Abstract
Anaerobic cultures capable of reductively dechlorinating 2,3,4,5-tetrachlorobiphenyl (CB) were enriched from three different sediments, one estuarine, one marine and one riverine. Two different electron donors were used in enrichments with the estuarine sediment (elemental iron or a mixture of fatty acids). The removal of doubly flanked meta and para chlorines to form 2,3,5-CB and 2,4,5-CB was observed in all cultures. Bacterial community analysis of PCR-amplified 16S rRNA gene fragments revealed different communities in these cultures, with the exception of one common population that showed a high phylogentic relatedness to Dehalococcoides species. No Dehalococcoides-like populations were ever detected in control cultures to which no PCBs were added. In addition, the dynamics of this Dehalococcoides-like population were strongly correlated with dechlorination. Subcultures of the estuarine sediment culture demonstrated that the Dehalococcoides-like population disappeared when dechlorination was inhibited with 2-bromoethanesulfonate or when 2,3,4,5-CB had been consumed. These results provide evidence that Dehalococcoides-like populations were involved in the removal of doubly flanked chlorines from 2,3,4,5-CB. Furthermore, the successful enrichment of these populations from geographically distant and geochemically distinct environments indicates the widespread presence of these PCB-dechlorinating, Dehalococcoides-like organisms.
Keywords: reductive dechlorination, polychlorinated biphenyls, Dehalococcoides
Introduction
Polychlorinated biphenyls (PCBs) are widespread environmental contaminants (Hutzinger & Veerkamp, 1981). Most PCB contamination is found in anaerobic sediments where reductive dechlorination (Brown et al., 1987; Quensen et al., 1988) is a possible approach for remediation. Little is known, however, about the microbiology of PCB dechlorination and about the diversity of these organisms. To date, there are only two reports in the literature of an isolated strain PCB-dechlorinating organism, Dehalococcoides ethenogenes strain 195, which dechlorinates two PCB congeners in the presence of perchloroethene (Fennell et al., 2004) and Dehalococcoides-like bacterium, DF-1, capable of respiring PCBs (Miller et al., 2005). Although field observations and experimental results have suggested the presence of multiple groups of PCB-dechlorinators (Morris et al., 1992; Ye et al., 1992; Brown et al., 1987; Wu & Wiegel, 1997; Cutter et al., 2001; Wu et al., 2002), efforts to categorically identify these microorganisms have not been successful.
Currently, an uncultured anaerobic PCB-dechlorinator, from a marine environment (Baltimore Harbor), has been putatively identified from highly enriched culture using the ribosomal RNA approach (Cutter et al., 2001). Phylogenetic analysis indicated that this organism, o-17, is distantly related to known Dehalococcoides species (Cutter et al., 2001), such as Dehalococcoides ethenogenes strain 195 (Maymó-Gatell et al., 1997) and Dehalococcoides spp. strain CBDB1 (Adrian et al., 2000). Although bacteria with identical 16S rRNA gene sequences can express substantially different physiologies (Jaspers & Overmann, 2004), all Dehalococcoides isolates studied to date appear to be obligate dehalorespirers, using halogenated aliphatic (Maymó-Gatell et al., 1997; He et al., 2003) or aromatic (Adrian et al., 2000; Bunge et al., 2003; Fennell et al., 2004) compounds as terminal electron acceptors. Therefore, there is evidence that at least three PCB dechlorinators are dehalorespirers and are phylogenetically related to Dehalococcoides species.
The identification of additional PCB-dechlorinating bacteria would provide not only more insight about the diversity of these organisms in various environments, but also additional critical information for evaluating remediation systems to treat PCB-contaminated sediments. Molecular-based techniques could be developed to quantify the abundance of PCB-dechlorinating microbes present at contaminated sites. This would help determine the feasibility of biostimulation at a particular site and the effectiveness of the stimulation strategy.
The goal of this research was to identify PCB-dechlorinators in three different sediments. Based on the hypothesis that PCB-dechlorinators would be enriched only when PCBs were present, the bacterial community structures of PCB-amended cultures were compared to the structures of cultures to which no PCBs were amended. It was found that the presence of Dehalococcoides-like populations were statistically correlated to the removal of doubly flanked (DF) chlorines from the PCB congener 2,3,4,5-tetrachlorobiphenyl (CB), and despite large differences in sediment characteristics and source, these Dehalococcoides-like populations and the concomitant removal of DF chlorines were observed in enrichment cultures derived from three very different sediments. This research should help to develop techniques to enrich for and enumerate these organisms in PCB-contaminated environments.
Materials and methods
Microcosm preparation and incubation
Sediment samples collected from Baltimore Harbor (BH), Palos Verdes (PV), and Hudson River (HR) were transported anaerobically and stored in an anaerobic glovebag (COY labs, Grass Lake, MI). An estuarine medium without sulfate (E-Cl) (Cutter et al., 1998) was prepared for BH and PV cultures and contained (per liter of deionized water): 0.001 g resazurin (redox indicator), 0.1 g MgCl2·6H2O, 0.1 g CaCl2·6H2O, 3.0 g Na2CO3, 0.5 g NH4Cl, 0.6 g Na2HPO4, 8.7 g K2HPO4, 10 mL vitamin solution, and 10 mL mineral solution (Wolin et al., 1963). Reduced anaerobic mineral medium (RAMM) was prepared for use with HR cultures as described by Shelton & Tiedje (1984) except that 2 mM L-cysteine was used as a reducing agent instead of Na2S. The pH of the E-Cl and RAMM media was adjusted to 7.0 with H3PO4 or NaOH. The media was sterilized by autoclaving and was transferred into an anaerobic glovebag. The media was reduced immediately prior to use with 20 mM cysteine.
Baltimore Harbor sediment cultures
Baltimore Harbor sediment microcosms were established in triplicate with either a mixture of fatty acids or Fe(0) as the electron donor. Treatments consisted of microcosms to which PCBs were added (PCB-fed) and control microcosms to which no PCBs were added (no-PCB control). Microcosms were set up in 160 mL serum bottles filled with 100 mL E-Cl medium and were opened only in the glovebag to avoid exposure to air. For PCB-fed treatments, 1 mL of hexane containing 10 μmol of 2,3,4,5-tetrachlorobiphenyl (2,3,4,5-CB) was added to the media in the bottles and the hexane was allowed to evaporate by leaving the reactors open in the glovebag (gas composition: 4% H2 and 96% N2) for about 10 min until the added hexane was no longer visible on top of the medium. Sediment (5 g dry weight equivalent) was added as inoculum to all bottles. A 1 mL mixture of acetate, propionate, and butyrate (20 mM of each, final concentration) was added to the fatty acid-fed bottles (in triplicate) using a pre-reduced (2 g L−1 cysteine) stock solution adjusted to a pH of 7.0. Fe(0) (20 mg iron powder, Fisher Scientific, Fairlawn, NJ, unwashed, 99% pure, 100 mesh) was added to the Fe(0)-fed bottles (in triplicate). All of the bottles were then capped with Teflon-lined stoppers and aluminum crimps and removed from the glovebag. The capped and crimped bottles were vigorously shaken on a wrist-action shaker for 2h to promote the homogeneous distribution of 2,3,4,5-CB. Control microcosms (no-PCB control) were set up in the glovebag as described above for both fatty acid-fed and Fe(0)-fed treatments, except that no 2,3,4,5-CB was introduced into the bottles. Hexane was not added to the no-PCB controls. Other than the initial time required to evaporate the added hexane (approximately 10 min), the no-PCB controls were handled identically to the PCB-fed treatments. All of the bottles were incubated in the dark on a rotating shaker (120 r.p.m.) over the course of the experiment. The bottles were sampled in the anaerobic glovebag to prevent contact with oxygen.
Baltimore Harbor sediment subcultures
Subcultures containing sterile BH sediment were established (in duplicate) from the PCB-dechlorinating BH cultures to further study the microbial community. To sterilize the BH sediment, it was first air dried and then autoclaved at 121 °C for 40 min on 2 consecutive days. Sterile sediment (2 g dry weight) was added to four 30 mL serum bottles. The congener 2,3,4,5-CB (2.5 μmol in 0.25 mL hexane) was then added to two of the sediment-containing bottles. The hexane was allowed to fully evaporate by placing the uncapped PCB-containing bottles in the glovebag for approximately 1 h, after which E-Cl medium (20 mL) and the mixture of fatty acids (as above) were added. Each of the four subcultures was then inoculated with 2 mL of well-mixed culture from a single actively-dechlorinating BH sediment microcosm (described above). The bottles were sealed, shaken and incubated in the same manner as described for the original BH sediment cultures.
Baltimore Harbor sediment-free subcultures
Sediment-free subcultures of the PCB-dechlorinating BH culture were also established (in triplicate) to investigate the effects of 2-bromoethanesulfonic acid (BESA), an inhibitor of methanogenesis (DiMarco et al., 1990) and dechlorination (Löffler et al., 1997; Ye et al., 1999; Chiu & Lee, 2001) on PCB dechlorination and the bacterial community. E-Cl medium (20 mL) and the fatty acid mixture (as above) were added into six 30 mL serum bottles before 2,3,4,5-CB (2.5 μmol in 0.25 mL hexane) was added. The hexane was again allowed to evaporate in the glovebag for approximately 1 h. BESA (3 mM, final concentration) was added to three of the six bottles, using a prereduced stock solution (pH = 7.0). Each of the six bottles was then inoculated with 2 mL of supernatant from one of the actively-dechlorinating BH sediment microcosms (described above). The bottles were then sealed, shaken and incubated in the same manner as described above.
Other sediment cultures
Additional sediment cultures were established as described above to investigate the effect of sediment source on dechlorination pattern and microbial community. These microcosms were set up as described above for the fatty acid-fed BH sediment cultures, except that RAMM medium was used for triplicate HR cultures (E-Cl medium was used for duplicate PV cultures). Again, control microcosms with no added 2,3,4,5-CB were set up.
Sample collection
Samples for PCB and microbial community analysis were collected every 20 to 60 days. Bottles were vigorously hand-shaken for 2 min before they were opened in the glovebag and approximately 1.5 mL of the sediment slurry was withdrawn with a sterile filed-off Pasteur pipette. For PCB analysis, samples were collected in a 20 mL serum vial and the exact weight of the sample was measured for PCB concentration (mg PCBs per kg sediment slurry) determination; samples were extracted immediately (see below). For microbial community analysis, culture samples were collected in sterile 1.6 mL microcentrifuge tubes, centrifuged at 10 000 g for 5 min, and the pellet was stored at −70 °C before analysis (see below).
PCB analysis
PCBs were extracted from samples in a manner similar to that described by Quensen et al. (1988). Briefly, sediment slurry samples were sequentially extracted, first with acetone (10 mL), then twice with hexane-acetone (1 : 1) (10 mL). The extracts were pooled and then purified in sequential steps with (1) a 2% (v/v) NaCl solution (10 mL), (2) a 30% (v/v) H2SO4 solution (4 mL), and (3) a 2% (v/v) NaCl solution (10 mL). The residual water in the extract was removed with anhydrous Na2SO4, and the extract was filtered through a Florisil-copper (25% copper w/w) column. PCBs were analyzed on a gas chromatograph (GC) (Hewlett Packard, Palo Alto, CA; 5890 series) equipped with an electron capture detector (ECD). PCB congeners (2 μL sample size) were separated on a HP-1 capillary column (25 m × 0.200 mm × 0.11 μm film thickness) with a carrier gas (He, UHP zero grade) flow rate of 3 mL min−1. Authentic standards (AccuStandard, New Haven, CT) were used to identify 2,3,4,5-CB and its dechlorination products and to construct 7-point external calibration curves for quantification. The method detection limit for 2,3,4,5-CB was 0.01 μM.
Biphenyl was analyzed on a Waters™ (Milford, MA) LC Module 1 Plus high performance liquid chromatograph (HPLC) equipped with a UV detector set at a wavelength of 254 nm. Biphenyl separation was accomplished on a C16 column (Discovery® RP Amide, 15 cm × 4.6 mm, 5 μm, Supelco) using gradient elution with acetonitrile and nanopure water (Millipore, Billerica, MA) as mobile phases (0.5 mL min−1). An external calibration curve for biphenyl was developed using authentic biphenyl standards [AccuStandard, 20 mg L−1 in AcCN : THF (1 : 1)] diluted in hexane. The identity of biphenyl was further confirmed in sediment extracts on an Agilent 6890 series GC (Palo Alto, CA) equipped with an Agilent 5973 network mass selective detector (MS). An Rtx-1 (Restek, Bellefonte, PA) column was used for separation (30 m × 0.32 mm × 5 μm).
Sediment characterization
Sediment characterization was performed by the Research Analytical Laboratory at the University of Minnesota (Table 1). The total N was measured according to the Dumas method, using a LECO FP-528 Nitrogen Analyzer (St Joseph, MI, USA) (Yeomans & Bremner, 1991). Sulfate was measured based on turbidity measurement using a Klett colorimeter following extraction by Ca(H2PO4)2·H2O and precipitation with BaCl2·H2O (Missouri Agricultural Experiment Station, 1998). Phosphate was extracted by NaHCO3, reduced by ascorbic acid, and the concentration was then determined using a Brinkmann PC 600D probe colorimeter (Westbury, NY) (Missouri Agricultural Experiment Station, 1998). Nitrate was extracted with CaSO4 and quantified colorimetrically with an Alpkem Rapid Flow Analyzer (Saskatoon, SK, Canada) (Willis & Gentry, 1987). The total organic carbon was determined by dry combustion and subsequent measurement of CO2 by IR spectrum absorption using a Skalar Primacs carbon furnace. Total inorganic carbon was measured by converting all of the inorganic carbon to CO2 with phosphoric acid, and then measuring the quantity of CO2 by IR spectrum absorption. Elemental analysis was conducted by inductively coupled plasma-atomic emission spectrometry (ICP-AES) (Dahlquist & Knoll, 1978) following extraction from the sediment using diethylenetriaminepentaacetic acid (DTPA) (US EPA, 1992).
Table 1.
Chemical characteristics of Baltimore Harbor (MD), Palos Verdes (CA), and Hudson River (MA) sediments
| Parameters | Baltimore Harbor | Hudson River | Palos Verdes |
|---|---|---|---|
| Total N (%) | 0.500 | 0.075 | 0.155 |
| Water pH | 7.7 | 7.1 | 8.0 |
| Sulfate (p.p.m.) | 141 | 11 | 123 |
| Phosphate (p.p.m.) | 40.0 | 10.0 | 39.5 |
| Nitrate (p.p.m.) | 1.7 | 1.6 | 1.6 |
| TOC (%) | 6.795 | 2.115 | 1.60 |
| Inorganic carbon (%) | 0.615 | ≤0.15 | 1.16 |
| Fe (p.p.b.) | 438.990 | 84.325 | 193.600 |
| Mn (p.p.b.) | 20.814 | 40.448 | 3.463 |
| Zn (p.p.b.) | 227.450 | 4.213 | 31.846 |
| Cu (p.p.b) | 47.725 | 1.830 | 8.861 |
| Pb (p.p.b.) | 101.360 | 13.967 | 12.330 |
| Ni (p.p.b.) | 5.547 | 0.231 | 1.852 |
| Cd (p.p.b.) | 1.155 | 0.076 | 1.947 |
| Cr (p.p.b.) | 0.241 | 0.028 | 0.207 |
Community analysis
A combination of sodium dodecyl sulfate treatment and bead-mill homogenization were adopted for cell lysis (Miller et al., 1999). Briefly, 450 μL phosphate buffer (100 mM, pH = 8) and 450 μL of SDS lysis mixture (100 mM NaCl, 500 mM Tris [pH = 8], 10% [w/v] SDS) were added to the sediment pellets. The samples were then incubated in a water bath at 70 °C for 90 min. Bead-beating and purification were conducted according to the manufacturer's instructions using a FastDNA kit for soil (Q-BIOgene, Irvine, CA, USA).
The procedure for denaturing gradient gel eletrophoresis of PCR-amplified 16S rRNA gene fragments (PCR-DGGE) was similar to that described previously (LaPara et al., 2000). Briefly, diluted genomic DNA was amplified by PCR using a primer pair targeting the highly variable V3 region of the domain Bacteria (PRBA338F (5′-ACT CCT ACG GGA GGC AGC AG-3′) (Lane, 1991) attached to a GC clamp (Muyzer et al., 1993) and PRUN518R (5′-ATT ACC GCC GCT GCT GG-3′) (Muyzer et al., 1993). Approximately equal quantities of PCR amplicons were then loaded onto a denaturing gradient gel (30–55%) for electrophoresis. Electrophoresis was performed on a D-Code apparatus (BioRad, Hercules, CA) in 0.5× TAE buffer at 60 °C, initially at 20 V for 20 min followed by 200 V for 180 min. The gel was then stained with SYBR Green I (Molecular Probes, Carlsbad, CA; diluted 1 : 5000 in 0.5× TAE) for 12 min before visualizing on an EC3 Bioimaging System (Ultra-Violet Products, Upland, CA). Bands with densities greater than 3% of the maximum intensity were catagorized as prominent.
Prominent bands were excised, incubated overnight in 20 μL of sterile water, and used as template for repeated cycles of PCR-DGGE until only a single band was detectable. The purified excised bands were then reamplified using primers PRBA338F (without the GC clamp) and PRUN518R. These PCR products were purified using a Geneclean Kit (Q-BIOgene) before sequencing. In several cases, vertically comigrating bands were analyzed from different gel lanes for quality assurance.
Nearly complete 16S rRNA genes targeting all members of the domain Bacteria were amplified by PCR using primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) (Edwards et al., 1989) and 1522R (5′-AAG GAG GTG ATC CAN CCR CA-3′) (Johnson, 1994). PCR was also used to amplify 16S rRNA gene fragments from Dehalococcoides-like populations using primers 27F and DeR (E. coli positions 1406–1422) (Cupples et al., 2003) from the PV samples. PCR products were purified using the Geneclean Kit (Q-BIOgene), ligated into the pGEM-T Easy cloning vector (Promega, Madison, WI), and transformed into competent E. coli DH5α cells. Transformants with inserts were selected by the blue/white screening method, and plasmids were extracted by the alkaline lysis method (Sambrook et al., 1989). Clones were initially screened by PCR-DGGE as described above to identify unique inserts; clones with unique inserts were then subjected to nucleotide sequence analysis as described below.
Nucleotide sequences were determined using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA; operated by the Advanced Genetic Analysis Center of University of Minnesota). PCR-DGGE bands were sequenced using primers PRBA338F and PRUN518R. Plasmid inserts were sequenced using primers 27F, 907F, 907R (Lane et al., 1991) and 1522r. Reported nucleotide sequences are the consensus of bi-directional sequence information. DNA sequences were screened for potential chimera using Chimera-Check from the Ribosomal Database Project II (Maidak et al., 2001). Sequences were then compared with sequences in the GenBank database (Benson et al., 2000) using the BLASTn program (Altschul et al., 1997) to search for their closest phylogenetic relatives. Sequences were deposited in the GenBank database under the accession numbers DQ080116 to DQ080206. The 16S rRNA gene sequences of Dehalococcoides strains, Bacteria DF-1 and o-17, and other dechlorinators were obtained from the Genbank database (Benson et al., 2000). These sequences, along with the Dehalococcoides-like and Chloroflexi clones of this study, were aligned by using ClustalW (Thompson et al., 1994). A bootstrapped phylogenetic tree (100 replicates, numbers at the nodes show percentage of replication) was constructed using Seqboot, Dnadist, Neighbor, Consensus, and Drawgram programs from the PHYLIP package (Felsenstein, 2002).
Statistical analysis
The correlation between 2,3,4,5-CB dechlorination and the microbial population dynamics was analyzed using the point-biserial correlation coefficient (rpb) (Chen & Popovich, 2002).
The dynamics of an individual band on a gel was represented by the variable Y, with its value as 1 or 0 when present or absent, respectively, at a certain sampling point. The dechlorination of 2,3,4,5-CB was represented by X, with its value as the average number of chlorines present per biphenyl molecule at a certain sampling point. The values of X̄1 and X̄0 are the mean values of X when Y = 1 and Y = 0, respectively. The value of p is equal to the number of X values when Y = 1 divided by the total number of X values (i.e. if there are five X values, and for three of them Y = 1 and for two of them Y = 0, p = 3/5). SX is the standard deviation of X. The P value was determined using the calculated rpb and corresponding tabulated P values (Chen & Popovich, 2002).
Results
Dechlorination in Baltimore Harbor sediment cultures
The removal of DF chlorines from 2,3,4,5-CB was the most prominent dechlorination activity observed in BH sediment cultures amended with either Fe(0) (Fig. 1a) or a mixture of fatty acids (Fig. 1b). The DF para chlorine of 2,3,4,5-CB was preferentially removed, resulting in the formation of 2,3,5-CB as a major dechlorination product. The addition of different electron donors (Fe(0) or a mixture of fatty acids) to the BH sediment cultures resulted in noticeable differences in the lag period prior to dechlorination and in the extent of dechlorination (Fig. 1); nevertheless, the prominent dechlorinating activity observed in the PCB-amended cultures was similar (i.e. DF chlorine removal).
Fig. 1.
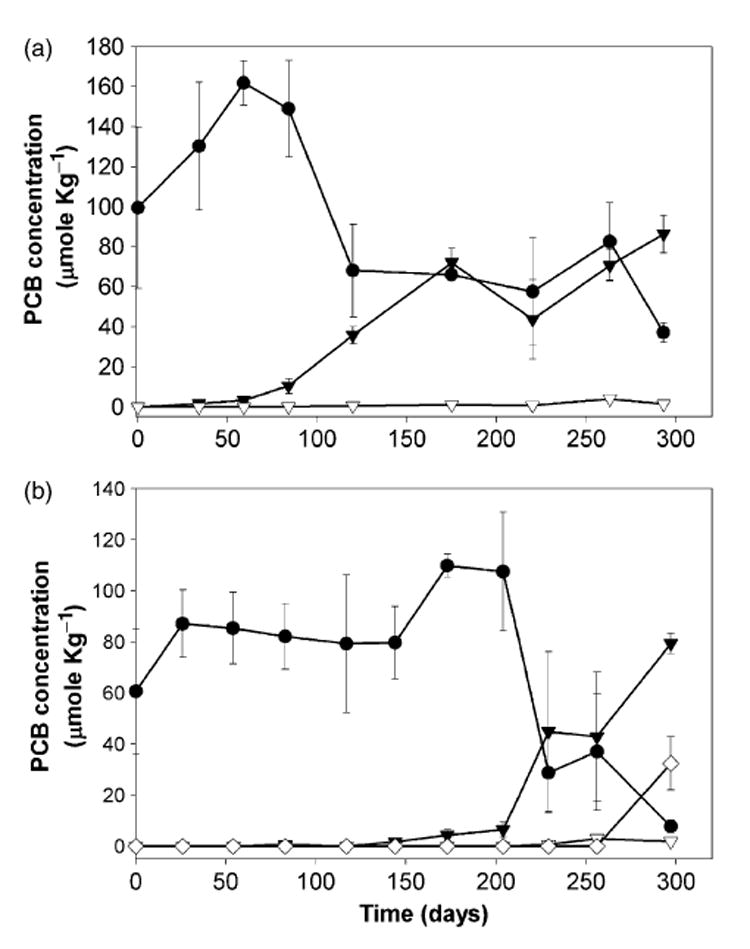
Dechlorination of 2,3,4,5-tetrachlorobiphenyl (CB) in cultures derived from Baltimore Harbor sediment amended with Fe(0) (a) and fatty acids (b). Symbols are 2,3,4,5-CB: black circle, 2,3,5-CB: black triangle, 2,4,5-CB: white triangle, and 2,5-CB: white diamond. The mean values of triplicate microcosms are shown in (a) and the mean values of duplicate microcosms are shown in (b); error bars represent the standard deviation of replicate microcosms.
Bacterial community dynamics in Baltimore Harbor sediment cultures
Bacterial community dynamics of the BH sediment cultures, both those to which 2,3,4,5-CB was added and control cultures that received no 2,3,4,5-CB, were investigated over time using PCR-DGGE (Fig. 2). Prominent bands were excised, purified, and sequenced; the phylogenetic affiliations of these bands were determined by comparison with sequences found in the GenBank database (Table 2). The communities in the PCB-amended cultures fed Fe(0) and fatty acids were almost completely different, with the exception of a single band, which was identical in the two cultures (Band BFE2 and Band BFA3). Nucleotide sequence analysis revealed that these two bands were 100% identical to each other, and to Dehalococcoides sp. CBDB1 over the fragment of the 16S rRNA gene that was analyzed (160 base pairs).
Fig. 2.
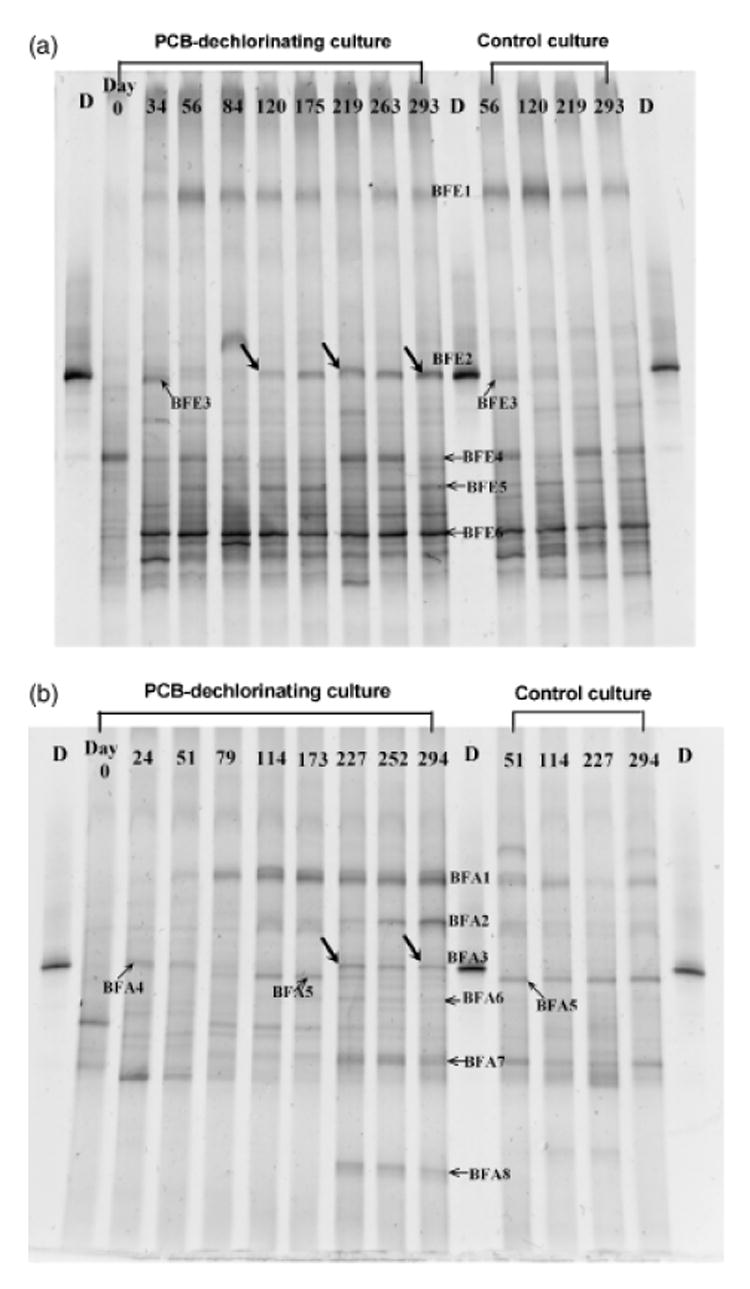
Microbial population dynamics, as determined by PCR-denaturing gradient gel electrophoresis, of 2,3,4,5-tetrachlorobiphenyl (CB)-dechlorinating Baltimore Harbor cultures amended with Fe(0) (a) and fatty acids (b). Lanes labeled D contain marker DNA for Dehalococcoides ethenogenes. Control cultures contained no added 2,3,4,5-CB but were otherwise incubated in an identical manner to the dechlorinating cultures. Bands indicated with an arrow (with or without a label) were excised and sequenced.
Table 2.
Phylogenetic affiliation of excised denaturing gradient gel electrophoresis bands from the Baltimore Harbor sediment culture as shown in Fig. 2
| Bands | Sequence length (bp) | Closest phylogenetic relative (accession no.) | Identity (%) | Phyla |
|---|---|---|---|---|
| BFE1 | 153 | Capnocytophaga sputigena (X67609) | 86.3 | Bacteroidetes |
| BFE2 | 164 | Dehalococcoides sp. CBDB1 (AF230641) | 100.0 | Chloroflexi |
| BFE3 | 157 | Clostridium lactatifermentans (AY033434) | 99.3 | Firmicutes |
| BFE4 | 159 | Uncultured bacterium SHA-15 (AJ249098) | 97.5 | Unknown |
| BFE5 | 156 | Uncultured bacterium clone DSBACT72 (AY762633) | 90.3 | Firmicutes |
| BFE6 | 156 | Clostridium cellulovorans (X73438) | 97.4 | Firmicutes |
| BFA1 | 163 | Epulopiscium sp. morphotype (M99574) | 84.7 | Firmicutes |
| BFA2 | 160 | Anaerolinea thermophila (AB046413) | 87.0 | Chloroflexi |
| BFA3 | 141 | Dehalococcoides sp. CBDB1 (AF230641) | 100.0 | Chloroflexi |
| BFA4 | 160 | Clostridium lactatifermentans (AY033434) | 96.3 | Firmicutes |
| BFA5 | 159 | Epulopiscium sp. morphotype (M99574) | 84.7 | Firmicutes |
| BFA6 | 159 | Pelospora glutarica (AJ251214) | 88.1 | Firmicutes |
| BFA7 | 159 | Syntrophomonas sp. TB-6 (AB098336) | 91.0 | Firmicutes |
| BFA8 | 158 | Uncultured Desulfosarcina sp. clone SIMO-1222 (AY710662) | 93.7 | δ-Proteobacterium |
Comparisons between PCB-amended and control (no PCB) cultures revealed several differences between the two cultures. In the Fe(0)-fed cultures in which only DF dechlorination of 2,3,4,5-CB occurred, BFE2 was the only population that was present in the PCB-amended cultures but absent in the control cultures. In the fatty acid-fed cultures, in which both DF dechlorination of 2,3,4,5-CB and further dechlorination to 2,5-CB occurred, Bands BFA3, BFA6 and BFA8 were present in the PCB-amended cultures but absent in the control cultures. The selective enrichment of a Dehalococcoides-like population (represented by Bands BFE2 and BFA3) in the 2,3,4,5-CB-amended cultures, coincident with the selective enrichment of DF dechlorination, strongly suggests the involvement of this population in 2,3,4,5-CB dechlorination to 2,3,5-CB and 2,4,5-CB. Nevertheless, selective enrichment of two additional populations (represented by Bands BFA6 and BFA8) was also observed in the fatty acid-amended cultures in which dechlorination past 2,3,5-CB and 2,4,5-CB to 2,5-CB occurred. Therefore, the involvement of these two non-Dehalococcoides-like populations in dechlorination, or their association with dechlorinators, cannot be precluded.
Population dynamics in the PCB-amended cultures, and their correlation with the dynamics of dechlorination, were also investigated. This correlation was based on the presence or absence of a band at a given time and the number of chlorines removed from 2,3,4,5-CB at that time. Based on the point-biserial correlation test, Band BFE2 (P = 0.068), BFA6 (P = 0.06), and Bands BFA3 and BFA8 (P = 1.3 × 10−8) were bands in which the population dynamics were most highly correlated with dechlorination (Fig. 2). No other bands were highly correlated with dechlorination (P ≤ 0.10).
Clone libraries from Baltimore Harbor sediment cultures
Clone libraries were constructed to provide longer 16S rRNA gene sequences for the populations identified in the 2,3,4,5-CB-fed cultures via PCR-DGGE. In the BH sediment cultures, a total of 57 clones were screened and sequenced to obtain 19 unique 16S rRNA gene sequences from one of the fatty acid-amended cultures (Day 294). Thirty clones were screened and sequenced to obtain eight unique 16S rRNA gene sequences from one of the Fe(0)-amended cultures (Day 293). The composition of these bacterial communities as detected by PCR-cloning was largely similar to that revealed by PCR-DGGE. In particular, two clones contained identical sequences to Bands BFE2 and BFA3, confirming the presence of these Dehalococcoides-like populations in the enrichment cultures, while simultaneously providing more sequence information. These two clones were identical over 1449 bases and were 99.7% identical to Dehalococcoides sp. CBDB1. The phylogenetic relationship between these two clones, representing populations BFE2 and BFA3 (clones BFE-P1 and BFA-K21), to other known Dehalococcoides species is shown in Fig. 3.
Fig. 3.
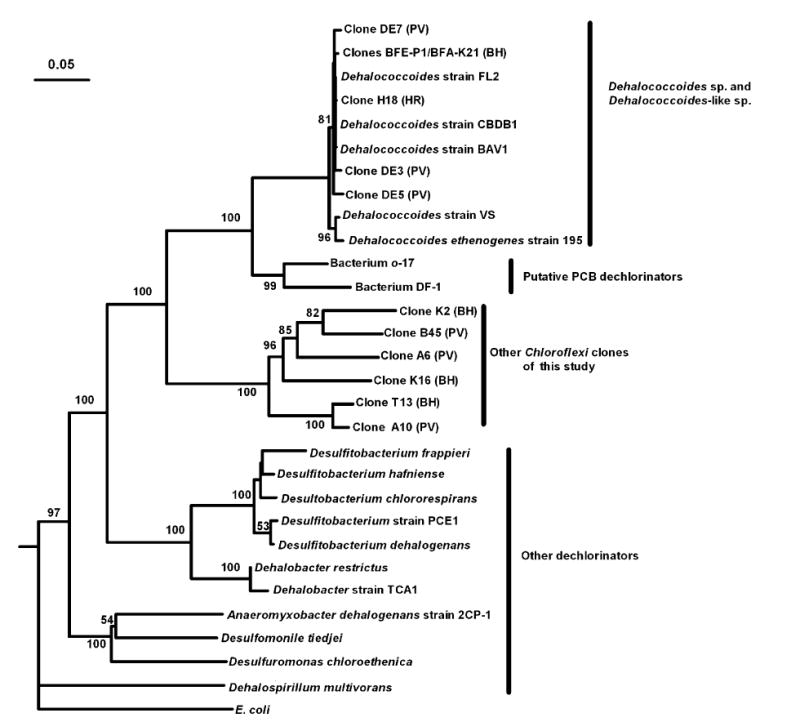
Phylogenetic tree for dechlorinators (adapted from Holliger et al., 1999), Dehalococcoides isolates (e.g. D. ethenogenes Strain 195 and Strain CBDB1), putative PCB dechlorinators (DF-1 and o-17) and the Dehalococcoides-like clones identified in this study. Sediment source is indicated in parentheses.
Baltimore Harbor subcultures
The fatty acid-amended BH dechlorinating culture was subcultured to investigate the effect of 2,3,4,5-CB withdrawl on the bacterial community structure. Positive control subcultures that received additional 2,3,4,5-CB sustained active dechlorination, and after 62 days 8.9 ± 3.7 (average ± standard deviation) mole percent 2,3,4,5-CB, 68.1 ± 16.3 mole percent 2,3,5-CB, 2.35 ± 0.1 mole percent 2,4,5-CB and 20.7 ± 19.9 mole percent 2,5-CB were present. Based on PCR-DGGE, the Dehalococcoides-like population was also present in these subcultures 62 days after the amendment of additional 2,3,4,5-CB (Fig. 4; lanes 1 and 2, indicated by arrows). In the subcultures to which no additional 2,3,4,5-CB was amended, the Dehalococcoides-like population, as determined by PCR-DGGE on Day 62, was lost (Fig. 4, lanes 3 and 4). This further suggests that the presence of 2,3,4,5-CB was important for the enrichment of this Dehalococcoides-like population, and this population was responsible for the DF dechlorination of 2,3,4,5-CB.
Fig. 4.
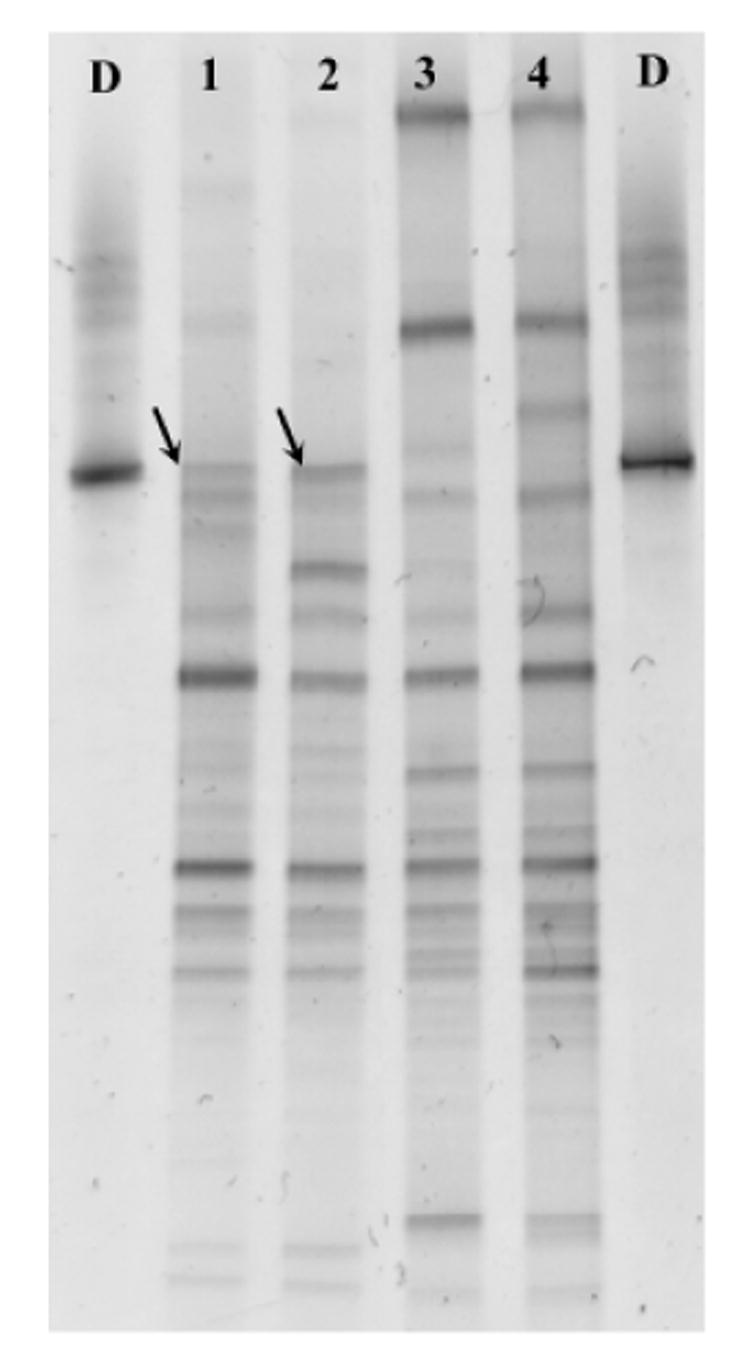
Effect of the addition or removal of 2,3,4,5-tetrachlorobiphenyl (CB) to Baltimore Harbor subcultures on the microbial communities present after 62 days, as determined by denaturing gradient gel electrophoresis. Lanes 1 and 2 represent duplicate subcultures in which additional 2,3,4,5-CB was amended; lanes 3 and 4 represent duplicate subcultures in which no additional 2,3,4,5-CB was amended. Lanes labeled D contain marker DNA for Dehalococcoides ethenogenes.
BESA-inhibition experiments were also performed on subcultures (sediment-free) established from the fatty acid-fed BH sediment cultures. BESA was previously thought to be a specific inhibitor of methanogens (DiMarco et al1990); more recent evidence, however, has shown that it can also inhibit dechlorinating bacteria (Löffler et al., 1997; Ye et al., 1999; Chiu & Lee, 2001). The concentration of the daughter products observed in the sediment-free subcultures (2,3,5-CB and 2,4,5-CB) is shown in Fig. 5a. As expected, the subcultures amended with BESA were inhibited, with no dechlorination of 2,3,4,5-CB (data not shown) and no formation of 2,3,5-CB and 2,4,5-CB (Fig. 5a). Dechlorination in the sediment-free subcultures to which no BESA was added primarily resulted in the formation of 2,3,5-CB (Fig. 5a). The bacterial communities in both treatments (BESA-amended and no-BESA) were finger-printed using PCR-DGGE on Day 194. A Dehalococcoides-like population, identical to the one identified in the BH sediment cultures, was detected in all of the dechlorinating replicates to which no BESA was added (Fig. 5b, lanes 1–3). In the BESA-amended treatments (in which dechlorination was inhibited), five populations disappeared (Fig. 5b), one of which was the Dehalococcoides-like population.
Fig. 5.
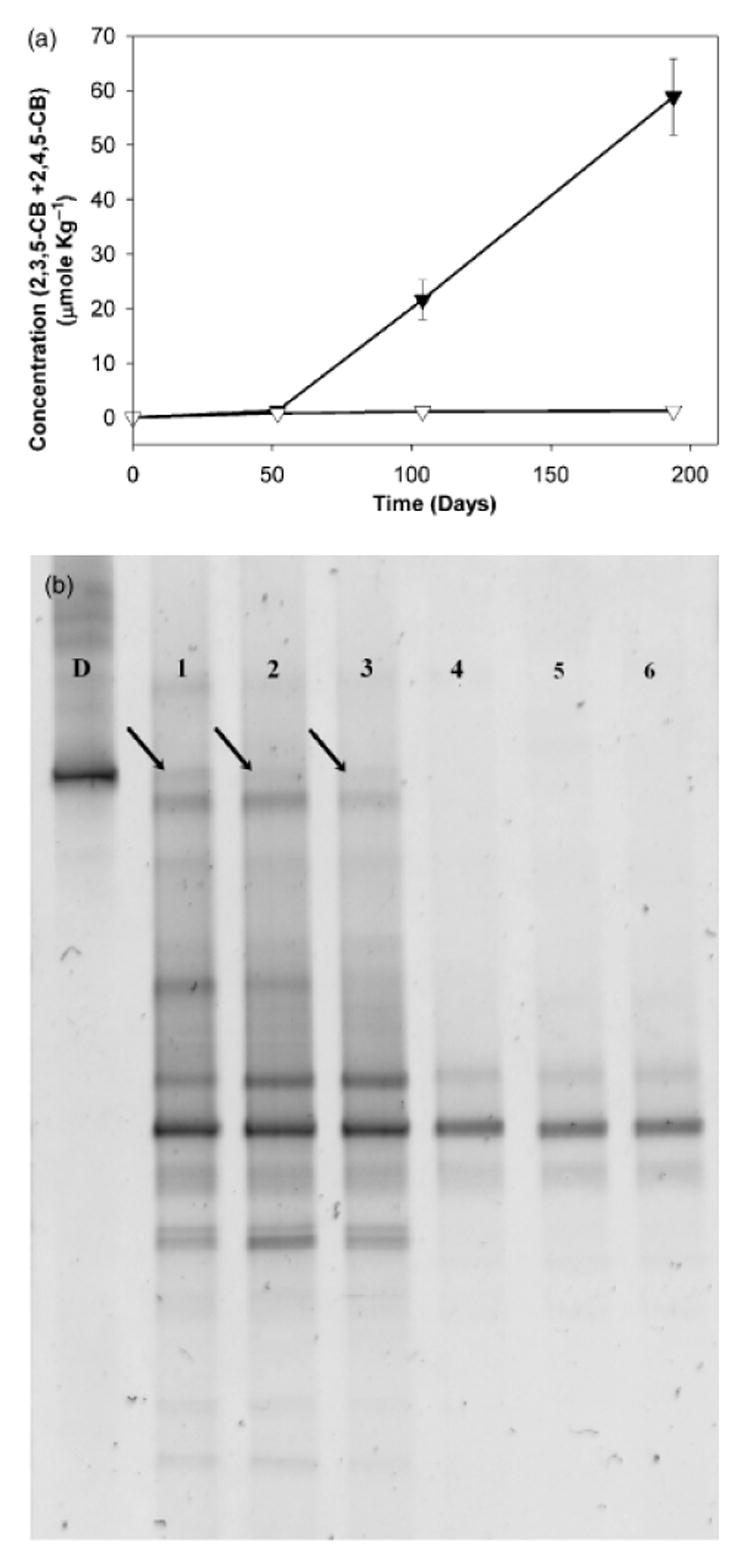
Inhibition of the dechlorination of 2,3,4,5-tetrachlorobiphenyl (CB) in sediment-free subcultures by 2-bromoethanesulfonic acid (BESA) (a) and the denaturing gradient gel electrophoresis pattern showing the bacterial communities in the subcultures on Day 194 (b). Black triangles show subcultures to which no BESA was added and white triangles show subcultures amended with BESA. Lane D contains DNA for Dehalococcoides ethenogenes as a marker; lanes 1–3 show the dechlorinating sediment-free cultures to which no BESA was added and lanes 4–6 show the sediment-free cultures to which BESA was added that were not capable of dechlorination. The arrows point to the bands corresponding to the Dehalococcoides-like population, which were excised and sequenced.
Dechlorination in other sediment cultures
Dechlorinating cultures were also enriched from HR and PV sediments. The three sediments used as inocula in this study (BH, HR, and PV) had significantly different characteristics (Table 1). BH and PV sediments had much higher sulfate contents than the HR sediment. The nutrient content was also quite different between the three sediments, with BH sediment containing the highest quantity of organic carbon, nitrogen, and trace metals, and HR sediment containing the lowest quantities of these nutrients (Table 1). As observed with BH sediment cultures, dechlorination in the HR and PV sediment cultures also began with the removal of DF chlorines (Fig. 6). The DF para chlorine of 2,3,4,5-CB was preferentially removed, resulting in the formation of 2,3,5-CB as a major dechlorination product. Further dechlorination of daughter products, including 2,3,5-CB, 2,4,5-CB and/or 3,4,5-CB, to 2,5-CB, 2,4-CB, 3,5-CB, and/or biphenyl was observed in all of the cultures (Fig. 6), but to different extents depending on the sediment source. PV cultures were capable of dechlorination to the greatest extent (biphenyl was observed in one of the duplicate cultures at concentrations of 0.55 and 28.3 μmol kg−1 on Days 297 and 327, respectively), whereas dechlorination in HR sediment cultures proceeded only to the di-chlorinated congener 2,5-CB. Nevertheless, the initial, prominent, and common dechlorination activity observed in all sediment cultures (BH, Fig. 1; PV and HR, Fig. 6) was DF chlorine removal.
Fig. 6.
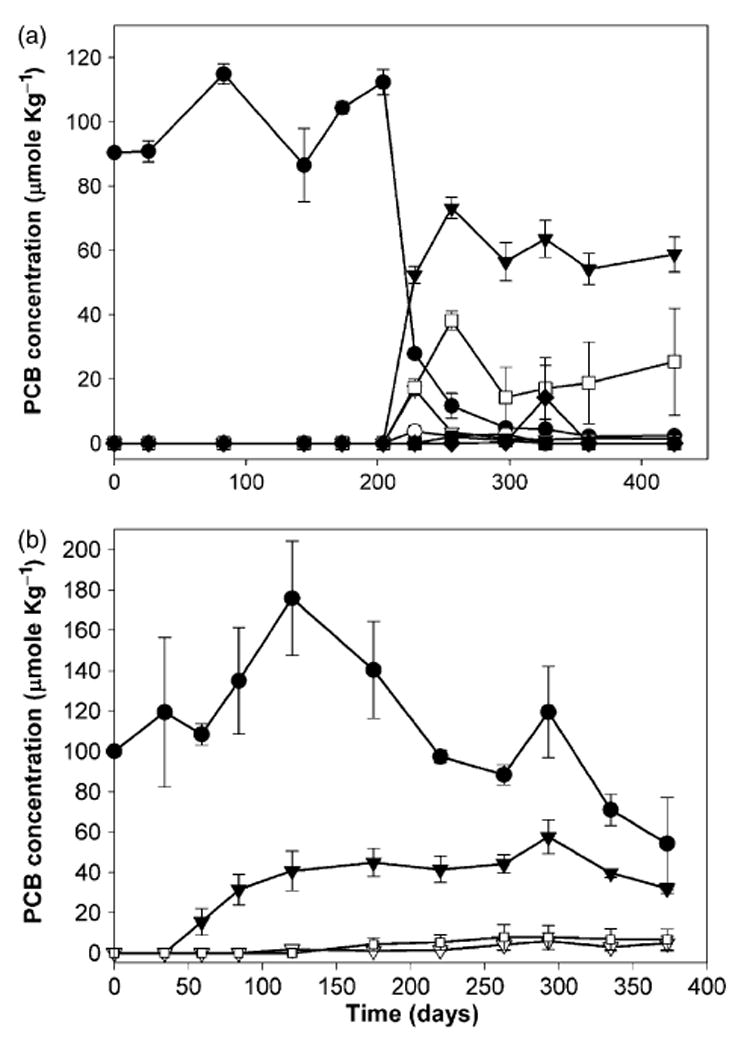
Dechlorination of 2,3,4,5-tetrachlorobiphenyl (CB) in cultures derived from Palos Vedes (a) and Hudson River (b) sediment amended with fatty acids. The mean values of duplicate microcosms are shown in (a) and the mean values of triplicate microcosms are shown in (b); error bars represent the standard deviation of replicate microcosms. Symbols are 2,3,4,5-CB: black circle, 2,3,5-CB: black triangle, 2,4,5-CB: white triangle, 3,4,5-CB: white circle, 3,5-CB: black square, 2,4-CB: white square, and biphenyl: black diamond.
Bacterial community dynamics in other sediment cultures
Bacterial community dynamics were also examined in both the PV (Fig. 7a) and HR (Fig. 7b) sediment cultures using PCR-DGGE. Prominent bands were excised from these gels and sequenced to determine the phylogenetic identity of these populations (Table 3). The bacterial populations detected in these microcosms were almost completely different from each other, and were also almost completely different from those identified in the BH sediment cultures (Table 2). Only two bands, PV3 and HR3 in PV and HR sediment cultures, respectively, shared an identical sequence, the closest phylogenetic relative of which was Dehalococcoides sp. CBDB1 (100% identity), based on the 16S rRNA gene fragment (160 base pairs) that was investigated. In contrast, no Dehalococcoides-like population was detected in the PV and HR control cultures that were not amended with 2,3,4,5-CB (Fig. 7). Other populations absent from the control (no-PCB) cultures included bands PV5, HR7, HR8 and HR10. The possible involvement of these populations in PCB dechlorination requires further investigation. Because dechlorination patterns other than DF chlorine removal were observed in both sediment cultures, it is possible that multiple dechlorinators, particularly non-Dehalococcoides-like organisms, were present in the microcosms amended with 2,3,4,5-CB.
Fig. 7.
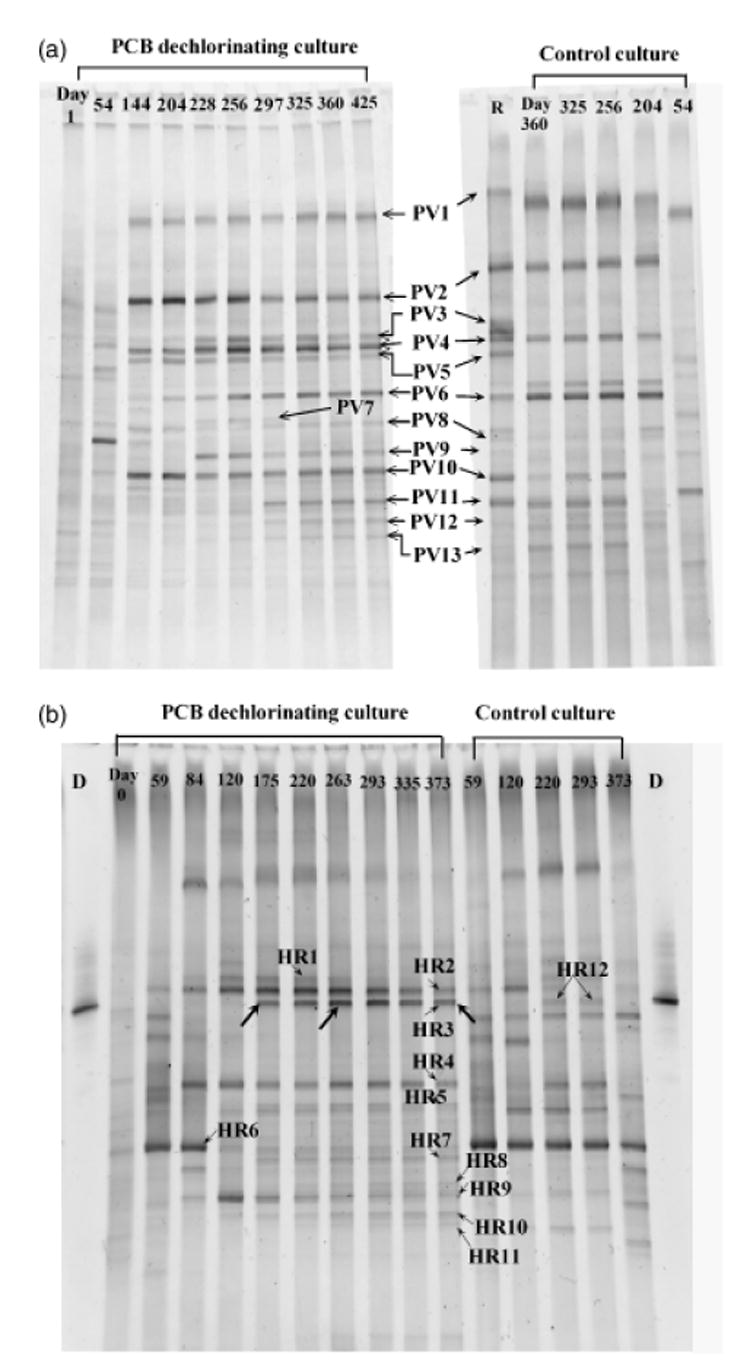
Microbial population dynamics, as determined by PCR-denaturing gradient gel electrophoresis (DGGE), of 2,3,4,5-tetrachlorobiphenyl (CB)-dechlorinating Palos Vedes (PV) cultures (a) and Hudson River cultures (b). Control cultures contained no added 2,3,4,5-CB but were otherwise incubated in an identical manner to the dechlorinating cultures. For PV cultures (a), the DGGE pattern of control culture was on a separate gel; a reference lane R, which is from PCB-dechlorinating culture (Day 297), illustrates the relative locations of bands. Bands indicated with an arrow (with or without a label) were excised and sequenced.
Table 3.
Phylogenetic affiliation of excised denaturing gradient gel electrophoresis bands from the Palos Vedes (PV) and Hudson River (HR) sediment cultures as shown in Fig. 7
| Bands* | Sequence length (bp) | Closest phylogenetic relative (accession no.) | Identity (%) | Phyla |
|---|---|---|---|---|
| PV1 | 181 | Uncultured bacterium (AJ306785) | 93.3 | Ambiguous |
| PV2 | 180 | Uncultured bacterium TSAC12 (AB186798) | 99.4 | Ambiguous |
| PV3 | 154 | Dehalococcoides sp. CBDB1 (AF230641) | 100 | Chloroflexi |
| PV4 | 174 | Uncultured bacterium SHA-94 (AJ306738) | 94 | Ambiguous |
| PV5 | 179 | Syntrophomonas sp. TB-6 (AB098336) | 94 | Firmicutes |
| PV6 | 153 | Uncultured bacterium SHD-245 (AJ278174) | 93 | Chloroflexi |
| PV7 | Chimeric | |||
| PV8 | 156 | Pelotomaculum sp. FP (AB159558) | 94.9 | Firmicutes |
| PV9 | 178 | Enterobacter sakazakii (AY752940) | 100 | γ-Proteobacteria |
| PV10 | 154 | Uncultured bacterium SJA-116 (AJ009487) | 96.7 | Chloroflexi |
| PV11 | 179 | Olavius algarvensis (AF328857) | 91.2 | δ-Proteobacteria |
| PV12 | 178 | Uncultured bacterium DSB-Dsa99-4 (AJ300510) | 92.5 | δ-Proteobacteria |
| PV13 | 172 | Cytophaga sp. (AB015525) | 93.3 | Bacteriodetes |
| HR1 | 153 | Capnocytophaga cynodegmi (X97245) | 91.1 | Bacteroidetes |
| HR2 | 174 | Capnocytophaga cynodegmi (X97245) | 92.1 | Bacteroidetes |
| HR3 | 155 | Dehalococcoides sp. CBDB1 (AF230641) | 100 | Chloroflexi |
| HR4 | 179 | Syntrophospora bryantii (M26491) | 92.1 | Firmicutes |
| HR5 | 179 | Syntrophospora bryantii (M26491) | 94.2 | Firmicutes |
| HR6 | 175 | Syntrophus acidotrophicus (U86447) | 90.8 | γ-Proteobacteria |
| HR7 | Sequence failed | |||
| HR8 | Sequence failed | |||
| HR9 | 157 | Clostridium sporogenes (AY442816) | 91.2 | Firmicutes |
| HR10 | 154 | Uncultured bacterium TDC-S1:29 (AF447148) | 100 | Unknown |
| HR11 | 173 | Flavobacterium mizutaii (AF361550) | 91.3 | Bacteroidetes |
| HR12 | 179 | Syntrophomonadaceae genomo sp. P1 (AY341821) | 85.3 | Firmicutes |
PV indicates bands derived from PV sediment cultures and HR indicates bands derived from HR sediment cultures.
Population dynamics in the PCB-amended cultures and their correlation with the dynamics of dechlorination were also investigated, as with the BH sediment cultures. Based on the point-biserial correlation test, Bands PV3 and PV9 were the ones most correlated with dechlorination in the PV sediment cultures (P = 7.5 × 10−7), with Bands PV1, PV2, PV4, PV5, PV6 (all P = 0.067) and PV11 (P = 0.008) also correlating with dechlorination, although less strongly. Bands HR3, HR7 and HR11 were the most correlated (P = 0.001) with dechlorination in the HR sediment cultures (Fig. 7), with Bands HR5 (P = 0.04) and HR10 (P = 0.006) also correlating with dechlorination. No other bands were highly correlated with dechlorination (P ≤ 0.10).
Clone libraries from other sediment cultures
Clone libraries were constructed for both HR and PV cultures to provide longer 16S rRNA gene sequences for the populations identified via PCR-DGGE. For the HR cultures, 39 clones were screened and sequenced to obtain 15 unique 16S rRNA gene sequences (Day 373). For the PV cultures (Day 297), 68 clones were screened and 13 unique sequences were obtained. Because no sequences corresponded to the Dehalococcoides-like population (Band PV3), a second clone library (three unique clones out 10 clones screened) was generated with this sample by targeting Dehalococcoides-like populations with a second primer set. The correspondence between the Dehalococcoides-like DGGE bands and the Dehalococcoides-like clones in both PV and HR cultures was established based on a perfect and exclusive match, indicating that the bands and the clones represented the same populations. The 16S rRNA gene sequences of the Dehalococcoides-like clones from PV and HR sediment were different from one another and were different from the Dehalococcoides-like clone enriched in the BH sediment microcosms, with 99.2–99.6% and 99.9% similarity between the PV clones and the HR clones to Dehalococcoides sp. CBDB1, respectively (based on 1336 and 1421 bases, respectively). The phylogenetic relationship between these four clones, representing populations PV3 (clones DE3, DE5 and DE7) and HR3 (H18), to other known Dehalococcoides species is shown in Fig. 3.
Discussion
Despite the potential biases of PCR-based molecular techniques (van Witzengerode et al., 1997; Head et al., 1998), these cultivation-independent methods have the ability to reveal microbial communities and community dynamics in complex environments (Amann et al., 1995). These techniques are particularly useful when the microorganisms of interest are difficult to isolate and when phylogenetic identification may provide critical information, such as when functional groupings correspond to phylogenetic groupings. Although little is known about dehalorespirers in general, a subset of these unique organisms appears to group phylogenetically (the Dehalococcoides-like dehalorespirers, Fig. 3). Indeed, 16S rRNA gene-based molecular techniques have been used to profile the microbial communities of PCB-dechlorinating cultures (Holoman et al., 1998), and to identify two putative PCB dechlorinators in highly-enriched cultures (Cutter et al., 2001; Wu et al., 2002). In this study, two PCR-based 16S rRNA gene analysis techniques, cloning and DGGE, were combined to identify putative PCB dechlorinators in cultures derived from different sources of sediments and subjected to different enrichment conditions.
In BH sediment cultures, BH subcultures, and BH sediment-free subcultures, the selective enrichment of a Dehalococcoides sp. CBDB1-like population was concomitant with active PCB dechlorination (DF chlorine removal). Indeed, all of the Dehalococcoides-like populations in the cultures investigated (BH, PV and HR) were correlated with PCB dechlorination at a confidence level of ≥93%, with the Dehalococcoides-like species in the fatty acid-fed BH, PV, and HR cultures correlating with dechlorination at a confidence level of >99%. Because PCBs are thought to be used as electron acceptors in energy conservation (Cutter et al., 2001; Wu et al., 2002; Cho et al., 2003), the absence of PCBs should result in the absence of PCB-dechlorinators. No Dehalococcoides-like populations developed in any of the control cultures to which no 2,3,4,5-CB was added (BH, PV and HR), presumably because no niche for these organisms existed without the presence of a chlorinated electron acceptor (Figs 2, 7). In addition, the disappearance of the Dehalococcoides-like populations occurred in BH subcultures when 2,3,4,5-CB was no longer available (Fig. 4). Finally, the inhibition of PCB dechlorination through the addition of BESA to sediment-free BH subcultures resulted in the disappearance of the Dehalococcoides-like population. Taken together, these data provide strong evidence that DF dechlorination of 2,3,4,5-CB was catalyzed by Dehalococcoides-like bacterial populations.
It is particularly interesting that functionally and phylogenetically similar Dehalococcoides-like organisms were present in geographically distant and geochemically distinct sediments. This finding suggests that these organisms were initially present in the sediment at low concentrations (below the PCR-DGGE detection limit) and enriched over time as a result of the niche provided by amended 2,3,4,5-CB. This finding is unlikely to be an artifact of cross-contamination, as (1) no Dehalococcoides-like species were ever detected in the no-PCB controls and these controls were opened in the glovebag with the same frequency as PCB-amended microcosms, (2) slight differences in the 16S rRNA gene sequences were present in the Dehalococcoides-like species present in the three different sediments used (BH, PV and HR) (Fig. 3) and (3) the detection of the Dehalococcoides-like populations statistically correlated with the onset of PCB dechlorination in these microcosms.
This is particularly exciting because it implies that PCB-dechlorinating Dehalococcoides-like organisms may be widespread in the environment. Studies on other chlorinated organic compounds have discovered a variety of Dehalococcoides-like organisms capable of dechlorinating a range of xenobiotic compounds. Fennell et al. (2004) reported that Dehalococcoides ethenogenes strain 195, isolated on tetrachloroethene (Maymó-Gatell et al., 1997), was capable of dechlorinating two PCB congeners when tetrachloroethene was used as a cosubstrate. Other chlorinated compounds that this Dehalococcoides strain is able to dechlorinate include 1,2,3,4-tetrachlorodibenzo-p-dioxin, 1,2,3,4-tetrachloronaphthalene, and hexachlorobenzene (Fennell et al., 2004). This versatility is not surprising, as 17 putative dehalogenase genes have recently been identified from the genome of Dehalococcoides ethenogenes strain 195 (Seshadri et al., 2005). In addition, another Dehalococcoides species, CBDB1, is also functionally versatile, dechlorinating both chlorinated benzenes (Adrian et al., 2000) and chlorinated dioxin (Bunge et al., 2003). Based on nearly complete 16S rRNA gene sequences, the phylogenetic relatedness between the Dehalococcoides-like populations detected in the three sediments reported herein and Dehalococcoides isolates (Maymó-Gatell et al., 1997; Adrian et al., 2000; He et al., 2003) is higher than 99% (Fig. 3). Although 16S rRNA gene-based phylogeny can only be used to infer physiology in very specific cases (Lonergan et al., 1996; Purkhold et al., 2000; Jaspers & Overmann, 2004), all Dehalococcoides isolates to date appear to be obligate dechlorinators. This suggests that in addition to other dechlorination processes previously linked to the presence of Dehalococcoides species (e.g. tetrachloroethene dechlorination), some PCB dechlorination patterns, notably DF dechlorination, also appear to be linked to the presence of Dehalococcoides-like species. With current knowledge of the physiology of Dehalococcoides species, the stimulation, enrichment and eventual isolation of Dehalococcoides-like PCB dechlorinators should now be more readily accomplished. In addition, the development and use of molecular-based techniques to assay contaminated sites for the presence and number of such organisms should now be feasible.
Acknowledgments
The authors gratefully acknowledge J. Meistrell and D. Cadien at the LA County Sanitary District for providing the Palos Verdes sediment samples, K. Sowers at the University of Maryland for providing the Baltimore Harbor sediment samples, D. Dzombak at Carnegie Mellon University for providing the Hudson River sediment samples and S. Zinder at Cornell University for providing Dehalococcoides ethenogenes DNA markers. Funding for this work was provided by the Office of Naval Research (Grant N00014-99-1-0923), the National Institute of Environmental Health Sciences (Grant ES12810-01), and the Hudson River Foundation (Graduate Fellowship). T. Y. was financially supported by a Sommerfeld fellowship from the University of Minnesota and by a Hudson River Foundation Graduate Fellowship (Grant GF/03/02).
References
- Adrian L, Szewzyk U, Wecke J, Görlsch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JF, Jr, Bedard DL, Brennan MJ, Carnahan JC, Feng H, Wagner RE. Polychlorinated biphenyl dechlorination in aquatic sediments. Science. 1987;236:709–712. doi: 10.1126/science.236.4802.709. [DOI] [PubMed] [Google Scholar]
- Bunge M, Adrian L, Kraus A, Opel M, Lorenz WG, Andreesen JR, Görisch H, Lechner U. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature. 2003;421:357–360. doi: 10.1038/nature01237. [DOI] [PubMed] [Google Scholar]
- Chen PY, Popovich PM. Correlation: Parametric and Nonparametric Measures. Sage Publication Inc; Thousand Oaks, CA: 2002. [Google Scholar]
- Chiu PC, Lee M. 2-bromoethanesulfonate affects bacteria in a trichloroethene-dechlorinating culture. Appl Environ Microbiol. 2001;67:2371–2374. doi: 10.1128/AEM.67.5.2371-2374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-C, Sokol RC, Frohnhoefer RC, Rhee GY. Reductive dechlorination of polychlorinated biphenyls. threshold concentration and dechlorination kinetics of individual congeners in Aroclor 1248. Environ Sci Technol. 2003;37:5651–5656. doi: 10.1021/es034600k. [DOI] [PubMed] [Google Scholar]
- Cupples AM, Spormann AM, McCarty PL. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter L, Sowers KR, May HD. Microbial dechlorination of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl Environ Microbiol. 1998;64:2966–2969. doi: 10.1128/aem.64.8.2966-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter LA, Watts JEM, Sowers KR, May HD. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ Microbiol. 2001;3:699–709. doi: 10.1046/j.1462-2920.2001.00246.x. [DOI] [PubMed] [Google Scholar]
- Dahlquist RL, Knoll JW. Inductively coupled plasma-atomic emission spectrometry: analysis of biological materials and soils for major, trace, and ultra-trace elements. Appl Spectrosc. 1978;32:1–30. [Google Scholar]
- DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. University of Washington; Seattle, WA: 2002. Phylogenetic inference program (PHYLIP) version 3.6. http://evolution.gs.washington.edu/phylip.html. [Google Scholar]
- Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol. 2004;38:2075–2081. doi: 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- He J, Ritalahti KM, Yang K-L, Koenigsberg S, Löffler FE. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- Head IM, Saunders JR, Pickup RW. Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1999;22:383–398. [Google Scholar]
- Holoman TRP, Elberson MA, Cutter LA, May HD, Sowers KR. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3359–3367. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzinger O, Veerkamp W. Microbial Degradation of Xenobiotics and Recalcitrant Compounds. Academic Press; New York: 1981. pp. 3–45. [Google Scholar]
- Jaspers E, Overmann J. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl Environ Microbiol. 2004;70:4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL. Similarity analysis of rRNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology. ASM Press; Washington, DC: 1994. pp. 683–700. [Google Scholar]
- LaPara TM, Nakatsu CH, Pantea L, Alleman JE. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl Environ Microbiol. 2000;66:3951–3959. doi: 10.1128/aem.66.9.3951-3959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; New York: 1991. pp. 115–175. [Google Scholar]
- Lonergan DJ, Jenter H, Coates JD, Phillips EJP, Schmidt TM, Lovely DR. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler FE, Ritalahti KM, Tiedje JM. Dechlorination of chloroethenes is inhibited by 2-bromoethanesulfonate in the absence of methanogens. Appl Environ Microbiol. 1997;63:4982–4985. doi: 10.1128/aem.63.12.4982-4985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymó-Gatell X, Chien Y-T, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Sowers KR, Milliken CE, May HD. Isolation and characterization of a PCB dechlorinating bacterium. Atlanta, GA. Poster presented at the 105th General Meeting of the American Society for Microbiology.2005. Jun 5–9, [Google Scholar]
- Missouri Agricultural Experiment Station. (1998) Recommended Chemical Soil Test Procedures for the North Central Region. North Central Regional Research Publication No. 221 (Revised).
- Morris PJ, Mohn WW, Quensen JF, III, Tiedje JM, Boyd SA. Establishment of a polychlorinated biphenyl-degrading enrichment culture with predominantly meta dechlorination. Appl Environ Microbiol. 1992;58:3088–3094. doi: 10.1128/aem.58.9.3088-3094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizer based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quensen JF, III, Tiedje JM, Boyd SA. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science. 1988;242:752–754. doi: 10.1126/science.242.4879.752. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Seshadri R, Adrian L, Fouts DE, et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- Shelton DR, Tiedje JM. General method for determining anaerobic biodegradation potential. Appl Environ Microbiol. 1984;47:850–857. doi: 10.1128/aem.47.4.850-857.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;1994(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis RB, Gentry CE. Automated method for determining nitrate and nitrite in water and soil extracts. Commun Soil Sci Plant Anal. 1987;18:625–36. [Google Scholar]
- van Witzengerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Wolin EA, Wolin MJ, Wolfe RS. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2882–2886. [PubMed] [Google Scholar]
- Wu Q, Watts JEM, Sowers KR, May HD. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl Environ Microbiol. 2002;68:807–812. doi: 10.1128/AEM.68.2.807-812.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wiegel J. Two anaerobic polychlorinated biphenyl-dehalogenating enrichments that exhibit different para-dechlorination specificities. Appl Environ Microbiol. 1997;63:4826–4832. doi: 10.1128/aem.63.12.4826-4832.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Quensen JF, III, Tiedje JM, Boyd SA. Anaerobic dechlorination of polychlorobiphenyls (Aroclor 1242) by pasteurized and ethanol-treated microorganisms from sediments. Appl Environ Microbiol. 1992;58:1110–1114. doi: 10.1128/aem.58.4.1110-1114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Quensen JF, III, Tiedje JM, Boyd SA. 2-bromoethanesulfonate, sulfate, molybdate, and ethanesulfonate inhibit anaerobic dechlorination of polychlorobiphenyls by pasteurized microorganisms. Appl Environ Microbiol. 1999;65:327–329. doi: 10.1128/aem.65.1.327-329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeomans JC, Bremner JM. Carbon and nitrogen analysis of soils by automated combustion techniques. Commun Soil Sci Plant Anal. 1991;22:843–850. [Google Scholar]
- US EPA. Test Methods of Evaluating Solid Waste, Physical/Chemical Methods, SW-846. 3. Environmental Monitoring and Support Laboratory, Office for Research and Development; Cincinnati, OH: 1992. Microwave assisted acid digestion of sediments, sludges, soils, and oils. [Google Scholar]


