Abstract
We show that a 2.6 kb fragment of the muscle myosin heavy-chain gene (Mhc) of Drosophila melanogaster (containing 458 base pairs of upstream sequence, the first exon, the first intron and the beginning of the second exon) drives expression in all muscles. Comparison of the minimal promoter to Mhc genes of ten Drosophila species identified putative regulatory elements in the upstream region and in the first intron. The first intron is required for expression in four small cells of the tergal depressor of the trochanter (jump) muscle and in the indirect flight muscle. The 3′ end of this intron is important for Mhc transcription in embryonic body wall muscle and contains AT–rich elements that are protected from DNase I digestion by nuclear proteins of Drosophila embryos. Sequences responsible for expression in embryonic, adult body wall and adult head muscles are present both within and outside the intron. Elements important for expression in leg muscles and in the large cells of the jump muscle flank the intron. We conclude that multiple transcriptional regulatory elements are responsible for Mhc expression in specific sets of Drosophila muscles.
Keywords: Drosophila melanogaster, muscle, myosin heavy chain, transcription, enhancer, gene regulation
Drosophila melanogaster is unusual in having a single muscle myosin heavy-chain gene (Mhc) (Bernstein et al., 1983; Rozek and Davidson, 1983), rather than a Mhc multigene family (see Emerson and Bernstein, 1987 for review). Alternative splicing of the primary transcripts from this gene yields multiple isoforms of the protein (Collier et al., 1990; George et al., 1989; Hastings and Emerson, 1991; Kazzaz and Rozek, 1989; Kronert et al., 1991; Zhang and Bernstein, 2001). Drosophila Mhc is expressed in all muscles at embryonic, larval, pupal and adult stages. Thus several stage- or muscle-specific enhancer elements may regulate its transcription.
Transcriptional regulatory regions for a number of Drosophila muscle genes have been mapped, resulting in the identification of both unique and shared cis-acting elements (Arredondo et al., 2001; Kelly et al., 2002; Marin et al., 2004; Mas et al., 2004; Meredith and Storti, 1993). The latter includes E-boxes that bind helix-loop-helix transcription factors and MEF2 sites that bind MEF2 protein (Bour et al., 1995; Lilly et al., 1995). E-boxes, MEF2 sites and their binding factors positively regulate vertebrate muscle gene expression as well (see Molkentin and Olson, 1996 for review).
The 5′ end of Drosophila Mhc (Wassenberg et al., 1987) shares a structural similarity with a number of other muscle genes. Each has a large first intron that interrupts the 5′ noncoding region or the first two codons of the gene. This structure is present in both Drosophila (Basi and Storti, 1986; Falkenthal et al., 1984; Geyer and Fyrberg, 1986; Marin et al., 2004; Mas et al., 2004; Parker et al., 1985) and vertebrate muscle genes (Baldwin et al., 1985; Chang et al., 1985; Fornwald et al., 1982; Nabeshima et al., 1984; Strehler et al., 1986). Transcriptional enhancer elements are located in the first intron of a number of such muscle genes (Konieczny and Emerson, 1987; Marin et al., 2004; Mas et al., 2004; Meredith and Storti, 1993; Ng et al., 1989; Yutzey et al., 1989).
To investigate the regulatory domains of Drosophila Mhc, we constructed a series of transgenic fly lines containing Mhc reporter constructs. We identified potential cis-acting transcriptional elements by searching for known enhancer sequences and by comparing putative regulatory regions to those of other Drosophila species. Further, we localized sites of nuclear protein binding by DNAse I footprinting. We demonstrate that multiple elements in Mhc, including regions of the first intron, regulate expression of the gene in specific somatic muscle types during embryonic and adult development.
1. Results and discussion
1.1 The 5′ end of Mhc is sufficient for muscle-specific expression in embryos and adults
We previously determined the transcription initiation site of Mhc by S1 nuclease mapping and primer extension experiments (Wassenberg et al., 1987). As in many promoter regions, there are CAAT box and TATA box homologies located within 60 nucleotides upstream of the transcription initiation site. Presumably these elements are important in regulating the efficiency and accuracy of transcription initiation. In contrast to the conserved position of such basal promoter sequences, enhancer elements can function at a considerable distance from the promoter and can be located 5′ or 3′ of the transcription start site (for review see Maniatis et al., 1987). Before searching for such elements via sequence comparisons, we narrowed down the region required for Mhc expression by examining transgenic lines containing Mhc sequences linked to a reporter gene.
We made a fusion construct, pπMHC-lacZ 1, to determine if sequences essential to muscle-specific Mhc expression are located within the 5′ region of the gene. This construct contains 2.8 kb of Mhc upstream sequence, exon 1, intron 1 and most of Mhc’s second exon linked in frame to the bacterial lacZ gene encoding ß-galactosidase (Fig. 1A). The translation start codon for the fusion protein is the normal translational start site of Mhc, located in exon 2. We analyzed flies transformed with a P element vector containing this construct by X-gal staining. In dissected flies, all identifiable muscle structures, but no non-muscle tissues, were stained (Fig. 2A). The indirect flight muscles located in the thorax are the largest, most prominently stained structures. Following disruption of the cuticle, leg muscles were stained with the same intensity as those in the thorax (not shown). In the head, staining was found in the muscle structures of the proboscis and antennae. The dorsal and ventral muscles of the abdominal segments were stained with less intensity. ß-galactosidase expression was found also along the gut, cardiac tissue and oviduct. In agreement with our results, Fernandes, et al. (1991) and Broadie and Bate (1991) studied muscle development using our pπMHC-lacZ1 transgenic line and observed ß-galactosidase expression in all adult muscles.
Figure 1. Structure of MHC-lacZ fusion and reporter gene constructs.
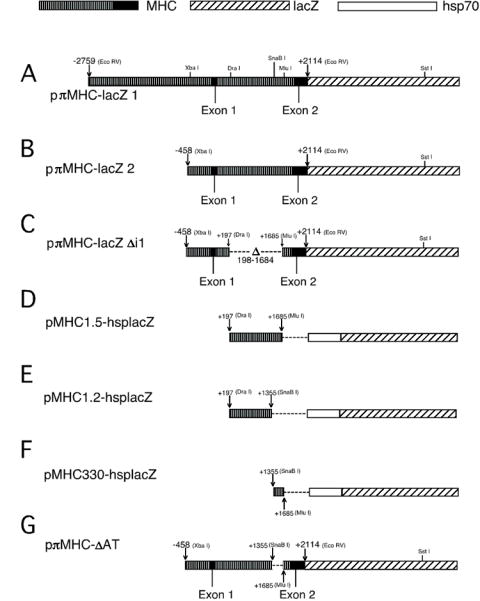
Constructs A, B, C and G were made by fusing the lacZ gene of pOX3 (Raghaven et al., 1986) in frame to the Mhc coding region. Constructs D, E and F were derived from pDM30 (Mismer and Rubin, 1987), which contains the Drosophila hsp70 minimal promoter linked to a lacZ reporter. The parental lacZ plasmids also provide a polyadenylation signal. All constructs were inserted into a P element plasmid vector prior to germline transformation. Details are given in Experimental Procedures.
Figure 2. Staining of organisms in X-gal solution reveals the relative level and tissue-specificity of MHC-lacZ fusion gene expression.
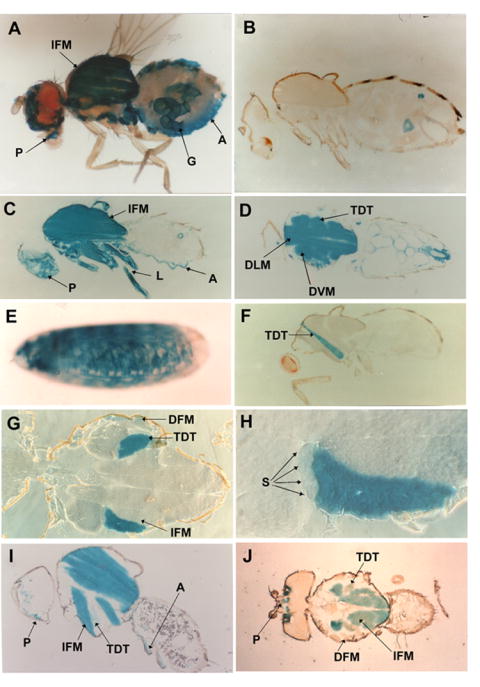
(A) Dissected adult containing pπMHC-lacZ 1 and stained in X-gal solution. Indirect flight muscles (IFM), proboscis muscles (P), abdominal body wall muscles (A) and gut muscles (G) are clearly stained. (B) Cryosection of an adult lacking a fusion gene. Only minor staining of abdominal structures is seen. (C) Parasagittal section of adult containing pπMHC-lacZ 1 shows ß-galactosidase activity in various muscle tissues including indirect flight muscles (IFM), proboscis muscles (P), abdominal body wall muscles (A) and leg muscles (L). (D) Horizontal section of adult carrying pπMHC-lacZ 1. Dorsal-lateral IFMs (DLM) and dorsal-ventral IFMs (DVM) are clearly stained, as is the tergal depressor of the trochanter muscle (TDT). (E) Whole mount of embryo carrying pπMHC-lacZ 1. All body wall muscles are stained. (F) Parasagittal section of fly transformed with pπMHC-lacZ Δi1, which deletes most of intron 1, reveals intense staining in the TDT. (G) Horizontal section of adult thorax expressing pπMHC-lacZ Δi1. The TDT is well stained, the direct flight muscles (DFM) exhibit clear staining, but the IFM shows extremely low levels of staining. (H) Higher magnification of (G) showing that the four smaller anterior cells (S) are poorly stained compared to the other cells of the TDT. (I) Parasagittal section of fly transformed with pMHC-1.5-hsplacZ, which contains intron 1, shows staining in the indirect flight muscles (IFM), proboscis muscles (P) and abdominal body wall muscles (A). The TDT barely stains. (J) Horizontal section of fly transformed with pMHC-1.5-hsplacZ shows staining in the indirect flight muscles (IFM) and proboscis muscles (P), but little or none in the TDT or direct flight muscles (DFM).
To provide a more reliable comparison of levels of reporter gene expression based on tissue staining intensity, we analyzed 15 μm thick cryosections of adult flies. Comparison of a large number of sections showed that thoracic and leg muscles stained somewhat more intensely than head muscles. Abdominal body wall and gut muscles stained least strongly. In both parasagittal (Fig. 2C) and horizontal (Fig. 2D) sections, all muscles of the thorax were stained with similar intensity. This suggests that the levels of Mhc expression in various muscles of the thorax are similar.
We also examined whole mounts of transformed late stage embryos. The predominant structures stained were the muscles of the body wall and the pharyngeal muscles (Fig. 2E). Drysdale et al. (1993) used our transgenic line in a genetic screen for muscle defects and also observed ß-galactosidase expression in all larval body wall muscles. Our results demonstrate that the 4.9 kb Mhc gene fragment of pπMHC-lacZ 1 contains regulatory elements responsible for high level expression in a muscle-specific manner.
To define more closely the location of elements necessary for tissue-specific expression of Mhc we modified pπMHC-lacZ 1 by deleting sequences upstream of position -458 (relative to the transcription initiation site). We designated the resulting construct pπMHC-lacZ 2 (Fig. 1B). Dissected or cryosectioned flies transformed with this construct as well as embryo preparations displayed an X-gal staining pattern that was indistinguishable from that of pπMHC-lacZ 1 transformed lines (not shown). Thus no major regulatory elements necessary for muscle-specific Mhc expression are located in the -455 to -2777 nucleotide region. We previously reported preliminary data on whole mount staining of adults expressing pπMHC-lacZ 1 and pπMHC-lacZ 2 (Hess et al., 1989).
1.2 Sequence analysis reveals putative regulatory elements in Mhc
We searched the -458 to +2114 minimal promoter region of Mhc for known muscle-specific regulatory elements (Supplementary Table 1). There are 10 E-box sequences. Three are upstream of the transcriptional start site and 7 are within the first intron. There are three MEF2 sites in the promoter region. All are within the first intron. The Pdp1 protein-binding element (RTTYYGTAAY; Lin et al., 1997) is not present.
To search for conserved regulatory elements, we used the sequence comparisons of Drosophila melanogaster Mhc to D. hydei Mhc (Miedema et al., 1994). The latter sequence is available beginning at 250 residues upstream of the transcription start site. Sequence identity is over 55% through the minimal promoter. The exons and surrounding regions show greater than 90% identity and there are numerous clusters of identity upstream of exon 1 and within the first intron. D. hydei Mhc contains 7 E-box sequences. One is upstream of exon 1 and is conserved in sequence and position with E-box 2 (-191) of D. melanogaster (Supplementary Table 1). The remaining 6 D. hydei E boxes are located within intron 1, at sites different from those of D. melanogaster. D. hydei Mhc has 4 MEF2 sites, all within the first intron. The second and third MEF2 sites of D. melanogaster are absolutely conserved in D. hydei Mhc. D. hydei Mhc has no Pdp1 sites.
To more thoroughly study variation of putative muscle regulatory elements of Drosophila Mhc across evolutionary time, we aligned the D. melanogaster Mhc sequence with those of nine other Drosophila genomes (Supplementary Table 1). In general, E-box sequences diverge from the D. melanogaster sequence and the consensus relatively rapidly. The only absolutely conserved E-box is at -191. In contrast, the three MEF2 sites of D. melanogaster show nearly absolute identity among the species (97%, 100% and 98%).
1.3 Regulatory elements are located in intron 1
Given the location of known muscle-specific transcription factor binding sites within the first intron of Mhc, we investigated the possibility that it contains regulatory elements important for tissue-specific and high-level transcription. We constructed a plasmid that has the same -458 bp Mhc 5′ end as pπMHC-lacZ 2 but has the majority of intron 1 deleted (Fig. 1C). The 1.5 kb deletion removes all 7 E-box elements of the intron and all three MEF2 sites of the promoter. Analysis of adults transformed with pπMHC-lacZ Δi1 revealed that the staining pattern of the thoracic musculature was dramatically altered (Figs. 2F, G) compared to adults with pπMHC-lacZ 2. The dorsal-ventral and dorsal-lateral indirect flight muscles now barely stain, while the tergal depressor of the trochanter (TDT) muscles (jump muscles) and the leg muscles continue to stain strongly, with the direct flight muscles staining somewhat less strongly. Proboscis and antennal muscles in the head and abdominal wall muscles stain well (not shown). The intensity of the strongly staining muscles was comparable to that in flies transformed with P elements that contain larger promoter fragments (Figs. 2C and D). Higher magnification cross-sections of the TDT muscle revealed that the ß-galactosidase expression within this muscle is not uniform (Fig. 2H). The weak staining intensity of four smaller cells at the anterior end is indistinguishable from that of the surrounding indirect flight muscles, while the 28 larger posterior cells stain strongly. Embryos containing pπMHC-lacZ Δi1 express ß-galactosidase in body wall and pharyngeal muscles (Fig. 3A, 3B), but at somewhat lower levels than in pπMHC-lacZ 2; further some embryos and some lines showed differential staining of muscles (Fig. 3C, 3D).
Figure 3. Staining of embryos in X-gal solution reveals the level and tissue-specificity of MHC-lacZ transgene expression.
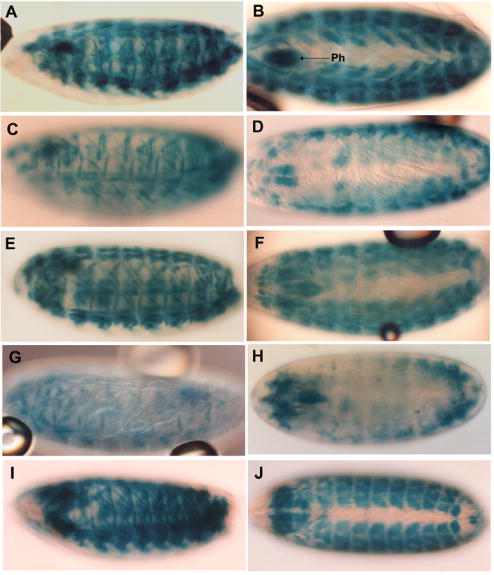
Lateral (left panels) and dorsal (right panels) views are shown. (A, B) Embryos containing pπMHC-lacZ Δi1, which lacks most of intron 1, express ß-galactosidase in body wall and pharyngeal muscles (Ph) in a similar manner to those expressing the minimal myosin promoter pπMHC-lacZ 2 (Fig. 2E). (C, D) Some pπMHC-lacZ Δi1 embryos show differential muscle staining. (E, F) Embryos expressing pMHC1.5-hsplacZ, which contains the intron 1 enhancer, show body wall and pharyngeal muscle staining similar to those expressing pπMHC-lacZ 2. (G, H) Deletion of the 330 terminal bp of pMHC1.5-hsplacZ to yield pMHC1.2-hsplacZ results in greatly reduced body wall muscle expression. (I, J) Expression of pπMHC-lacZ ΔAT, which was produced by deletion of the 330 bp intron fragment from pπMHC-lacZ 2, allows high levels of embryonic ß-galactosidase expression.
To test the transcriptional enhancer function of the 1.5 kb intron 1 region, we placed the sequence from +198 to +1684 in a hsp70 promoter–lacZ vector. This vector is designed to allow inserted enhancer elements to activate lacZ expression following P element-mediated transformation. Flies transformed with pMHC1.5-hsplacZ (Fig. 1D) displayed muscle-specific ß-galactosidase expression in their thoraces in a pattern complementary to that seen for pπMHC-lacZ Δi1. Staining of TDT muscle, direct flight muscles and leg muscles is absent or barely detectable, while staining of indirect flight muscle is relatively strong (Fig. 2I, 2J). Abdominal body wall muscles, proboscis and antennal muscles stain (Fig. 2I, 2J). In embryos (Fig. 3E, F), staining of body wall muscles and pharyngeal muscles expressing pMHC1.5-hsplacZ is qualitatively similar but slightly less intense than embryos containing the minimal myosin promoter (pπMHC-lacZ 2).
Our results show that tissue-specific regulatory elements that govern Mhc expression in the indirect flight muscle are located in the first intron. Elements that permit expression in the 28 large cells of the TDT, leg muscles and direct flight muscles are located outside the 1.5 kb intron region. Elements that enhance expression in embryonic body wall and pharyngeal muscles, adult abdominal muscles, proboscis muscles and antennal muscles are present both within the 1.5 kb intron segment and in the flanking regions.
1.4 DNase I footprinting using Drosophila embryo nuclear protein extract shows binding to AT-repeat sequences within the first intron of Mhc
To identify the targets of putative muscle-specific trans-acting factors within the first intron of Mhc, we performed DNase I footprinting assays on intron restriction fragments using 10 to 16 hour Drosophila embryo nuclear extracts. This corresponds to the period during which Mhc is first expressed. We detected several strong footprints between +1360 to +1550 (Fig. 4). When 0 to 12 hour embryo nuclear extracts were used in a DNase I footprint assay, no footprint was observed at these sites (not shown). The footprinted regions encompass several AT repeat sequences, some of which share identity with the cf2 binding site (RTATATRTA; Gogos et al., 1992). There are four canonical cf2 elements within the Mhc promoter region (Supplementary Table 1). These overlap with each other and with the footprint at +1404. Most of the footprinted regions contain AT or AC repeats and show varying degrees of conservation (58-97%) with other Drosophila Mhc genes (Supplementary Table 1).
Figure 4. DNAse footprint reaction of proteins from 10-16 hour old Drosophila embryos.
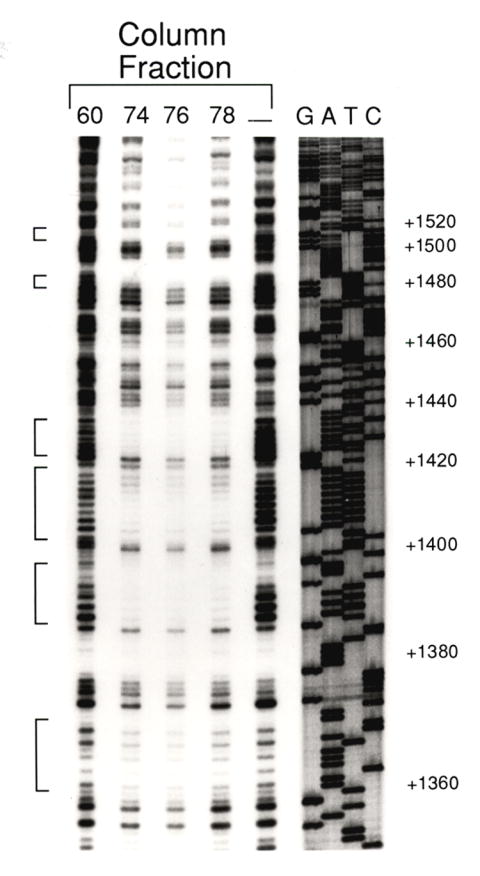
Sephacryl S400 HR column fractions of nuclear extracts were reacted with an end-labeled Mhc gene intron sequence (+1355 to +1685). Following DNAse digestion and gel electrophoresis, AT-rich regions were shown to be protected (brackets) by column fractions 74, 76 and 78, compared to column fraction 60 or a no-protein sample (-).
We tested the importance of the region containing the footprints by deleting the 330 bp fragment containing these elements from the pMHC1.5-hsplacZ plasmid. This deletion also removes two of the three MEF2 sites. The resulting pMHC1.2-hsplacZ reporter construct (Fig. 1E) produced adult expression consistent with its parent construct. ß-galactosidase accumulated in indirect flight muscles, abdominal body wall muscles, proboscis muscles and antennal muscles (not shown). However, the pattern of expression in embryonic muscles was greatly reduced, with fewer body wall muscles staining (Fig. 3G, H). We tested whether the 330 bp fragment alone showed enhancer activity by inserting it into the reporter construct to yield pMHC330-hsplacZ (Fig. 1F). This fragment yielded no muscle expression. Further, deletion of the 330 bp fragment from pπMHC-lacZ 2 to produce pπMHC-lacZ ΔAT did not affect adult (not shown) or embryonic (Fig. 3I, J) ß-galactosidase expression, indicating that embryonic elements are present outside the 330 bp intron fragment. We conclude that the 330 bp intron fragment is not important for adult muscle expression, but that elements within the 330 bp intron fragment can work in conjunction with elements within the 1.2 kb intron fragment to enhance embryonic muscle Mhc expression.
1.5 Concluding comments
Our results demonstrate that Drosophila Mhc contains multiple positive elements required for its muscle-specific expression (summarized in Table 1). Elements within intron 1 are important for indirect flight muscle expression and for transcription in the small cells of the TDT muscle. At least two elements within intron 1 regulate expression in embryonic body wall muscles, but there are sequences outside the intron that permit expression in these muscles as well. Likewise, sequences responsible for Mhc expression in pharyngeal, adult abdominal body wall, proboscis and antennal muscles are located both within the intron and in flanking regions. The flanking regions have elements that are important for expression in leg, large cells of the TDT and direct flight muscles. Efforts to more precisely map the locations of elements within flanking regions using an enhancer-reporter vector did not yield positive results (K.T., unpublished data), suggesting that the function of these enhancers is dependent upon maintaining their position within Mhc. Hanke and Storti (1988) noted that a 26 bp element beginning at -76 of the Drosophila Mhc gene is quite similar to sequences upstream of the Drosophila tropomyosin I and II genes. This sequence could serve as a position-dependent enhancer in all three genes.
Table 1.
Summary of transgenic lines and expression patterns
| Construct | # of lines | Embryo
body wall |
Pharyngeal | Antennal | Proboscis | Indirect
flight |
Direct
flight |
TDT | Leg | Adult
body wall |
|---|---|---|---|---|---|---|---|---|---|---|
| A: pπMHC-lacZ 1 | 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| B: pπMHC-lacZ 2 | 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| C: pπMHC-lacZ Δi1 | 3 | ++ | ++ | ++ | ++ | +/- | ++ | +++ | +++ | ++ |
| D: pMHC1.5-hsplacZ | 8 | ++ | ++ | ++ | ++ | ++ | - | - | - | + |
| E: pMHC1.2-hsplacZ | 5 | + | + | ++ | ++ | ++ | - | - | - | + |
| F: pMHC330-hsplacZ | 11 | - | - | - | - | - | - | - | - | - |
| G: pMHC- ΔAT | 6 | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | ++ |
Other Drosophila muscle genes are interrupted by an intron near their 5′ ends (Basi and Storti, 1986; Falkenthal et al., 1984; Geyer and Fyrberg, 1986; Marin et al., 2004; Mas et al., 2004; Parker et al., 1985), and these introns often contain transcriptional activation sequences. The ß3-tubulin gene has a visceral muscle-specific enhancer element in its first intron (Gasch et al., 1989), as well as a somatic muscle element (Hinz et al., 1992). The first intron of the tropomyosin I gene has elements that permit expression in all muscles of the adult and embryo (Schultz et al., 1991). The tropomyosin II gene is similar in this respect and deletion of a 454 bp intron region eliminates indirect flight and TDT muscle expression (Meredith and Storti, 1993). The first intron of the troponin T (Mas et al., 2004) and troponin I (Marin et al., 2004) genes contain enhancer element that are largely redundant compared to upstream enhancers. However the intron element in the troponin T gene is more important for expression in adult muscle, particularly indirect flight muscle. Enhancer redundancy in the troponin genes is proposed to be important for yielding proper levels of gene expression (Marin et al., 2004; Mas et al., 2004), as may be the case for redundant elements in Mhc.
It is particularly striking that removal of the first intron from Mhc allows transcription in the 28 large cells of the TDT, but not the 4 small cells. This result is intriguing in that we previously documented another difference between the two TDT cell types in Mhc7 and Mhc9 mutants (O’Donnell et al., 1989). The mutants display normal numbers of thick filaments in the TDT muscle large cells but a reduced number in the small cells. All other muscles appear normal, except for the indirect flight muscles, which fail to accumulate thick filaments. Our present results indicate that such phenotypes could arise from the mutation of an enhancer element that is specific to the indirect flight muscles and the small cells of the TDT. Another intriguing result in this regard is that the contractile protein make-up of the large and small cells of the TDT muscle differs. Different isoforms of MHC are present in these cell types (Kronert et al., 1991) and different Z-band proteins accumulate (Vigoreaux et al., 1991). Given these disparities between the two TDT cell types, it is likely that they serve distinct contractile functions.
Three MEF2 sites are present in the D. melanogaster Mhc minimal promoter. All are evolutionarily conserved, but deletion of the intron region that contains these sites does not disable Mhc expression in many muscle tissues. Likewise, two adult-specific muscle actin genes accumulate normal levels of transcripts when levels of MEF2 are severely reduced (Baker et al., 2005). Further, the miniparamyosin adult-specific promoter lacks a MEF2 site (Arredondo et al., 2001). On the other hand, MEF2 sites are important for expression of some Drosophila muscle genes. The MEF2 site in the Actin57B gene is essential for cardiac, skeletal, and visceral muscle expression in embryos (Kelly et al., 2002). Severe reduction of MEF2 expression in adults results in a more than four-fold decrease in Actin57B gene expression (Baker et al., 2005). Expression of the ß-3 tubulin gene in embryonic somatic muscles is dependent upon a MEF2 site (Damm et al., 1998). In the paramyosin gene, a MEF2 element is important for high-level expression in larval muscles (Arredondo et al., 2001). In the tropomyosin I gene, Lin et al. (1996) showed that two MEF2 sites are necessary to yield high level expression in most muscle types. However, both this group and Kelly et al. (2002) showed that a MEF2 element by itself is not capable of inducing muscle-specific expression.
Embryonic nuclear proteins that bind to the 3′ end of the first intron of Mhc could be important for Mhc expression in embryos. However the muscle-specificity and positive action of the bound factors remain to be demonstrated. The high level of identity of some of the protected DNA elements among various Drosophila species (Supplementary Table 1) suggests that they serve a functional role. The 330 bp region containing the footprinted elements clearly is important for Mhc regulation since its deletion greatly reduces the ability of the remaining intron fragment to drive expression in embryos. However, since the deleted region is unable to drive Mhc expression independently, the DNAse insensitive elements and the two MEF2 elements in the 330 bp region are not sufficient to promote Mhc expression in a position-independent fashion.
The multiplicity of elements that regulate Mhc expression reflects the fact that the gene must be expressed at multiple stages of development in numerous muscle types. Our identification of minimal promoter elements has proved important for studies of muscle lineages (Broadie and Bate, 1991; Bernard et al., 2006; Farrell et al., 1996; Fernandes et al., 1991) and for expression of Mhc transgenes designed to study the function of particular myosin domains (reviewed by Swank et al., 2000). The mapping of additional tissue-specific elements will permit an understanding of the basis of muscle gene regulation and provide useful tools for transgene expression in Drosophila.
2. Experimental Procedures
2.1 Plasmid construction
P element constructs with the designation pπ are modifications of pπ18, which derives from pUChsneo (Steller and Pirrotta, 1985). pπ18 has Sph I, Xba I, Kpn I and Sst I restriction sites added to the polylinker. To produce fusion genes a modified ß-galactosidase (ß-gal) gene (Raghaven et al., 1986) was inserted into the polylinker of pπ18 to yield construct pπlacZ. Correct processing at the 3′ end of the transcripts was assured by the presence of a polyadenylation site at the 3′ end of lacZ. To produce each test construct, a particular Mhc promoter fragment was ligated into the Sma I site of the polylinker of pπlacZ. Joining the Eco RV site located in the second exon of the Mhc gene and the Sma I site of the polylinker in pπlacZ permitted the in-frame fusion of the two genes. For the construction of pπMHC-lacZ 1, the Eco RV site 2.7 kb upstream of the transcription start site of the Mhc gene was used (Fig. 1A). The 5′ end of pπMHC-lacZ 2 is the Xba I site 458 bp upstream of the transcription start site (Fig. 1B). Both constructs contain exon 1, intron 1 and exon 2 up to the Eco RV site. The internal deletion in pπMHC-lacZ Δi1 was generated by recombining the 5′ Xba I - Dra I fragment starting at -458 and ending at +197 with the Mlu I - Eco RV Mhc fragment containing the 3′ end of intron 1 and most of exon 2 (Fig. 1C). In a second step this fusion was introduced into the Sma I site of pπlacZ. To construct pπMHC-ΔAT (Fig. 1G), the SnaB I - Mlu I fragment was removed from a subcloned portion of pπMHC-lacZ 2 and the remaining fragment was blunt-ended and religated. A Kpn I - Eco RV fragment from the amplified plasmid was isolated and substituted into pπMHC-lacZ 2 cut at the same restriction sites.
P element constructs containing a Drosophila hsp70 promoter–lacZ reporter were used to detect in vivo enhancer activity from Mhc DNA cloned immediately upstream of the hsp70 promoter. The P element construct is a derivative of pDM30 (rosy+) (Mismer and Rubin, 1987) into which a Drosophila hsp70 promoter–lacZ reporter has been inserted. Various fragments of Mhc (see Fig. 1D, E, F) were gel isolated, blunt ended and ligated into a filled-in Not I site in the reporter vector.
2.2 P element transformation
Canton S embryos were injected with 1μg/μl P element (neor) vector DNA and 0.5 μg/μl helper plasmid p25.7wc (Karess and Rubin, 1984) in injection buffer (100 mM sodium phosphate, 5 mM KCl, pH 7.2) as described by Rubin and Spradling (1982). G1 embryos expressing neor were selected on instant food (Formula 4-24, Carolina Biologicals Supply Company) containing G418 (Gibco) at a concentration of 1 mg/ml (0.5 mg/ml active substance). For rosy+P element vectors containing the hsp70 promoter–lacZ reporter, injection was into ry- embryos and the G1 generation was screened for wild-type eye color.
2.3 Chromosomal mapping
Single males from each neor transformed line were mated to several virgin females of the stock R13 (b Df(2L)A47 cn bw/CyO; MKRS, Sb ry/TM2, Ubx130ry). Offspring were selected on G418-containing medium. The presence of only female offspring indicated P element insertion on the X chromosome. For mapping insertions on other chromosomes, male CyO; Sb offspring were mated to virgin Canton S (wild-type) females and the progeny were selected on G418 and scored for the segregation of the CyO and Sb markers. The absence of CyO progeny indicated a P element insertion on chromosome 2, while the absence of the Sb marker denoted integration on chromosome 3. Segregation of CyO and Sb independent of G418 resistance indicated P element insertion on chromosome 4, or integration of elements on more than one chromosome. Stocks were balanced by standard genetic crosses. Transgenes containing ry+ were mapped in an analogous way using the stock CyO/+; Ser, ry506, e/ ry506.
2.4 Cryosections
Sectioning was performed as described by Hafen and Levine (1986). The procedure for section staining was modified from Liu et al. (1988). The sections were fixed in 0.5% gluteraldehyde in PBS (10 mM sodium phosphate, pH 7.0, 140 mM NaCl, 2.5 mM KCl) for 5 min and incubated in staining solution (20 mM potassium ferricyanide, 20 mM potassium ferro-cyanide, 1 mM MgCl2, 0.1% X-gal, in PBS) for 10 min. The staining solution was removed by 3 washes of 5 min each in PBS and one wash in H20 for 2 min. The dried sections were embedded in Polymount (American Scientific).
2.5 Whole mount staining
Flies were dissected with a razor blade and tissue was fixed for 5 min in dissecting buffer (0.1 M sodium phosphate, pH 7.5, 1 mM MgCl2, 0.5% gluteraldehyde). Tissue was stained in 0.1 M sodium phosphate, pH 7.5, 1 mM MgCl2, 0.2 % X-gal for 30 - 60 min and then transferred into dissecting buffer for inspection (Simon and Lis, 1987). Embryo whole mount staining was performed in a manner similar to that of Singh et al. (1995). In brief, embryos were collected on yeasted grape-agar plates, rinsed, dechorionated in 50% bleach, rinsed, sprayed with heptane and then fixed in phosphate-buffered 4 % formaldehyde solution containing 50% heptane. After washing, embryos were incubated in X-gal solution containing Triton X-100 to detect ß-galactosidase.
2.6 Preparation of Drosophila embryo nuclear protein extracts
Drosophila Canton S embryos were collected between 10 and 16 hours after fertilization and were stored for up to three days at 4°C. Extracts were prepared according to the protocol of Soller et al. (1988) and Kerrigan et al. (1991). All operations were performed at 4°C. Eighty to 100 grams of dechorionated embryos were homogenized with a motorized teflon pestle, filtered through Miracloth, and nuclei were pelleted in a GSA rotor at 8000 rpm (10,400 × g). Nuclei were resuspended in buffer using a glass dounce homogenizer with a B pestle. Nuclei were lysed by addition of ammonium sulfate to a final concentration of 0.364 M (9% saturation). Lysed nuclei were pelleted at 142,000 × g in a 45 Ti rotor for one hour. The supernatant was recovered and nuclear proteins were precipitated by addition of solid ammonium sulfate to a concentration of 2.26 M (56% saturation at 20°C). The nuclear protein precipitate was pelleted in an SS-34 rotor at 15,000 rpm (27,000 × g) for 20 min. The pellet was resuspended in buffer and the nuclear protein extract was applied to a Sephadex G25 SF desalting column. Desalted nuclear extracts were fractionated on a Sephacryl S400 HR column to remove phosphatase and nuclease that would interfere with subsequent DNase I footprinting assays. Peak protein fractions were quick frozen in liquid nitrogen and stored in aliquots at -80°C.
2.7 DNase I footprinting assay of Drosophila embryo nuclear protein extracts
Drosophila Mhc DNA approximately 170 to 400 base pairs in length was subcloned into pBluescript (Stratagene Cloning Systems). Mhc DNA labeled with 32P at one end only was synthesized by polymerase chain reaction using a T3 and T7 primer, one of which was labeled using polynucleotide kinase and γ–32P–ATP. 32P–labeled Mhc DNA PCR products were purified on non-denaturing 5% polyacrylamide gels. DNase I footprinting reactions were performed essentially as described by Kerrigan et al. (1991). 32P–labeled Mhc DNA (10 to 25 fmoles, 10,000 to 20,000 cpm) was mixed on ice with 20 to 25 μg of Drosophila embryo nuclear extract and 10 A260 units of poly dI-dC. The mixture was incubated on ice for 15 min. 0.5 units of DNase (RNase free) (Promega Biotech) was added to the protein–DNA mixture in 5 mM MgCl2, 2.5 mM CaCl2, incubated at room temperature for two min and the reaction was stopped by addition of EDTA/SDS solution, phenol/chloroform extraction and ethanol precipitation. Footprinted DNA was electrophoresed on a denaturing 6% polyacrylamide sequencing gel, and the dried gels were autoradiographed for one to five days.
2.8 DNA sequence analysis
Drosophila genome sequences from the database developed by the Eisen laboratory at the University of California, Berkeley (http://rana.lbl.gov/drosophila/alignments_eisenlab.html) were aligned using the program Multi-LAGAN (Brudno et al., 2003) and kindly provided by Dr. Venky Iyer. Sequence alignments were analyzed using the Jalview Java Alignment Editor (Clamp et al., 2004).
Supplementary Material
Acknowledgments
We appreciate the excellent technical assistance of Cecilia Gutierrez, Douglas Allen, Anthony Ferkich, William Kronert, Walter Callen, and Venetia Collier. We thank Elliot Meyerowitz (California Institute of Technology) for providing the lacZ gene, James Posakony (University of California, San Diego) for providing the R13 fly stock and Kevin Moses (Howard Hughes Medical Institute) for providing the pDM30 hsp70-lacZ plasmid. We appreciate the help of K. David Becker with genetic analysis. We thank William Kronert and Corey Dambacher for help with figure preparation and Venky Iyer (University of California, Berkeley) and Lauren Aguado for aiding with the sequence comparisons. We thank Dianne Hodges, K. David Becker, William Kronert and Richard Cripps (University of New Mexico) for their critical comments on the manuscript. N.K.H. was supported by a fellowship from the American Heart Association, California Affiliate. K.T. was supported by NIH NIGMS MBRS Program Grant GM 58906. S.I.B. served as an Established Investigator of the American Heart Association. This project was funded by NIH grant GM32443.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arredondo JJ, Ferreres RM, Maroto M, Cripps RM, Marco R, Bernstein SI, Cervera M. Control of Drosophila paramyosin/miniparamyosin gene expression. Differential regulatory mechanisms for muscle-specific transcription. J Biol Chem. 2001;276:8278–8287. doi: 10.1074/jbc.M009302200. [DOI] [PubMed] [Google Scholar]
- Baker PW, Tanaka KK, Klitgord N, Cripps RM. Adult myogenesis in Drosophila melanogaster can proceed independently of myocyte enhancer factor-2. Genetics. 2005;170:1747–1759. doi: 10.1534/genetics.105.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS, Jr, Kittler ELW, Emerson CP., Jr Structure, evolution, and regulation of a fast skeletal muscle troponin I gene. Proc Natl Acad Sci USA. 1985;82:8080–8084. doi: 10.1073/pnas.82.23.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi GS, Storti RV. Structure and DNA sequence of the tropomyosin I gene from Drosophila melanogaster. J Biol Chem. 1986;261:817–827. [PubMed] [Google Scholar]
- Bernard F, Dutriaux A, Silber J, Lalouette A. Notch pathway repression by vestigial is required to promote indirect flight muscle differentiation in Drosophila melanogaster. Dev Biol. 2006;295:164–177. doi: 10.1016/j.ydbio.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Broadie KS, Bate M. The development of adult muscles in Drosophila: ablation of identified muscle precursor cells. Development. 1991;113:103–118. doi: 10.1242/dev.113.1.103. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, NISC Comparative Sequencing Program. Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KS, Rothblum KM, Schwartz RJ. The complete sequence of the chicken alpha-cardiac actin gene: a highly conserved vertebrate gene. Nucleic Acids Res. 1985;13:1223–1237. doi: 10.1093/nar/13.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Collier VL, Kronert WA, O’Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- Damm C, Wolk A, Buttgereit D, Loher K, Wagner E, Lilly B, Olson EN, Hasenpusch-Theil K, Renkawitz-Pohl R. Independent regulatory elements in the upstream region of the Drosophila 3 tubulin gene (Tub60D) guide expression in the dorsal vessel and the somatic muscles. Dev Biol. 1998;199:138–149. doi: 10.1006/dbio.1998.8916. [DOI] [PubMed] [Google Scholar]
- Drysdale R, Rushton E, Bate M. Genes required for embryonic muscle development in Drosophila melanogaster: a survey of the X chromosome. Roux’s Arch Dev Biol. 1993;202:276–295. doi: 10.1007/BF00363217. [DOI] [PubMed] [Google Scholar]
- Emerson CP, Jr, Bernstein SI. Molecular genetics of myosin. Ann Rev Biochem. 1987;56:695–726. doi: 10.1146/annurev.bi.56.070187.003403. [DOI] [PubMed] [Google Scholar]
- Falkenthal S, Parker VP, Mattox WW, Davidson N. Drosophila melanogaster has only one myosin alkali light-chain gene which encodes a protein with considerable amino acid sequence homology to chicken myosin alkali light chains. Mol Cell Biol. 1984;4:956–965. doi: 10.1128/mcb.4.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell ER, Fernandes J, Keshishian H. Muscle organizers in Drosophila: the role of persistent larval fibers in adult flight muscle development. Dev Biol. 1996;176:220–229. doi: 10.1006/dbio.1996.0129. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Bate M, VijayRaghaven K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Fornwald JA, Kuncio G, Peng I, Ordahl CP. The complete nucleotide sequence of the chick alpha-actin gene and its evolutionary relationship to the actin gene family. Nucleic Acids Res. 1982;10:3861–3876. doi: 10.1093/nar/10.13.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A, Hinz U, Renkawitz-Pohl R. Intron and upstream sequences regulate expression of the Drosophila beta 3-tubulin gene in the visceral and somatic musculature, respectively. Proc Natl Acad Sci USA. 1989;86:3215–3218. doi: 10.1073/pnas.86.9.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EL, Ober MB, Emerson CP., Jr Functional domains of the Drosophila muscle myosin heavy chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9:2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Fyrberg EA. 5′-flanking sequence required for regulated expression of a muscle-specific Drosophila melanogaster actin gene. Mol Cell Biol. 1986;6:3388–3396. doi: 10.1128/mcb.6.10.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Hsu T, Bolton J, Kafatos FC. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992;257:1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- Hafen E, Levine M. The localization of RNAs in Drosophila tissue sections by in situ hybridization. In: Roberts DB, editor. Drosophila a Practical Approach. IRL; Oxford: 1986. pp. 139–158. [Google Scholar]
- Hanke PD, Storti RV. The Drosophila melanogaster tropomyosin II gene produces multiple proteins by use of alternative tissue-specific promoters and alternative splicing. Mol Cell Biol. 1988;8:3591–3602. doi: 10.1128/mcb.8.9.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings GA, Emerson CP., Jr Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J Cell Biol. 1991;114:263–276. doi: 10.1083/jcb.114.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess N, Kronert WA, Bernstein SI. Transcriptional and post-transcriptional regulation of Drosophila myosin heavy chain gene expression. In: Kedes LH, Stockdale FE, editors. Cellular and Molecular Biology of Muscle Development. A.R Liss; New York: 1989. pp. 621–631. [Google Scholar]
- Hinz U, Wolk A, Renkawitz-Pohl R. Ultrabithorax is a regulator of beta 3 tubulin expression in the Drosophila visceral mesoderm. Development. 1992;116:543–554. doi: 10.1242/dev.116.3.543. [DOI] [PubMed] [Google Scholar]
- Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Kazzaz JA, Rozek CE. Tissue-specific expression of the alternately processed Drosophila myosin heavy-chain messenger RNAs. Dev Biol. 1989;133:550–561. doi: 10.1016/0012-1606(89)90057-2. [DOI] [PubMed] [Google Scholar]
- Kelly KK, Meadows SM, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110:39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- Kerrigan LA, Croston GE, Lira LM, Kadonaga JT. Sequence–specific transcriptional antirepression of the Drosophila Krüppel gene by the GAGA factor. J Biol Chem. 1991;266:574–582. [PubMed] [Google Scholar]
- Kronert WA, Edwards KA, Roche ES, Wells L, Bernstein SI. Muscle-specific accumulation of Drosophila myosin heavy chains: a splicing mutation in an alternative exon results in an isoform substitution. EMBO J. 1991;10:2479–2488. doi: 10.1002/j.1460-2075.1991.tb07787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny SF, Emerson CP., Jr Complex regulation of the muscle-specific contractile protein (troponin I) gene. Mol Cell Biol. 1987;7:3065–3075. doi: 10.1128/mcb.7.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schluz RA, Olson EN. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci USA. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lin MH, Horvath P, Reddy KL, Storti RV. PDP1, a novel Drosophila PAR domain bZIP transcription factor expressed in developing mesoderm, endoderm and ectoderm, is a transcriptional regulator of somatic muscle genes. Development. 1997;124:4685–4696. doi: 10.1242/dev.124.22.4685. [DOI] [PubMed] [Google Scholar]
- Liu X, Lorenz L, Yu Q, Hall JC, Rosbash M. Spatial and temporal expression of the period gene in Drosophila melanogaster. Genes Dev. 1988;2:228–238. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Marin MC, Rodriguez JR, Ferrus A. Transcription of Drosophila troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol Biol Cell. 2004;15:1185–1196. doi: 10.1091/mbc.E03-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas JA, Garcia-Zaragoza E, Cervera M. Two functionally identical modular enhancers in Drosophila troponin T gene establish the correct protein levels in different muscle types. Mol Biol Cell. 2004;15:1931–1945. doi: 10.1091/mbc.E03-10-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith J, Storti RV. Developmental regulation of the Drosophila tropomyosin II gene in different muscles is controlled by muscle-type-specific intron enhancer elements and distal and proximal promoter control elements. Dev Biol. 1993;159:500–512. doi: 10.1006/dbio.1993.1259. [DOI] [PubMed] [Google Scholar]
- Miedema K, Harhangi H, Mentzel S, Wilbrink M, Akhmanova A, Hooiveld M, Bindels P, Hennig W. Interspecific sequence comparison of the muscle-myosin heavy-chain genes from Drosophila hydei and Drosophila melanogaster. J Molec Evol. 1994;39:357–368. doi: 10.1007/BF00160268. [DOI] [PubMed] [Google Scholar]
- Mismer D, Rubin GM. Analysis of the promoter of the ninaE opsin Gene in Drosophila melanogaster. Genetics. 1987;116:565–578. doi: 10.1093/genetics/116.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Fujii-Kuriyama Y, Muramatsu M, Ogata K. Alternative transcription and two modes of splicing result in two myosin light chains from one gene. Nature. 1984;308:333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Gunning P, Liu S-H, Leavitt J, Kedes L. Regulation of the human beta-actin promoter by upstream and intron domains. Nucleic Acids Res. 1989;17:601–615. doi: 10.1093/nar/17.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–1246. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- Parker VP, Falkenthal S, Davidson N. Characterization of the myosin light-chain-2 gene of Drosophila melanogaster. Mol Cell Biol. 1985;5:3058–3068. doi: 10.1128/mcb.5.11.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghaven KV, Crosby MA, Mathers PH, Meyerowitz EM. Sequences sufficient for correct regulation of Sgs-3 lie close to or within the gene. EMBO J. 1986;5:3321–3326. doi: 10.1002/j.1460-2075.1986.tb04646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozek CE, Davidson N. Drosophila has one myosin heavy chain gene with three developmentally regulated transcripts. Cell. 1983;32:23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila using transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tansey T, Gremke L, Storti RV. A muscle-specific intron enhancer required for rescue of indirect flight muscle function regulates Drosophila tropomyosin I gene expression. Mol Cell Biol. 1991;11:1901–1911. doi: 10.1128/mcb.11.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lis JT. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nucleic Acids Res. 1987;15:2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kango M, Sinha P. An improved method for chemical devitellinization of X-gal stained Drosophila embryos. Indian J Exp Biol. 1995;33:150–152. [PubMed] [Google Scholar]
- Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Steller H, Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985;4:167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler EE, Strehler-Page M-A, Perriard J-C, Periasamy M, Nadal-Ginard B. Complete nucleotide and encoded amino acid sequence of a mammalian myosin heavy chain gene. Evidence against intron-dependent evolution of the rod. J Mol Biol. 1986;190:291–317. doi: 10.1016/0022-2836(86)90003-3. [DOI] [PubMed] [Google Scholar]
- Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech. 2000;50:430–442. doi: 10.1002/1097-0029(20000915)50:6<430::AID-JEMT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Vigoreaux JO, Saide JD, Pardue ML. Structurally different Drosophila striated muscles utilize distinct variants of Z-band-associated proteins. J Muscle Res Cell Motil. 1991;12:340–354. doi: 10.1007/BF01738589. [DOI] [PubMed] [Google Scholar]
- Wassenberg DR, II, Kronert WA, O’Donnell PT, Bernstein SI. Analysis of the 5′ end of the Drosophila muscle myosin heavy chain gene: alternatively spliced transcripts initiate at a single site and intron locations are conserved compared to myosin genes of other organisms. J Biol Chem. 1987;262:10741–10747. [PubMed] [Google Scholar]
- Yutzey KE, Kline RL, Konieczny SF. An internal regulatory element controls troponin I gene expression. Mol Cell Biol. 1989;9:1397–1405. doi: 10.1128/mcb.9.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


