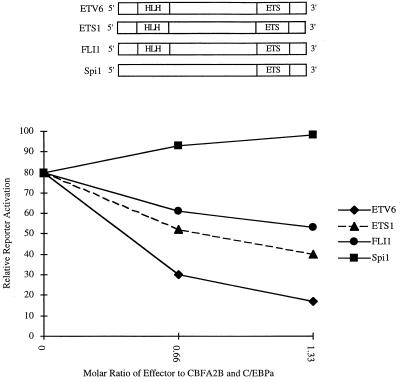
We were intrigued by the ability of not only ETV6/CBFA2, but also ETV6 to inhibit the activation of the MCSFR promoter. We were curious whether this phenomenon was specific to ETV6, or whether homologous proteins shared this property. We therefore cotransfected other ETS family member proteins, ETS1, FLI1, and spi1 (PU.1), with the MCSFR promoter and CBFA2B and C/EBPa. A binding site for PU.1 has been identified about 20 bp 3′ of the CBF binding site (32). This site has been shown to be necessary for the transcriptional activation of the promoter by PU.1. We found that both FLI1 and ETS1, but not spi1 (PU.1), had the ability to inhibit reporter gene activation (Fig. 6). The inhibition by FLI1 and ETS1 was not as dramatic as the inhibition by ETV6 but was consistent and only seen if C/EBPa was included in the cotransfection assay (data not shown). The inhibitory effects of FLI1 and ETS1 were also apparent when the 29-bp hexamer promoter was used as the reporter gene (data not shown). Spi1 did not have an effect on the 29-bp hexamer reporter. None of these proteins had any effect when cotransfected with a reporter gene containing the thymidine kinase minimal promoter alone. Interestingly, both FLI1 and ETS1, but not spi1, share homology in the 5′ HLH region of ETV6.
Figure 6.
Effect of ETS family members on the activation of the MCSFR reporter gene. (Upper) Diagrammatic representation of the ETS family members used in the cotransfection experiments. All constructs share 3′ homology in the ETS domain. Only ETV6, ETS1, and FLI1 share homology in the 5′ HLH domain. Like ETV6, ETS1 and FLI1 inhibit reporter gene activation in the presence of both CBFA2B and C/EBPa. Spi1 (PU.1) shows additional activation of the reporter gene.
EMSAs.
To determine the ability of ETV6/CBFA2 to bind DNA, we in vitro translated the cDNA and performed gel shift assays using a probe with a specific CBF site. As reported previously, in vitro translated ETV6/CBFA2 binds very poorly to a probe containing the CBF binding consensus (14) (data not shown). However, when we repeated the assays with nuclear extracts from ETV6/CBFA2 infected cells we detected a higher migrating band not present in the vector-alone infected cells indicating that in vivo expressed ETV6/CBFA2 is able to recognize and bind a CBF probe (Fig. 7).
Figure 7.
EMSA results using stably infected 32D cell nuclear extracts. The first lane represents probe alone. The second lane contains vector-alone infected 32D cells and lane 3 ETV6/CBFA2 infected 32D cells. Arrowhead, location of a possible ETV6/CBFA2 migrating band.
DISCUSSION
The purpose of this study was to characterize ETV6 and ETV6/CBFA2 in the regulation of the MCSFR promoter. This promoter has several binding sites for transcription factors, which are necessary for its activation (26, 29, 31, 32). Two of these factors are CBF and C/EBPa, which synergistically activate the promoter 80-fold. These two sites are 3 bp apart. In this study, we used the normal MCSFR promoter and a synthetic promoter containing six tandem copies of a 29-bp region containing the CBF and C/EBPa binding sites. In addition, we used promoters in which one or the other of the two sites had been mutated. We tested the promoters with ETV6 and ETV6/CBFA2 and with several deletion mutants of the two proteins lacking specific functional domains.
Our results confirm that activation of the promoter by CBFA2 and C/EBPa requires both proteins and both intact sites. In addition, our results indicate that ETV6/CBFA2 inhibits activation of the normal promoter. The region necessary for the inhibition must contain intact sites for both CBFA2 and C/EBPa, as we determined using the synthetic and mutated promoters. Because inhibition by ETV6/CBFA2 is observed in the presence of both CBFA2 and C/EBPa and because both sites must be intact, we hypothesize that ETV6/CBFA2 destroys the synergy between the two proteins. By using ETV6/CBFA2 deletional mutants, we determined that removal of the HLH domain will decrease but not eliminate the inhibition. Furthermore, an ETV6/CBFA2 deletional mutant lacking the activation domain of CBFA2 has a stronger inhibitory effect, suggesting that this domain has a positive role in promoter regulation by this fusion protein. This is in sharp contrast to what is observed in the regulation of the T cell receptor b promoter by ETV6/CBFA2 (14).
Whether ETV6/CBFA2 inhibits the promoter by actually binding at the promoter site is still an unresolved issue. Our EMSA results strongly suggest that if binding occurs, it is minimal. However, the protein we used was translated in vitro, and we cannot disregard the fact that posttranslation modifications are needed to increase the affinity of the protein for DNA. Another possibility is that additional proteins are necessary for recognition and/or binding of ETV6/CBFA2 to DNA. Our EMSA results using nuclear extracts from ETV6/CBFA2 expressing cells support this possibility.
We also examined the role of ETV6 in the regulation of the MCSFR promoter. ETV6 is a strong repressor of the promoter, and requires an intact ETS domain. As for ETV6/CBFA2, the effect of ETV6 is observed only if CBFA2 and C/EBPa sites are intact and both CBFA2 and C/EBPa are present. Other HLH–ETS proteins such as FLI1 and ETS1 repress the promoter and have the same requirements for repression (i.e., they require intact sites for CBFA2 and C/EBPa and both proteins). This would suggest that ETV6 and other HLH–ETS proteins belong to a family of repressors whose function could be used to direct tissue-specific regulation by factors (such as CBFA2), which are nonspecifically expressed in most tissues. This type of function is similar to that observed for the Drosophila tissue-specific repressor Yan-Pok, which has homology to ETV6 (33).
This paper shows that the fusion of the first kilobase of ETV6 sequence to CBFA2 results in a fusion protein with a novel function. It has been shown that CBFA2 and C/EBPa are important for the differentiation of specific hematopoietic lineages. We have shown that ETV6/CBFA2 can modify the regulation of the MCSFR promoter by CBFA2B and C/EBPa. Thus, the inappropriate expression of the ETV6/CBFA2 fusion gene after the occurrence of a t(12;21) would also alter the expression pattern in a cell, resulting in a leukemogenic phenotype.
Acknowledgments
We gratefully acknowledge Dr. Eldad Dann’s gift of the ETV6/CBFA2 expressing cell line. We also acknowledge Dr. Stephan Bohlander for the ETV6 cDNA construct, Dr. Hiebert for the ETV6/CBFA2 cDNA construct, and Dr. Friedman for the C/EBPa cDNA. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation (J.D.R.), Department of Energy Grant DE-FG02–86ER60408 (J.D.R.), Charlotte Geyer Foundation (G.N.), National Cancer Institute of Canada (Y.B.-D.), and National Institutes of Health Grants CA 42557 (J.D.R.) and CA 67189 (G.N.). S.F. was supported by National Institutes of Health Graduate Training Grant HD-07009. G.N. is Special Fellow of the Leukemia Society of America. Y.B.D. is a Research Scholar of the National Cancer Institute of Canada.
ABBREVIATIONS
- ETS
E26-transforming specific
- HLH
helix–loop–helix
- CBF
core binding factor
- MCSFR
monocyte colony stimulating factor receptor
- EMSAs
electrophoretic mobility-shift assays
References
- 1.Raimondi S C, Williams D L, Callihan T, Peiper S, Rivera G K, Murphy S B. Blood. 1986;68:69–75. [PubMed] [Google Scholar]
- 2.Cavé H, Gérard B, Martin E, Guidal C, Devaux I, Weissenbach J, Elion J, Vilmer E, Grandchamp B. Blood. 1995;86:3869–3875. [PubMed] [Google Scholar]
- 3.Höglund M, Johansson B, Pedersen-Bjergaard J, Marynen P, Mitelman F. Blood. 1996;87:324–330. [PubMed] [Google Scholar]
- 4.Wlodarska I, Marynen P, La Starza R, Mecucci C, Van den Berghe H. Cytogenet Cell Genet. 1996;72:229–235. doi: 10.1159/000134197. [DOI] [PubMed] [Google Scholar]
- 5.Shurtleff S A, Buijs A, Behm F G, Rubnitz J E, Raimondi S C, Hancock M L, Chan G C-F, Pui C-H, Grosveld G, Downing J R. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 6.Romana S P, Poire H, LeConiat M, Flexor M A, Mauchauffe M, Jonveaux P, Macintyre E A, Bernard O A. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 7.Fears S, Vignon C, Bohlander S K, Smith S, Rowley J D, Nucifora G. Genes Chromosomes Cancer. 1996;17:127–135. doi: 10.1002/(SICI)1098-2264(199610)17:2<127::AID-GCC8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 8.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romana S P, Mauchauffe’M, LeConiat M, Chukamov I, LePaslier D, Berger R, Bernard O A. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 10.Romana S P, Le Contiat M, Poirel H, Marynen P, Bernard O A, Berger R. Leukemia. 1996;10:167–170. [PubMed] [Google Scholar]
- 11.Stegmaier K, Pendse S, Barker G F, Bray-Ward P, Ward D C, Montgomery K T, Krauter K S, Reynolds C, Sklar J, Donnelly M, Bohlander S K, Rowley J D, Sallan S E, Gilliland D G, Golub T R. Blood. 1995;86:38–44. [PubMed] [Google Scholar]
- 12.Raynaud S, Cavé H, Baens M, Bastard C, Cacheux V, Grosgeorge J, Guidal-Giroux C, Guo C, Vilmer E, Marynen P, Grandchamp B. Blood. 1996;87:2891–2899. [PubMed] [Google Scholar]
- 13.Kim D-H, Moldwin R L, Vignon C, Bohlander S K, Suto Y, Giordano L, Gupta R, Fears S, Nucifora G, Rowley J D, Smith S D. Blood. 1996;88:785–794. [PubMed] [Google Scholar]
- 14.Hiebert S, Sun W, Davis N, Golub T, Shurtleff S, Buijs A, Downing J, Grosveld G, Roussel M, Gilliland D G, Lenny N, Meyers S. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson P, Gao J, Chang K-S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J D, Drabkin H. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 16.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daga A, Karlovich C A, Dumstrei K, Banerjee U. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- 18.Zaiman A L, Lewis A F, Crute B E, Speck N A, Lenz J. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levanon D, Negreanu B V, Ghozi M C, Bar-Am I, Aloya R, Goldenberg D, Lotem J, Groner Y. DNA Cell Biol. 1996;15:175–185. doi: 10.1089/dna.1996.15.175. [DOI] [PubMed] [Google Scholar]
- 21.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 24.Nuchprayoon I, Meyers S, Scott L M, Suzow J, Hiebert S, Friedman A D. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wotton D, Ghysdael J, Wang S, Speck N A, Owen M J. Mol Cell Biol. 1994;14:840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D-E, Fujioka K-I, Hetherington C J, Shapiro L, Chen H-M, Look T, Tenen D G. Mol Cell Biol. 1996;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez-Munain C, Krangel M S. Mol Cell Biol. 1995;15:3090–3099. doi: 10.1128/mcb.15.6.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wargnier A, Legros-Maida S, Bosselut R, Bourge J F, Lafaurie C, Ghysdael J, Sasportes M, Paul P. Proc Natl Acad Sci USA. 1995;92:6930–6934. doi: 10.1073/pnas.92.15.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D-E, Hetherington C J, Meyers S, Rhoades K L, Larson C L, Chen H-M, Hiebert S W, Tenen D G. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zent C, Kim N, Hiebert S, Zhang D-E, Tenen D G, Rowley J D, Nucifora G. Top Microbiol Immunol. 1996;211:243–252. doi: 10.1007/978-3-642-85232-9_24. [DOI] [PubMed] [Google Scholar]
- 31.Reddy M A, Yang B-S, Yue X, Barnett C J K, Ross I L, Swee M J, Hume D A, Ostrowski M C. J Exp Med. 1994;180:2309–2319. doi: 10.1084/jem.180.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D-E, Hetherington C J, Chen H-M, Tenen D G. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golub T R, Barker G F, Lovett M, Gilliland D G. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]