Summary
The LDL receptor-related protein 1B (LRP1B) is a putative tumor suppressor homologous to LRP1. Both LRP1 and LRP1B contain cytoplasmic tails with several potential endocytosis motifs. Although the positions of these endocytic motifs are similar in both receptors, LRP1B is internalized at a 15-fold slower rate than LRP1. To determine whether the slow endocytosis of LRP1B is due to the utilization of an endocytosis motif other than the YATL motif used by LRP1, we tested minireceptors with mutations in each of the five potential motifs in the LRP1B tail. Only mutation of both NPXY motifs together abolished LRP1B endocytosis, suggesting LRP1B can use either of these motifs for internalization. LRP1B contains a unique insertion of 33 amino acids not present in LRP1 that could lead to altered recognition of trafficking motifs. Surprisingly, deletion of this insertion had no effect on the endocytosis rate of LRP1B. However, replacing either half of the LRP1B tail with the corresponding LRP1 sequence markedly accelerated LRP1B endocytosis. From these data, we propose that both halves of the LRP1B cytoplasmic tail contribute to a unique global conformation, which results in less efficient recognition by endocytic adaptors and a slow endocytosis rate.
Keywords: LRP1B, LRP1, endocytosis, tumor suppressor
Introduction
The LDL receptor-related protein 1 (LRP1)1 is an endocytic receptor of the low density lipoprotein receptor (LDLR) family. LRP1 is a 600 kDa, type I, single-pass transmembrane protein that is cleaved by the endopeptidase furin in the trans-Golgi network to generate a non-covalently associated heterodimer of 515 and 85 kDa [1, 2]. The 515 kDa subunit consists of four extracellular cysteine-rich ligand binding domains that are interspersed with EGF and YWTD repeats. The 85 kDa subunit contains the transmembrane domain and 100 amino acid-cytoplasmic tail. Through its ligand binding domains, LRP1 binds and facilitates the internalization of numerous structurally and functionally distinct ligands including proteases, protease inhibitors, growth factors, lipoproteins, and viral and bacterial proteins [3]. LRP1 undergoes constitutive and rapid endocytosis with a t1/2 of ~0.5 minutes, consistent with its predominant distribution in clathrin coated structures and endosomes [4]. Following endocytosis, LRP1 is recycled back to the cell surface while internalized ligands are primarily trafficked to the lysosome for degradation [4]. By modulating the metabolism of its ligands, LRP1 plays roles in coagulation, lipid homeostasis, wound healing, and long-term potentiation of memory, as well as in the pathogenesis of Alzheimer’s disease [3]. The cytoplasmic tail of LRP1 contains several potential endocytosis motifs, including two NPXY motifs (X=any amino acid), one YXXØ motif (Ø= a bulky hydrophobic residue), and two dileucine motifs. The dominant endocytosis motif within the cytoplasmic tail of LRP1 is the tyrosine-based YATL sequence located 63–66 amino acids from the transmembrane domain, although the distal dileucine motif also contributes to rapid endocytosis [5]. Both YXXØ and dileucine motifs are known to interact with the AP-2 adaptor complex, which links receptor tails to the structural coat protein clathrin. Nevertheless, it has recently been shown that multiple and redundant factors are required for the endocytosis of LRP1, suggesting that LRP1 endocytosis is somewhat unique [6].
LRP1B, a close homologue of LRP1, was initially discovered as a putative tumor suppressor in non-small cell lung carcinoma, and inactivating mutations have since been identified in multiple types of cancer [7–11]. LRP1 and LRP1B have very similar domain structures and are 59 percent identical at the amino acid level. The two features that distinguish LRP1B from LRP1 are the presence of an extra ligand binding repeat in the fourth ligand binding domain and a unique insertion of 33 amino acids within the cytoplasmic tail. Although the biological functions of LRP1B have not yet been defined, LRP1B has been shown to bind several of LRP1’s ligands, and therefore may also play roles in a wide variety of cellular processes [12]. Despite the many similarities between LRP1 and LRP1B, the endocytosis rate of an LRP1B minireceptor is much slower (t1/2 ~ 8 min) than that of a similar LRP1 minireceptor (t1/2 ~ 0.5 min) [12]. This difference in endocytic rate could have important implications for the biological and tumor suppressor functions of LRP1B, as LRP1 and LRP1B may compete for binding to ligands while exerting opposite effects on their clearance from the cell surface [13]. Since the cytoplasmic tail of LRP1B contains the same potential endocytosis motifs as LRP1, we sought to uncover the mechanism underlying the distinct rates of endocytosis between LRP1 and LRP1B. We hypothesized that sequences within the LRP1B tail inhibit interactions between the endocytosis motifs in the tail and the endocytic machinery. We were specifically interested in the 33 amino acid insertion, which resides between potential endocytosis motifs and may have cytoplasmic binding partners that would sterically prevent endocytic adaptors from binding the adjacent motifs. In this study, we constructed a series of chimeric and mutant minireceptors in order to determine which sequences are critical for the slower endocytic rate of LRP1B.
Materials and Methods
Materials
Human recombinant RAP was expressed as a glutathione S-transferase fusion protein and isolated as described previously [14]. Mouse anti-HA antibody was from Covance (HA.11). Carrier-free Na125I was purchased from PerkinElmer Life Sciences. Proteins were iodinated using the IODO-GEN method as described previously [5]. Mirus Trans-IT CHO transfection kit was from Mirus. Restriction enzymes were from New England Biolabs. QuikChange mutagenesis kit and Pfu ultra DNA polymerase were from Stratagene. All oligonucleotides were synthesized by IDT.
Cell Culture and Transfection
The LRP1-null Chinese hamster ovary (CHO) cell line (kindly provided by David FitzGerald, National Institutes of Health; see [15]) was cultured in Ham’s F-12 medium as described [15]. Transient and stable transfection into LRP1-null CHO cells was achieved by transfection of 2 μg of plasmid DNA in 6-well plates using the Mirus Trans-IT CHO kit according to the manufacture’s instructions. Endocytosis assays following transient transfection were performed 24 hours after transfection and were performed at least twice. Stable transfectants were selected using 700 μg/ml G418 and maintained with 350 μg/ml G418. Clonal purity was assayed by Western blotting and flow cytometry using anti-HA IgG. At least two independently derived clones were used for each assay.
Construction of LRP1B minireceptors
The domain IV-containing LRP1B minireceptor (mLRP1B4) and the domain IV-containing LRP1 minireceptor (mLRP4) have been described previously [5, 12]. All constructs were sequenced prior to their transfection into CHO cells. mLRP1B4Δins was constructed according to standard cloning methods. mLRP4-LRP1Btail, mLRP1B-LRP1tail, mLRP4+1Bins, mLRP1B-LRP1COOH tail, mLRP1B-LRP1NH2 tail, mLRP4-1B(1–29LRP1)tail, mLRP4-1B(30–54LRP1)tail, mLRP4-1B(55–76LRP1)tail, and mLRP4-1B(77–100LRP1)tail were all generated using the enzymeless cloning method and the Quik Change site-directed mutagenesis kit as described [16]. The Quik Change kit was also used to generate the following point mutants: mLRP1B4 C105S, mLRP4-1B Y27A, mLRP4-1B L42AL43A, mLRP4-1B N92A, mLRP4-1B L98A, mLRP4-1B L118AL119A, mLRP4-1B double LL-AA, and mLRP4-1BY27A,N92A.
Kinetic Analysis of Endocytosis
Kinetic analysis of endocytosis was performed according to previously published methods [17, 18]. Transiently or stably transfected CHO cells were plated in 12-well plates at a density of 2 × 105 cells/well and used after overnight culture. Cells were rinsed twice in cold ligand binding buffer (DMEM containing 0.6% BSA), and thereafter incubated with 125I-RAP (5 nM) or 125I-anti HA IgG (1 nM) in cold ligand binding buffer (0.5 ml/well) at 4°C for 40 min with gentle rocking. Unbound ligand was removed by washing cell monolayers three times with cold ligand binding buffer. Cold strip solution (PBS, pH 2.0) was added to one set of plates without warming up. The remaining plates were then placed in a 37°C water bath, and 0.6 ml of ligand binding buffer that had been prewarmed to 37°C was quickly added to the monolayers to initiate internalization. After each time point, the plates were quickly placed on ice and the ligand binding buffer was replaced with cold strip solution. Cell surface ligand was stripped by incubation of cell monolayers with cold strip solution for a total of 20 min (0.5 ml for 10 min, twice) and counted. Cell monolayers were then solubilized with low SDS lysis buffer (62.5 mM Tris-HCl, pH 6.8, 0.2% SDS, 10% v/v glycerol) and counted. The sum of ligand that was internalized plus that which remained on the cell surface after each assay was used as the maximum potential internalization. The fraction of internalized ligand after each time point was calculated and plotted.
Immunoelectron Microscopy
CHO-mLRP4 and CHO-mLRP1B4 cells were fixed in a mixture of 2% paraformaldehyde and 0.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for 1 h and then stored in 1% paraformaldehyde in 0.1 M phosphate buffer until use. Cryosections of 50–100 nm were picked up from the diamond knife in a sucrose/methylcellulose mixture and sequentially incubated with mouse monoclonal anti HA antibody and 10-nm protein A-conjugated gold particles [19].
Results
LRP1B undergoes slow endocytosis and localizes to the cell surface
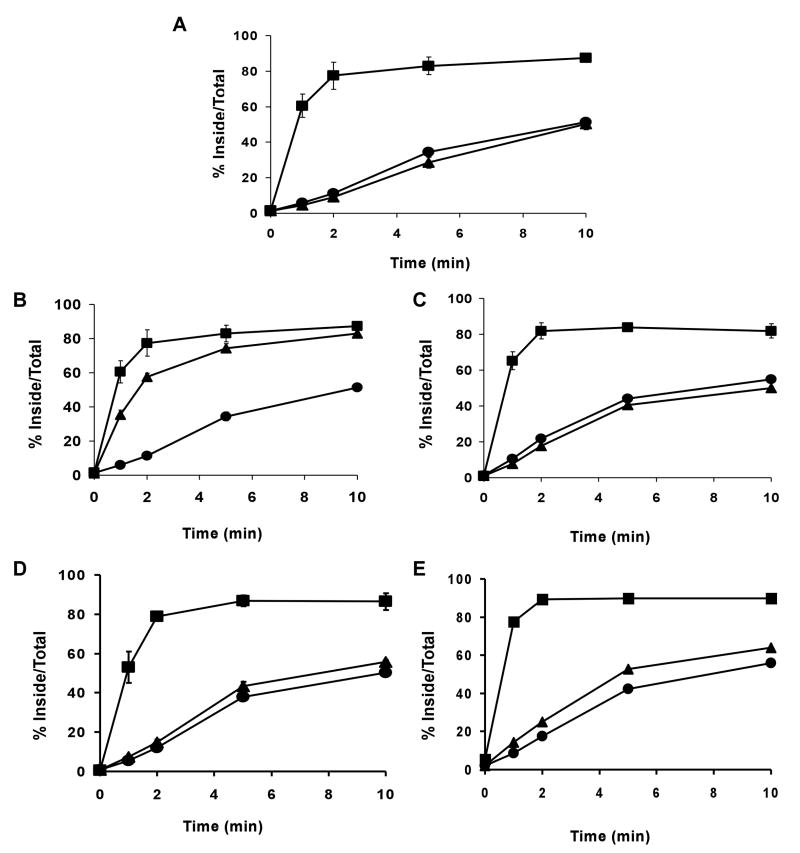
The cytoplasmic tail of LRP1 contains 5 potential endocytosis motifs: two NPXY, one YXXØ, and two dileucine motifs, and the rapid endocytosis of LRP1 is primarily dependent on the YXXØ and the dileucine motif distal to the plasma membrane [5]. Although all of these motifs are present in the LRP1B tail within the same local context as the LRP1 tail, the endocytosis rate of LRP1B is more than 15-fold slower than that of LRP1 (Figure 2A) [12]. In order to study the functional differences between LRP1 and LRP1B, we previously constructed minireceptors consisting of an amino-terminal HA epitope, the fourth ligand binding domain, and the entire small subunit of either LRP1 (mLRP4) or LRP1B (mLRP1B4), and have stably transfected them into CHO cells that lack endogenous LRP1 (see Figure 1 for construct list) [5, 12]. Members of the LDL receptor family have been described as constitutively recycling receptors that undergo endocytosis in the presence or absence of ligand [20]. To confirm that the endocytosis rates of mLRP4 and mLRP1B4 are independent of ligand binding we compared the endocytosis rate measured using anti-HA IgG and ligand (recombinant RAP) (Figure 2A) [14]. As expected, the endocytosis rates of mLRP4 and mLRP1B4 are not affected by the presence of ligand. Importantly, although the endocytosis rate of mLRP1B4 is markedly slower than that of mLRP4, it is faster than that of mLRP4 tailless, an LRP1 minireceptor lacking a cytoplasmic tail. In order to determine the steady-state localization of mLRP1B4, we performed immuno-EM localization studies of the two minireceptors. As shown in Figure 2B, both minireceptors were found in clathrin-coated structures, consistent with the idea that both receptors undergo clathrin-mediated endocytosis. Consistent with our endocytosis findings, mLRP4 was more heavily distributed in endosomal compartments while mLRP1B4 was primarily localized to the cell surface. Taken together, these data demonstrate that mLRP1B4 internalizes more slowly than mLRP4, resulting in a higher proportion of cell surface relative to endosomal receptors.
Figure 2. mLRP1B4 internalizes via clathrin-mediated endocytosis.
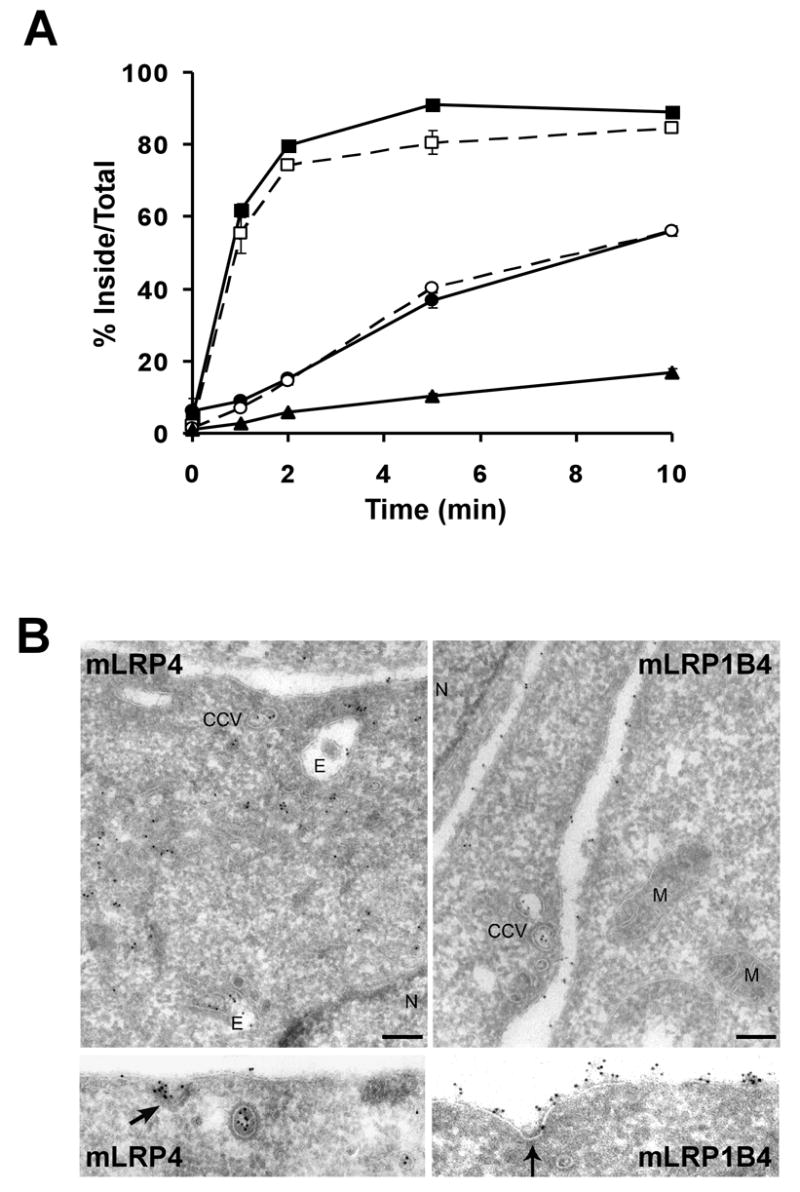
A) CHO LRP1 null cells stably transfected with mLRP4 (squares) mLRP1B4 (circles), or mLRP4 tailless (triangles) were incubated with 5 nM 125I-labled RAP (closed symbols) or 1 nM 125I-labeled anti-HA antibody (open symbols) for 40 minutes at 4°C. The cells were shifted to 37°C for different amounts of time to initiate receptor endocytosis and then immediately placed on ice. The percent of ligand internalized at each time point is equal to the amount of ligand internalized divided by the total cell-associated ligand (the sum of internalized and cell surface ligand). Values are the average of triplicate wells with the S.D. indicated by error bars. This experiment is representative of two such experiments performed with similar results. In this and all subsequent endocytosis assays, a minimum of two independently derived stable clones were analyzed for each construct. B) Cryosections of CHO LRP1 null cells stably transfected with mLRP4 or mLRP1B4 were immunogold labeled with anti-HA antibodies and 10 nm gold particles. mLRP4 localizes primarily to clathrin coated structures and to endosomes, whereas mLRP1B4 is distributed mainly on the cell-surface. Bar = 200 nm. CCV=clathrin coated vesicle, N=nucleus, E=endosome, M=mitochondria
Figure 1. Endocytosis rate of mLRP1B4.
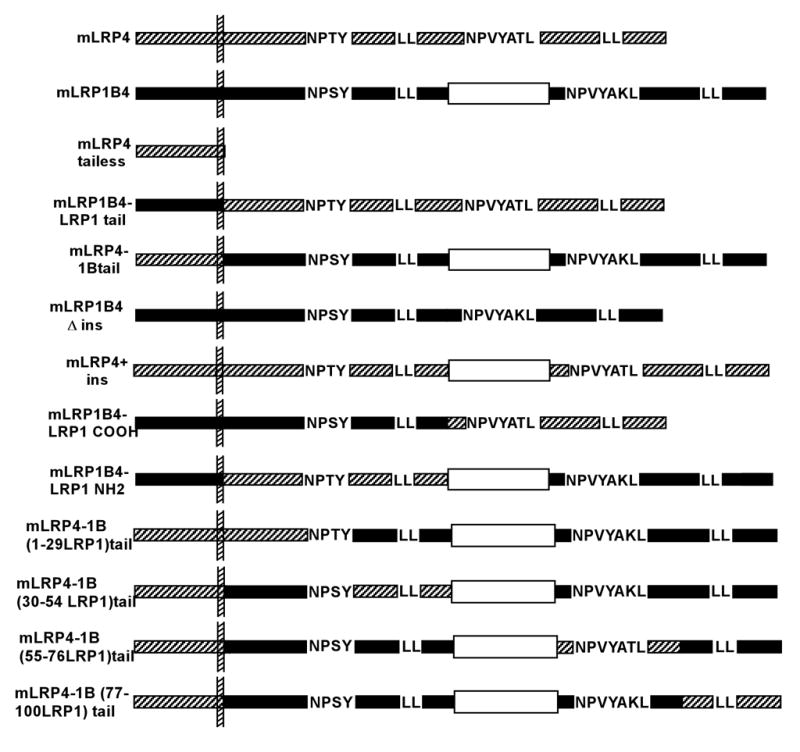
Schematic of LRP1 and LRP1B minireceptor constructs. The 33 amino acid insert is represented by an open rectangle. LRP1 sequences are represented by hatched bars, while LRP1B sequences are shown as dark bars.
Determinants for the differential endocytosis rates of LRP1 and LRP1B reside exclusively within the cytoplasmic tail regions
In order to determine whether the extracellular and/or transmembrane domain of LRP1B contributes to its slow rate of endocytosis, we constructed two chimeric minireceptors. mLRP4-LRP1B tail consisted of the extracellular and transmembrane domain of mLRP4 followed by the cytoplasmic tail of LRP1B (see Figure 1). Conversely, mLRP1B-LRP1 tail consisted of the extracellular and transmembrane domain of LRP1B fused to the cytoplasmic tail of LRP1. As shown in Figure 3, the endocytosis rate of these chimeric minireceptors was dictated by the identity of the cytoplasmic tail; i.e. mLRP4-LRP1B tail had the same endocytosis rate as mLRP1B4 and mLRP1B4-LRP1 tail displayed the same endocytosis rate as mLRP4. These data clearly demonstrate that the endocytosis rates of both LRP1 and LRP1B are dictated exclusively by information within their respective cytoplasmic domains.
Figure 3. The information for endocytic rate resides exclusively within the cytoplasmic tails of LRP1 and LRP1B.
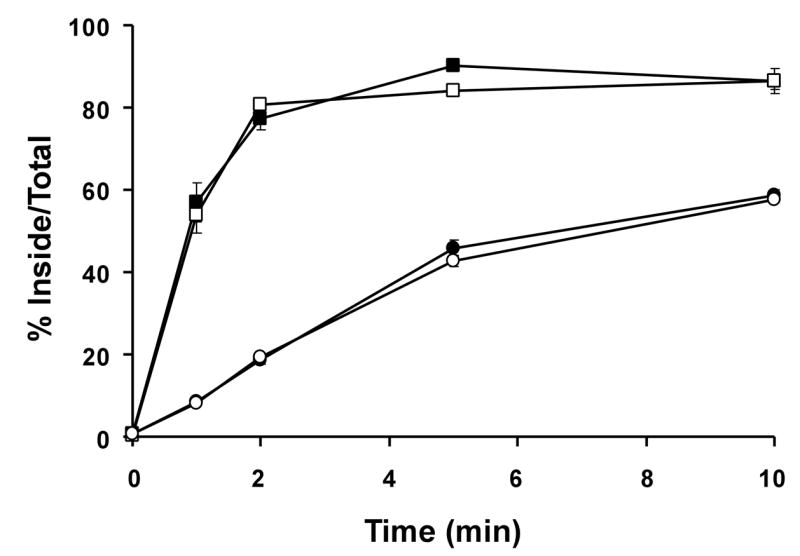
CHO LRP1 null cells stably expressing mLRP4 (■), mLRP1B4 (●), mLRP4-1Btail (○), or mLRP1B4-LRPtail (□) were subjected to a kinetic analysis of endocytosis using 1 nM 125I-labeled anti-HA antibody as described in Figure 2 and experimental procedures. Values are the average of triplicate wells with the S.D. indicated by error bars. This experiment is representative of two such experiments performed with similar results.
Endocytosis of mLRP1B4 requires one of two NPXY motifs
Because the rate of LRP1B endocytosis is similar to the rates of ApoER2 and VLDLR [21], which use the NPXY-binding clathrin adaptors ARH and Dab2 [22], we sought to determine whether one of the two NPXY motifs within the LRP1B cytoplasmic tail might be the dominant endocytosis motif. In contrast, the rapid internalization of LRP1 is dependent on the AP-2-binding YXXØ and distal acidic-cluster dileucine motifs. To address which of the five potential endocytosis motifs in the LRP1B tail is essential for internalization, we generated point mutations in each within the context of the mLRP4-1Btail construct (displays identical endocytosis rate to mLRP1B4, see below and Figure 3). The resulting constructs, mLRP4-1B Y27A, mLRP4-1B L42AL43A, mLRP4-1B N92A, mLRP4-1B L98A, and mLRP4-1B L118AL119A (see Figure 4A), were transiently transfected into CHO-LRP1 null cells, and the endocytosis rate conferred by each was measured. In sharp contrast to a similar analysis of the LRP1 tail [5], no mutation in any of the five motifs alone resulted in a decreased endocytosis rate (Figure 4B and 4C). We thought it possible that the NPXY or the dileucine motifs might function in a redundant manner, such that mutation of both motifs together would be required for defective endocytosis. To test this, we generated a chimeric minireceptor harboring mutations in both NPXY motifs (mLRP4-1B Y27, N92A), as well as one with alanine substitutions at both dileucine motifs (mLRP4-1B double LL-AA). Transient transfection of these constructs followed by endocytosis assay demonstrated that mutation of both NPXY motifs causes the minireceptor to be internalized with a rate consistent with mLRP4 tailless, which lacks a cytoplasmic tail (Figure 4D). In contrast, mutation of both dileucine motifs did not alter the endocytosis rate of mLRP1B (Figure 4D). These data suggest that the NPXY motifs in the LRP1B tail are able to functionally compensate for one another. Therefore, it is likely that LRP1 and LRP1B utilize different adaptor proteins for clathrin-mediated endocytosis.
Figure 4. LRP1B uses redundant NPXY motifs for internalization.
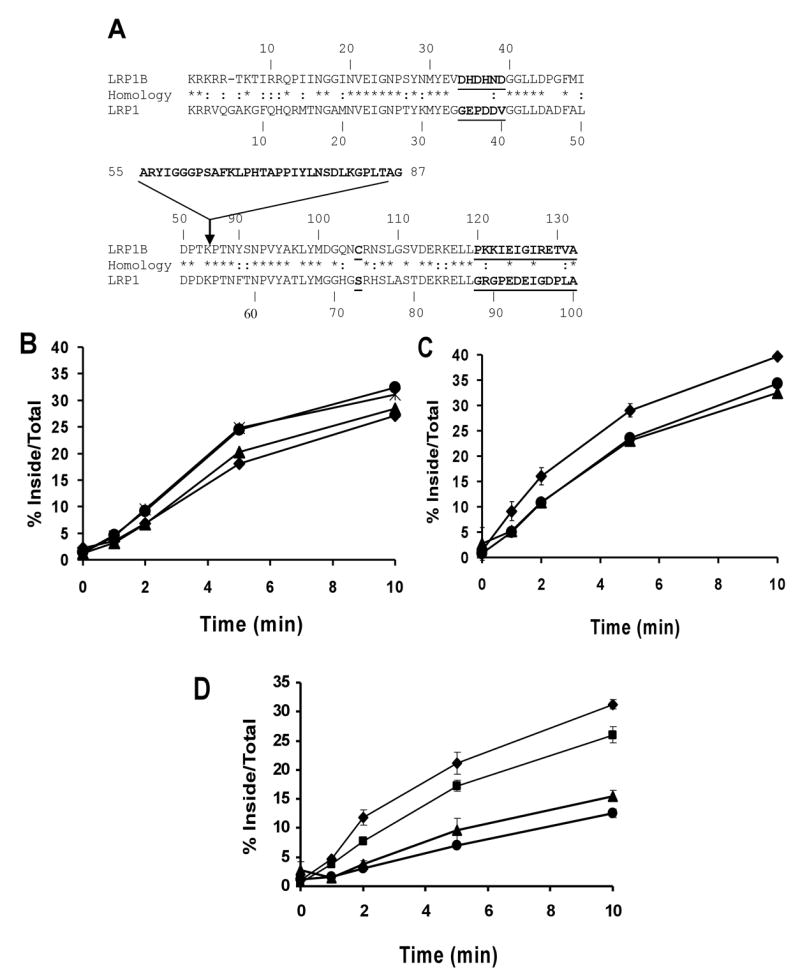
A) Comparison between the cytoplasmic tails of LRP1 and LRP1B. The identical (*) and similar (:) amino acids are marked. The inserted 33-residue sequence in LRP1B tail and the two divergent regions between LRP1 and LRP1B (last 13 amino acids and amino acids 34–39) are in bold type. CHO LRP1 null cells were transiently transfected with B) mLRP4-1Btail (●), mLRP4-1B Y27A (▲), mLRP4-1B N92A (◆), or (*) mLRP4-1BL98A; C) mLRP4-1Btail (●), mLRP4-1B L42A,L43A (▲), or mLRP4-1B L118A,L119A (◆); D) mLRP4-1Btail (■), mLRP4 tailess (●), mLRP4-1B Y27A, N92A (▲), or mLRP4-1B double dileucine mutant (◆), seeded into triplicate wells of twelve well plates, and assayed in a kinetic analysis of endocytosis using 1 nM 125I-labeled anti-HA antibody as described in Figure 2 and experimental procedures. Values are the average of triplicate wells with the S.D. indicated by error bars. This experiment is representative of two such experiments performed with similar results.
The 33 amino acid tail insertion of LRP1B does not inhibit efficient endocytosis
The cytoplasmic tail of LRP1B contains a unique stretch of 33 amino acids that is situated between potential endocytosis motifs (Figure 4A). We sought to determine whether the presence of this insert is the major inhibitory factor preventing rapid endocytosis of LRP1B. To this end, we constructed two new minireceptors: mLRP1B4 lacking the 33 amino acid insert (mLRP1BΔins) and mLRP4 that contains the 1B insertion in the corresponding position of the LRP1 tail (mLRP4+ins) (see Figure 1). These constructs were stably transfected into CHO cells and tested in a kinetic analysis of endocytosis. Most surprisingly, mLRP1BΔins had the same rate of endocytosis as mLRP1B4 (Figure 5A). Consistent with this result, the introduction of the LRP1B insert into the LRP1 tail resulted in a marginal reduction of the endocytosis rate of mLRP4 (Figure 5B). Although mLRP4+ins had a t1/2 of about 1.5 minutes, 3-fold slower than that of mLRP4, it still internalizes five times faster than mLRP1B4. The observed decrease in endocytosis rate could be due to the increased distance of the primary endocytosis motifs of LRP1 from the plasma membrane within this construct, rather than the specific 33 amino acid sequence. Together, these results demonstrate that the LRP1B 33 amino acid insert is neither necessary nor sufficient for the slow endocytosis rate of LRP1B.
Figure 5. The 33 amino acid insert of LRP1B is not responsible for its slow endocytosis rate.
CHO LRP1 null cells stably expressing mLRP4 (■) and mLRP1B4 (●), along with A) mLRP1B4 ins (▲), B) mLRP4+ins (▲), C) mLRP1B4-LRP13CT (▲) D) mLRP1B4-LRP34–39 (▲) or E) mLRP1B4 C105S (▲), were subjected to a kinetic analysis of endocytosis using 1 nM 125I-labeled anti-HA antibody as described in Figure 2 and experimental procedures. Values are the average of triplicate wells with the S.D. indicated by error bars. These experiments are representative of two such experiments performed with similar results.
Two divergent regions of the LRP1B tail do not inhibit efficient endocytosis
In order to identify amino acid sequences within the amino- and carboxyl-terminal halves of the LRP1B tail responsible for the slow endocytosis rate, we aligned the tail sequences of LRP1 and LRP1B minus the 33 amino acid insertion (Figure 4A). Of the 98 amino acids that can be aligned, 50% are identical, ~20% are similar, and ~28% are different between the two tails. We selected two clusters of highly divergent amino acids, one from position 34 to 39 of the LRP1B tail (DHDHND) and one consisting of the last 13 amino acids of the LRP1B tail (PKKIEIGIRETVA), and mutated them to the corresponding LRP1 sequence (GEPDDV and GRGPEDEIGDPLA, respectively). These constructs, termed mLRP1B4-LRP1 34–39 and mLRP1B4-LRP1 13CT, respectively, were stably transfected into CHO-LRP1 null cells and assayed for endocytosis rate. As shown in Figure 5C and 5D, the replacement of either of these clusters of amino acids with the corresponding LRP1 sequence did not increase the endocytosis rate of mLRP1B4.
Elimination of a potential cysteine palmitoylation site in the LRP1B tail does not alter endocytosis
It has been shown that mutation of two palmitoylated cysteines within the transferrin receptor cytoplasmic tail significantly enhances transferrin internalization [23]. We observed that serine 73 of the LRP1 cytoplasmic tail corresponds to a cysteine residue (C105) in the LRP1B tail. In order to determine whether palmitoylation of this cysteine in the LRP1B tail might hinder its efficient internalization, we generated a point mutant minireceptor, mLRP1B4 C105S. CHO-LRP1 null cells stably expressing this minireceptor did not display a significantly altered rate of endocytosis compared to mLRP1B4-expressing cells (Figure 5E). Although we did not study whether this residue can, in fact, be palmitoylated, these data demonstrate that the C105S mutation does not affect mLRP1B4 endocytosis.
Both halves of the LRP1B tail contain inhibitory elements for endocytosis
Having examined the most obvious difference between the sequences of the LRP1 and LRP1B tails, we next addressed whether the inhibitory elements within the LRP1B tail were present in the amino-terminal or carboxyl-terminal half of the LRP1B tail by generating two additional chimeric minireceptors. mLRP1B4-LRP1 NH2 tail substituted the first half of the LRP1 tail into mLRP1B4, and mLRP1B4-LRP1 COOH tail consisted of mLRP1B4 with the carboxyl-terminal half of the LRP1 tail (cut point at the position of the 33 amino acid insert, see Figure 4A). These two constructs were stably transfected into CHO-LRP1 null cells and were tested in a kinetic analysis of endocytosis. As shown in Figure 6, the presence of either half of the LRP1 tail significantly accelerated the endocytosis rate of mLRP1B4, although mLRP1B4-LRP1 COOH tail internalized faster (t1/2 ~ 1 min) than mLRP1B4-LRP1 NH2 tail (t1/2 ~ 3 min). One possible interpretation of these data is that the endocytosis rate of the chimeric minireceptors is dictated by the endocytosis motifs within the LRP1 sequences (i.e., the proximal NPXY motif in mLRP1B4-LRP1 NH2 tail and the YATL motif in mLRP1B4-LRP1 COOH tail). However, when the endocytosis rate of mLRP1B4-LRP1NH2 tail was directly compared to that of mLRP4T59 (mLRP4 truncated after amino acid 59, just before the distal dileucine motif), we found that the former was significantly faster, suggesting that the LRP1B sequence at least contributes to, if not entirely determines, the endocytosis rate of this receptor (data not shown). These data suggest that the LRP1B tail harbors potentially strong endocytosis motifs, but that the two halves of the tail together mask this signal in vivo.
Figure 6. Both halves of the LRP1B tail contain inhibitory signals for endocytosis.
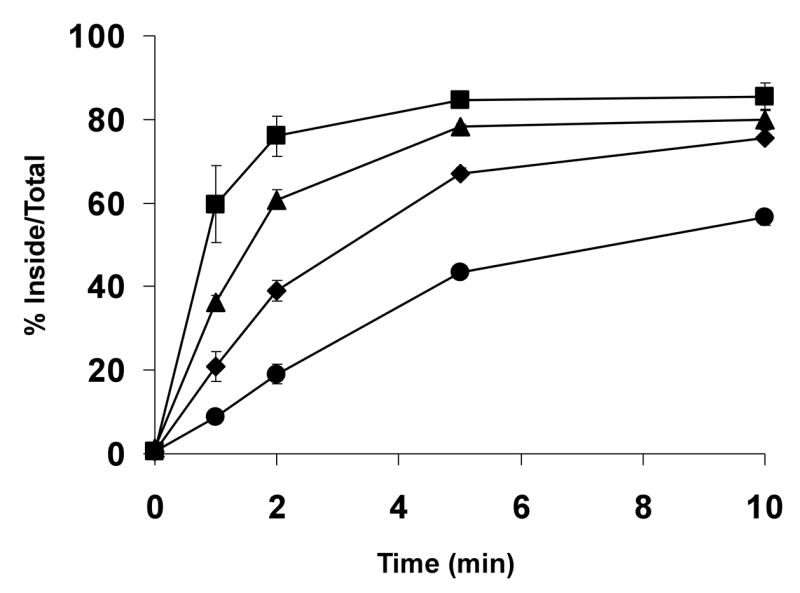
CHO LRP1 null cells stably expressing mLRP4 (■), mLRP1B4 (●), mLRP1B4-LRP1 NH2 tail (◆), or mLRP1B4-LRP1 COOH tail (▲) were subjected to a kinetic analysis of endocytosis using 1 nM 125I-labeled anti-HA antibody as described in Figure 2 and experimental procedures. Values are the average of triplicate wells with the S.D. indicated by error bars. This experiment is representative of two such experiments performed with similar results.
In order to further investigate which specific sequences serve to inhibit rapid endocytosis of LRP1B, we constructed four additional chimeric minireceptor constructs, each containing one quarter of the LRP1 intracellular domain in place of the corresponding LRP1B sequence, mLRP4-1B(1–29LRP1)tail, mLRP4-1B(30–54LRP1)tail, mLRP4-1B(55–76LRP1)tail, and mLRP4-1B(77–100LRP1)tail, see Figure 1. Each of these constructs was transiently transfected into CHO-LRP1 null cells, and a kinetic analysis of endocytosis using 125I-αHA was performed 24 hours later. As shown in Figure 7A and 7B, the replacement of each quarter of the LRP1B tail alone did not accelerate the endocytosis rate of LRP1B. This suggests that the LRP1B tail adopts a complex structure that requires disruption at multiple sites in order for the endocytosis motifs in the LRP1B tail to be made accessible for adaptor protein binding.
Figure 7. None of the four quarters of the LRP1B tail contain inhibitory signals for endocytosis.
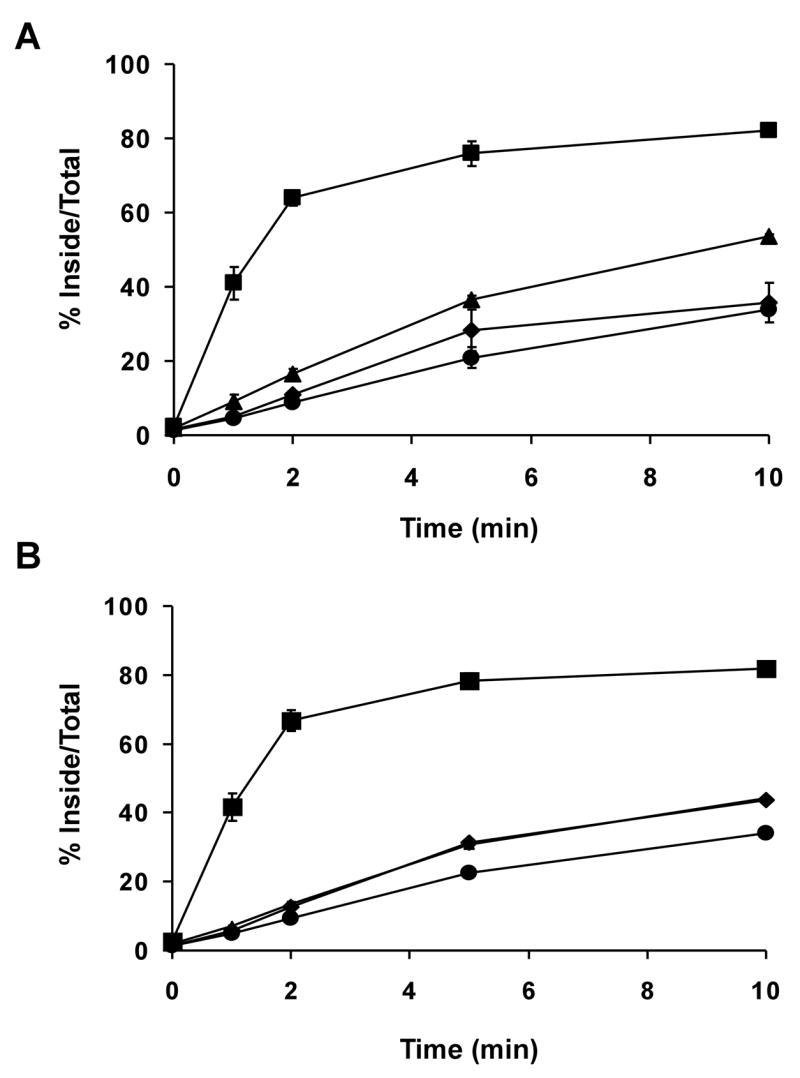
CHO LRP1 null cells were transiently transfected with A) mLRP4 (■), mLRP4-1B tail (●), mLRP4-1B(1–29LRP1)tail (▲), or mLRP4-1B(30–54LRP1)tail (◆); B) mLRP4 (■), mLRP4-1B tail (●), mLRP4-1B(55–76LRP1)tail (▲), or mLRP4-1B(77–100LRP1)tail (◆), seeded into twelve-well plates, and assayed for endocytosis rate using 1 nM 125I-labeled anti-HA antibody as described in Figure 2 and experimental procedures. Values are the average of triplicate wells with the S.D. indicated by error bars. This experiment is representative of two such experiments performed with similar results.
Discussion
The cytoplasmic domains of LRP1 and LRP1B are highly homologous, yet the endocytosis rate of LRP1B is markedly slower. The present study aimed to identify sequences within the LRP1B cytoplasmic tail that are responsible for this difference in endocytosis rate. Our data suggest that one factor influencing this difference could be the utilization of different clathrin adaptors for internalization. Whereas LRP1 uses the YXXØ and distal LL motifs for rapid endocytosis, LRP1B can use either of its NPXY motifs. Typically, YXXØ motifs in receptor tails interact with the μ2 subunit of the AP-2 clathrin adaptor complex, which serves to link the clathrin structural scaffold to the receptor cargo within budding vesicles[24, 25]. Although LRP1 had been assumed to internalize in an AP-2 dependent manner via its YATL motif, a recent report suggests that LRP1 endocytosis requires several additional cytosolic factors, including Hsc70, CRMP-2 and synaptojanin 1 [6]. Therefore, dissection of the adaptor protein binding spectrum of LRP1 via traditional GST-pulldown assays is not straightforward (data not shown). Given that LRP1B uses either NPXY motif for internalization, it seems likely that several regions of the LRP1B cytoplasmic tail interact with one another or with other soluble or cyotskeletal proteins, thereby hindering AP-2 from efficiently binding to the YXXØ motif. Such competition has been shown to occur between AP-2 and the 47-kDa tail-interacting protein (TIP47) for access to the cytoplasmic tail of the cation-independent mannose-6-phosphate receptor (CI-MPR)[26]. NPXY motifs are recognized by phosphotyrosine binding (PTB) domain (PTB)-containing proteins, such as Dab2 and ARH. These proteins have been shown to mediate the internalization of the LDL receptor[27, 28], megalin [29, 30], and ApoER2 [22]. Therefore, the finding that LRP1B uses NPXY motifs for internalization is only surprising in light of the similarity between the LRP1 and LRP1B cytoplasmic tails.
It had been previously assumed that the 33 amino acid insert within the LRP1B tail was responsible for the slow endocytosis rate of LRP1B [13]. We hypothesized that removal of this major structural element from the LRP1B tail would alter adaptor protein binding, allowing it to internalize as rapidly as LRP1. Surprisingly, we found that removal of the insert sequence from the LRP1B tail did not result in an enhanced endocytic rate. We also examined two additional regions of LRP1B that are highly divergent from the corresponding LRP1 sequences: the last 13 amino acids of the LRP1B tail, and amino acids 34–39 of the LRP1B tail, and found that neither of these regions alone is responsible for the slow endocytosis rate of LRP1B. However, we have shown that each half of the LRP1B tail contains elements that together contribute to slow endocytosis, which raises the possibility of the inhibitory sequences within each half interacting with each other. In addition, the fact that these sequences could not be pinpointed in the quarter-tail chimeric minireceptor constructs suggests that these elements may be three-dimensional sequences requiring non-adjacent sections of each half of the tail for proper folding, rather than linear sequences. Indeed, a study of the sorting signals within the tail of the lysosomal protein tyrosinase provides evidence that such structures do exist in other receptor tails [31].
Using LRP1-LRP1B chimeras has enabled us to gain valuable insight into the determinants of the slow endocytosis rate of LRP1B. However, our results with the quarter tail replacement chimeric receptors strongly suggests that individual amino acid residues required for the unique conformation of LRP1B will be difficult, if not impossible, to identify. Rather, structural analyses, such as nuclear magnetic resonance imaging or crystallization studies could provide a more in-depth understanding of the determinants of the different structures of the LRP1 and LRP1B tails.
Members of the LDLR family are classically thought of as endocytic receptors. LRP1B, however, is unique both in its slow rate of endocytosis and its genetic linkage to cancer. Several recent genetic studies of human non-small cell lung carcinoma, urothelial cancer, esophageal squamous cell carcinoma, and endocervical adenocarcinoma have revealed that the LRP1B gene is frequently inactivated in these types of cancer, suggesting that LRP1B may have inhibitory effects on some stage(s) of tumorigenesis [7–10]. LRP1 and LRP1B are highly homologous and presumably have identical ligand-binding spectra, yet only LRP1B has characteristics of a tumor suppressor. The obvious functional difference in the endocytosis rates of LRP1 and LRP1B may represent a critical link to the tumor suppressor function of LRP1B. Among the ligands known to bind LRP1 are matrix metalloproteinases (MMPs), thrombospondins, and trimeric complexes of urokinase plasminogen activator (uPA), plasminogen activator inhibitor type 1 (PAI-1), and uPA receptor, in addition to growth factors such as platelet-derived growth factor (PDGF), insulin-like growth factor binding protein-3 (IGFBP3), connective tissue growth factor (CTGF), transforming growth factor β (TGFβ), and midkine, all of which play important roles in tumor biology [32–39]. One potential mechanism by which LRP1B expression could prevent tumors is to alter the levels or activity of such ligands at the cell surface, thereby impacting the behavior of tumor cells. We have previously shown that the expression of an LRP1B minireceptor in CHO cells results in the retention of inactive uPA at the cell surface and a corresponding decrease in migration rate [13]. These results are consistent with a role for LRP1B in inhibiting the migration necessary for cancer cells to metastasize to distant sites. Recent data have also suggested an inhibitory role for LRP1B in the initial stages of tumorigenesis, as expression of an LRP1B minireceptor inhibits focus formation in an esophageal squamous cell carcinoma cell line lacking endogenous LRP1B [10]. Furthermore, LRP1B has recently been shown to undergo processing by γ-secretase, followed by translocation of the LRP1B cytoplasmic tail to the nucleus, suggesting that the 33 amino acid insert might play a role in transcriptional regulation [40].
In summary, our data suggest that the 33 amino acid insert of LRP1B is not required for the slow endocytic rate of LRP1B. Rather, both the amino- and carboxyl- terminal regions of the receptor’s cytoplasmic tail contribute sequences that exclude LRP1B from the rapid endocytic route utilized by LRP1. However, the precise mechanism underlying this inhibition and the implications for the tumor suppressor function of LRP1B remain to be elucidated.
Acknowledgments
We thank Dr. Michelle Schlief and Dr. Stuart Kornfeld for their critical reading of this manuscript. We are also grateful to Dr. Maria Isabel Yuseff and members of the Bu lab for helpful discussion of this work. This work was supported by National Institutes of Health Grant R01 CA100520 and by a grant from the Alzheimer’s Association (to G.B.). G.B. is an Established Investigator of the American Heart association. Y.L. is partially supported by a grant from the American Heart Association (0330118H). J.M.K. is partially supported by a fellowship from the Washington University Siteman Cancer Center Cancer Biology Pathway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herz J, Kowal RC, Goldstein JL, Brown MS. Proteolytic processing of the 600 kd low density lipoprotein receptor-related protein (LRP) occurs in a trans-Golgi compartment. EMBO J. 1990;9:1769–76. doi: 10.1002/j.1460-2075.1990.tb08301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willnow TE, Moehring JM, Inocencio NM, Moehring TJ, Herz J. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem J. 1996;313(Pt 1):71–6. doi: 10.1042/bj3130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 2002;71:405–34. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- 4.Bu G, Maksymovitch EA, Geuze H, Schwartz AL. Subcellular localization and endocytic function of low density lipoprotein receptor-related protein in human glioblastoma cells. J Biol Chem. 1994;269:29874–82. [PubMed] [Google Scholar]
- 5.Li Y, Marzolo MP, van Kerkhof P, Strous GJ, Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. J Biol Chem. 2000;275:17187–94. doi: 10.1074/jbc.M000490200. [DOI] [PubMed] [Google Scholar]
- 6.Miwako I, Schmid SL. A cell-free biochemical complementation assay reveals complex and redundant cytosolic requirements for LRP endocytosis. Exp Cell Res. 2006;312:1335–44. doi: 10.1016/j.yexcr.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Liu CX, Musco S, Lisitsina NM, Forgacs E, Minna JD, Lisitsyn NA. LRP-DIT, a putative endocytic receptor gene, is frequently inactivated in non-small cell lung cancer cell lines. Cancer Res. 2000;60:1961–7. [PubMed] [Google Scholar]
- 8.Langbein S, Szakacs O, Wilhelm M, Sukosd F, Weber S, Jauch A, Lopez Beltran A, Alken P, Kalble T, Kovacs G. Alteration of the LRP1B gene region is associated with high grade of urothelial cancer. Lab Invest. 2002;82:639–43. doi: 10.1038/labinvest.3780458. [DOI] [PubMed] [Google Scholar]
- 9.Hirai Y, Utsugi K, Takeshima N, Kawamata Y, Furuta R, Kitagawa T, Kawaguchi T, Hasumi K, Noda ST. Putative gene loci associated with carcinogenesis and metastasis of endocervical adenocarcinomas of uterus determined by conventional and array-based CGH. Am J Obstet Gynecol. 2004;191:1173–82. doi: 10.1016/j.ajog.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Sonoda I, Imoto I, Inoue J, Shibata T, Shimada Y, Chin K, Imamura M, Amagasa T, Gray JW, Hirohashi S, Inazawa J. Frequent silencing of low density lipoprotein receptor-related protein 1B (LRP1B) expression by genetic and epigenetic mechanisms in esophageal squamous cell carcinoma. Cancer Res. 2004;64:3741–7. doi: 10.1158/0008-5472.CAN-04-0172. [DOI] [PubMed] [Google Scholar]
- 11.Smith DI, Zhu Y, McAvoy S, Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 12.Liu CX, Li Y, Obermoeller-McCormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem. 2001;276:28889–96. doi: 10.1074/jbc.M102727200. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Knisely JM, Lu W, McCormick LM, Wang J, Henkin J, Schwartz AL, Bu G. Low density lipoprotein (LDL) receptor-related protein 1B impairs urokinase receptor regeneration on the cell surface and inhibits cell migration. J Biol Chem. 2002;277:42366–71. doi: 10.1074/jbc.M207705200. [DOI] [PubMed] [Google Scholar]
- 14.Bu G, Maksymovitch EA, Schwartz AL. Receptor-mediated endocytosis of tissue-type plasminogen activator by low density lipoprotein receptor-related protein on human hepatoma HepG2 cells. J Biol Chem. 1993;268:13002–9. [PubMed] [Google Scholar]
- 15.FitzGerald DJ, Fryling CM, Zdanovsky A, Saelinger CB, Kounnas M, Winkles JA, Strickland D, Leppla S. Pseudomonas exotoxin-mediated selection yields cells with altered expression of low-density lipoprotein receptor-related protein. J Cell Biol. 1995;129:1533–41. doi: 10.1083/jcb.129.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–90. 92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- 17.York SJ, Arneson LS, Gregory WT, Dahms NM, Kornfeld S. The rate of internalization of the mannose 6-phosphate/insulin-like growth factor II receptor is enhanced by multivalent ligand binding. J Biol Chem. 1999;274:1164–71. doi: 10.1074/jbc.274.2.1164. [DOI] [PubMed] [Google Scholar]
- 18.Vilhardt F, Nielsen M, Sandvig K, van Deurs B. Urokinase-type plasminogen activator receptor is internalized by different mechanisms in polarized and nonpolarized Madin-Darby canine kidney epithelial cells. Mol Biol Cell. 1999;10:179–95. doi: 10.1091/mbc.10.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleijmeer MJ, Raposo G, Geuze HJ. Characterization of MHC Class II Compartments by Immunoelectron Microscopy. Methods. 1996;10:191–207. doi: 10.1006/meth.1996.0095. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Lu W, Marzolo MP, Bu G. Differential functions of members of the low density lipoprotein receptor family suggested by their distinct endocytosis rates. J Biol Chem. 2001;276:18000–6. doi: 10.1074/jbc.M101589200. [DOI] [PubMed] [Google Scholar]
- 22.Cuitino L, Matute R, Retamal C, Bu G, Inestrosa NC, Marzolo MP. ApoER2 is endocytosed by a clathrin-mediated process involving the adaptor protein Dab2 independent of its Rafts’ association. Traffic. 2005;6:820–38. doi: 10.1111/j.1600-0854.2005.00320.x. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez E, Girones N, Davis RJ. Inhibition of the receptor-mediated endocytosis of diferric transferrin is associated with the covalent modification of the transferrin receptor with palmitic acid. J Biol Chem. 1990;265:16644–55. [PubMed] [Google Scholar]
- 24.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 25.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–32. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orsel JG, Sincock PM, Krise JP, Pfeffer SR. Recognition of the 300-kDa mannose 6-phosphate receptor cytoplasmic domain by 47-kDa tail-interacting protein. Proc Natl Acad Sci U S A. 2000;97:9047–51. doi: 10.1073/pnas.160251397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, Barnes R, Cohen JC, Hobbs HH. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–8. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 28.Keyel PA, Mishra SK, Roth R, Heuser JE, Watkins SC, Traub LM. A Single Common Portal for Clathrin-mediated Endocytosis of Distinct Cargo Governed by Cargo-selective Adaptors. Mol Biol Cell. 2006;17:4300–17. doi: 10.1091/mbc.E06-05-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer ME, Cooper JA. Endocytosis of megalin by visceral endoderm cells requires the Dab2 adaptor protein. J Cell Sci. 2005;118:5345–55. doi: 10.1242/jcs.02650. [DOI] [PubMed] [Google Scholar]
- 30.Nagai M, Meerloo T, Takeda T, Farquhar MG. The adaptor protein ARH escorts megalin to and through endosomes. Mol Biol Cell. 2003;14:4984–96. doi: 10.1091/mbc.E03-06-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmen T, Schmidt A, Hunziker W, Beermann F. The tyrosinase tail mediates sorting to the lysosomal compartment in MDCK cells via a di-leucine and a tyrosine-based signal. J Cell Sci. 1999;112:45–53. doi: 10.1242/jcs.112.1.45. [DOI] [PubMed] [Google Scholar]
- 32.Hahn-Dantona E, Ruiz JF, Bornstein P, Strickland DK. The low density lipoprotein receptor-related protein modulates levels of matrix metalloproteinase 9 (MMP-9) by mediating its cellular catabolism. J Biol Chem. 2001;276:15498–503. doi: 10.1074/jbc.M100121200. [DOI] [PubMed] [Google Scholar]
- 33.Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–21. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 34.Nykjaer A, Petersen CM, Moller B, Jensen PH, Moestrup SK, Holtet TL, Etzerodt M, Thogersen HC, Munch M, Andreasen PA, et al. Purified alpha 2-macroglobulin receptor/LDL receptor-related protein binds urokinase. plasminogen activator inhibitor type-1 complex. Evidence that the alpha 2-macroglobulin receptor mediates cellular degradation of urokinase receptor-bound complexes. J Biol Chem. 1992;267:14543–6. [PubMed] [Google Scholar]
- 35.Mikhailenko I, Kounnas MZ, Strickland DK. Low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor mediates the cellular internalization and degradation of thrombospondin. A process facilitated by cell-surface proteoglycans. J Biol Chem. 1995;270:9543–9. doi: 10.1074/jbc.270.16.9543. [DOI] [PubMed] [Google Scholar]
- 36.Loukinova E, Ranganathan S, Kuznetsov S, Gorlatova N, Migliorini MM, Loukinov D, Ulery PG, Mikhailenko I, Lawrence DA, Strickland DK. Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP). Evidence for integrated co-receptor function betwenn LRP and the PDGF. J Biol Chem. 2002;277:15499–506. doi: 10.1074/jbc.M200427200. [DOI] [PubMed] [Google Scholar]
- 37.Huang SS, Ling TY, Tseng WF, Huang YH, Tang FM, Leal SM, Huang JS. Cellular growth inhibition by IGFBP-3 and TGF-beta1 requires LRP-1. FASEB J. 2003;17:2068–81. doi: 10.1096/fj.03-0256com. [DOI] [PubMed] [Google Scholar]
- 38.Segarini PR, Nesbitt JE, Li D, Hays LG, Yates JR, 3rd, Carmichael DF. The low density lipoprotein receptor-related protein/alpha2-macroglobulin receptor is a receptor for connective tissue growth factor. J Biol Chem. 2001;276:40659–67. doi: 10.1074/jbc.M105180200. [DOI] [PubMed] [Google Scholar]
- 39.Muramatsu H, Zou K, Sakaguchi N, Ikematsu S, Sakuma S, Muramatsu T. LDL receptor-related protein as a component of the midkine receptor. Biochem Biophys Res Commun. 2000;270:936–41. doi: 10.1006/bbrc.2000.2549. [DOI] [PubMed] [Google Scholar]
- 40.Liu CX, Ranganathan S, Robinson S, Strickland DK. gamma-Secretase-mediated release of the low density lipoprotein receptor-related protein 1B intracellular domain suppresses anchorage-independent growth of neuroglioma cells. J Biol Chem. 2007;282:7504–11. doi: 10.1074/jbc.M608088200. [DOI] [PubMed] [Google Scholar]